Overexpression of the Phosphoserine Phosphatase-Encoding Gene (AtPSP1) Promotes Starch Accumulation in Lemna turionifera 5511 under Sulfur Deficiency
Abstract
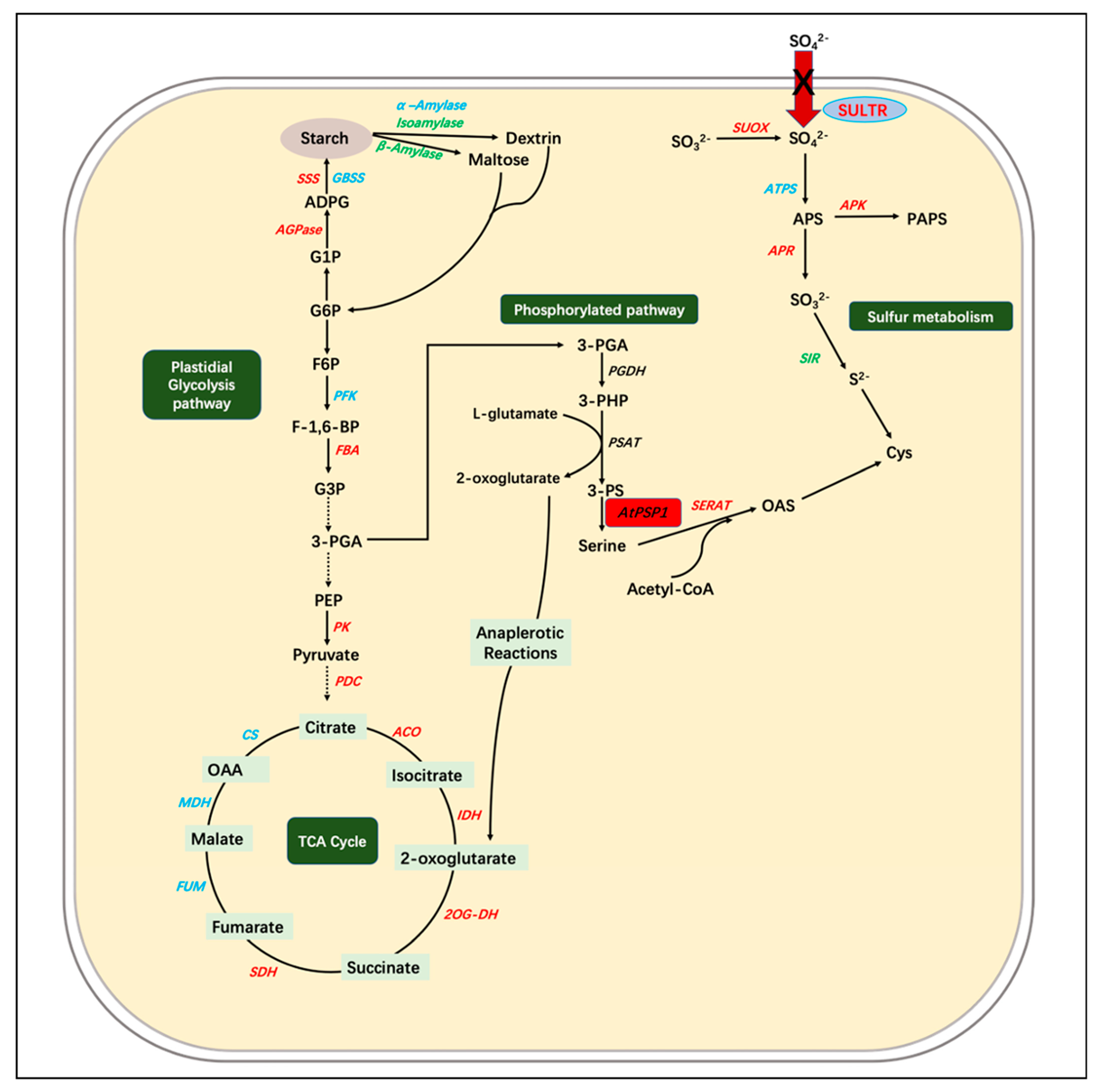
1. Introduction
2. Results
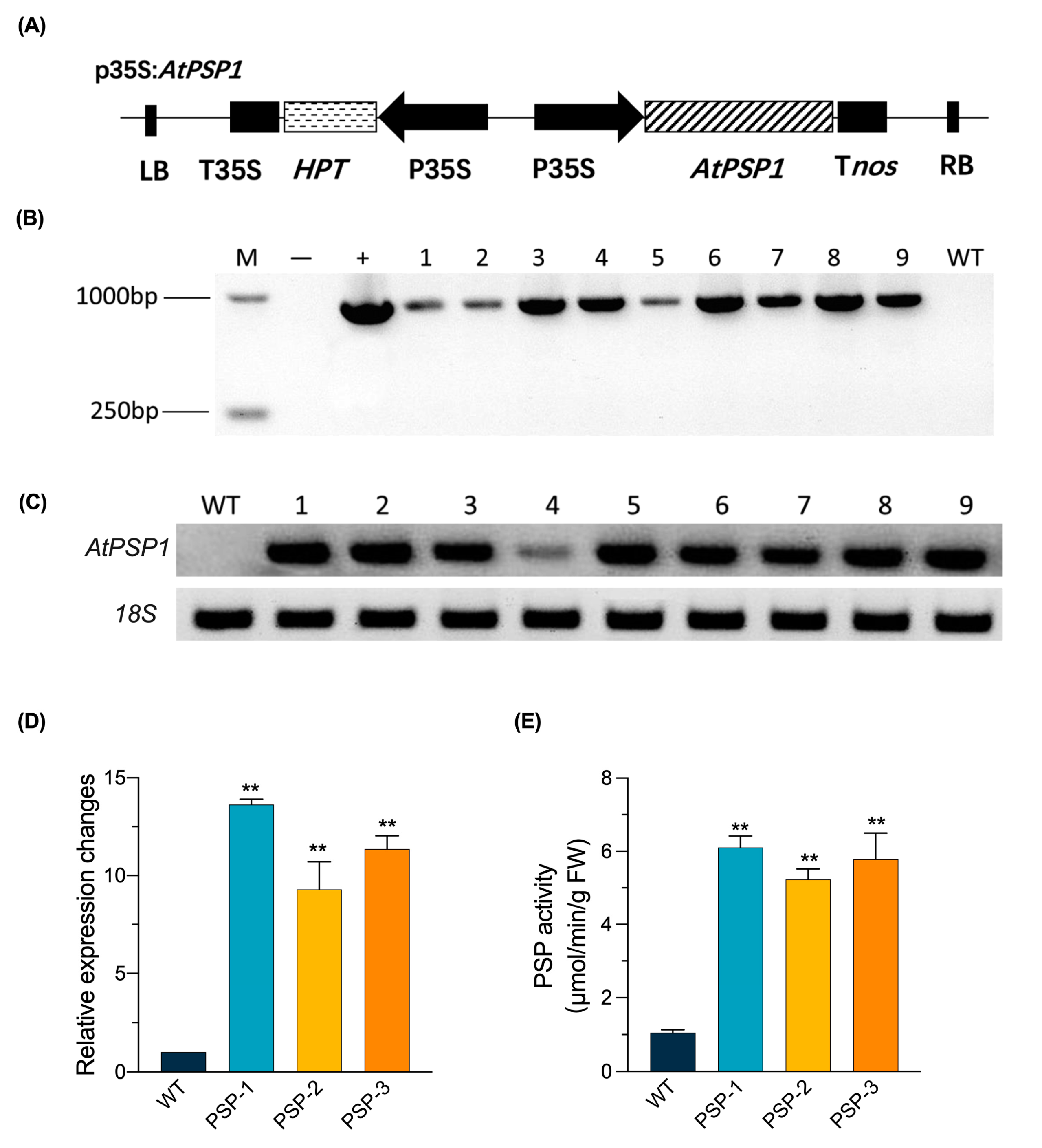
2.1. Generation of AtPSP1 Overexpressing Transgenic Lines
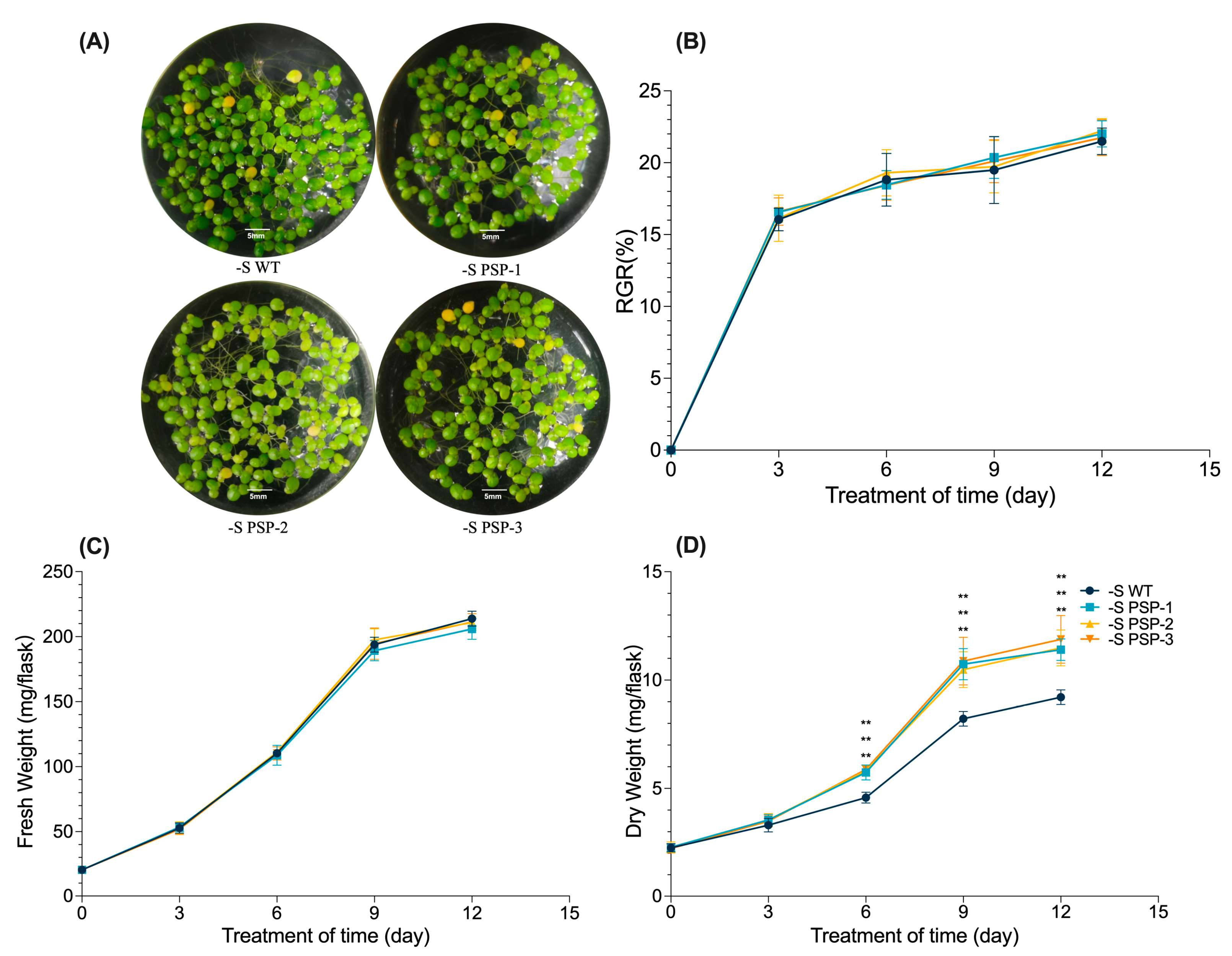
2.2. Overexpression of AtPSP1 Increased Dry Weight under Sulfur Deficiency
2.3. The Effect of the Overexpression of AtPSP1 on Photosynthetic Pigment and Chlorophyll Fluorescence Parameters under Sulfur Deficiency
2.4. The Effect of the Overexpression of AtPSP1 on Starch, Sugar and Soluble Protein Contents under Sulfur Deficiency
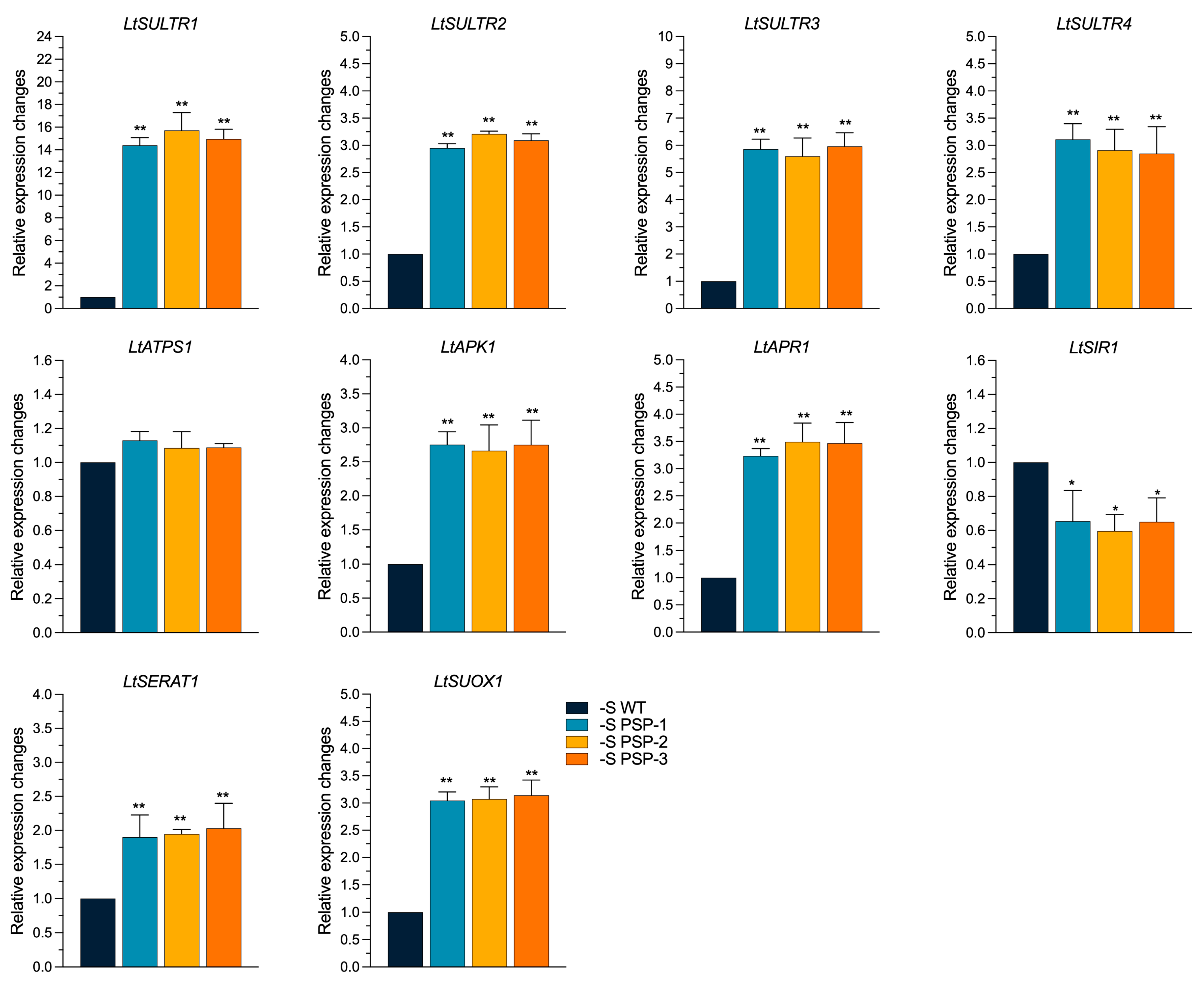
2.5. The Effect of the Overexpression of AtPSP1 on the Expression of Sulfur Assimilation-Related Genes under Sulfur Deficiency
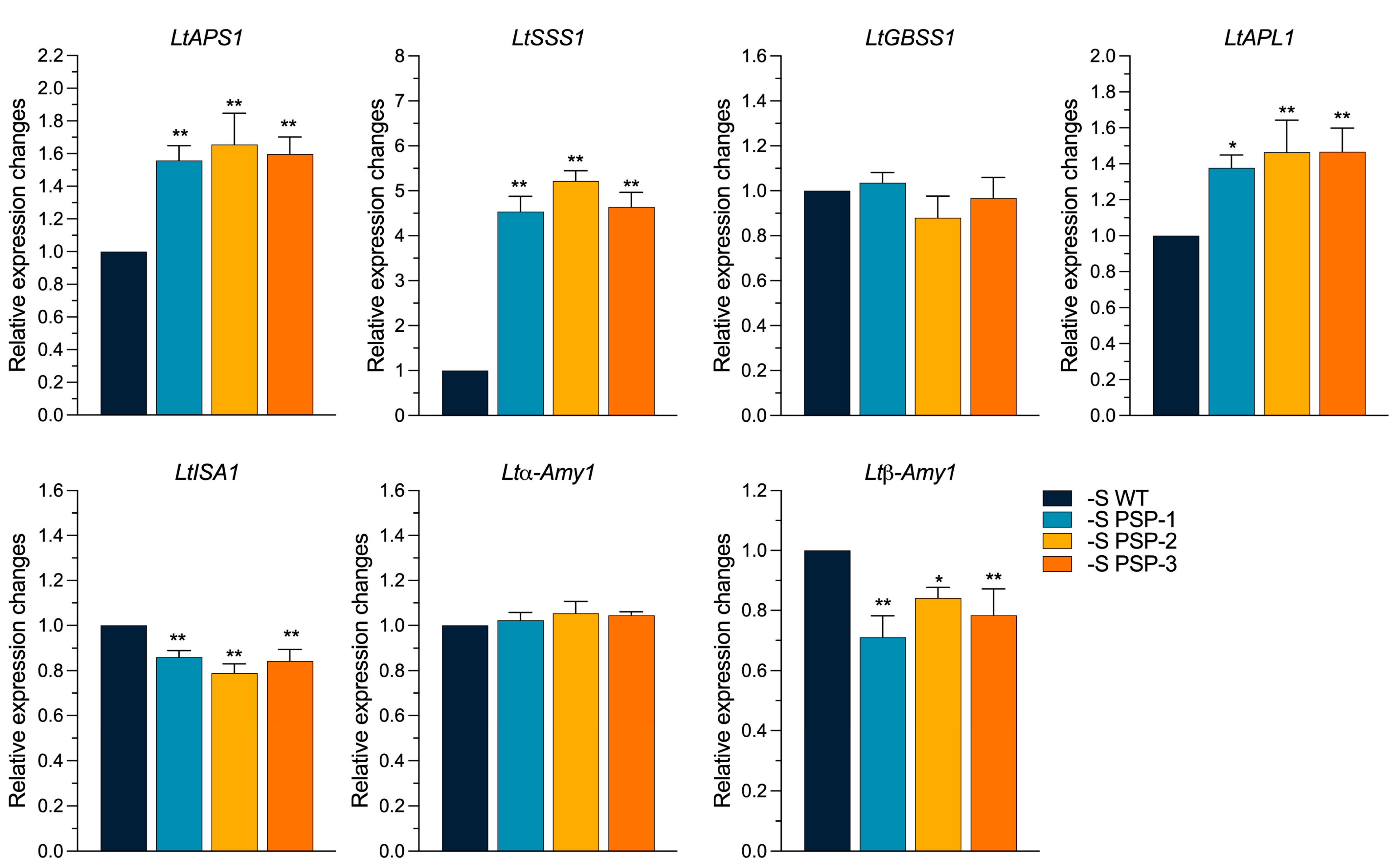
2.6. The Effect of the Overexpression of AtPSP1 on the Expression of Starch Metabolism-Related Genes under Sulfur Deficiency
2.7. The Effect of the Overexpression of AtPSP1 on the Expression of Carbon Metabolism-Related Genes under Sulfur Deficiency
3. Discussion
3.1. The Overexpression of AtPSP1 Prompts Growth and Starch Accumulation in Duckweed under Sulfur Deficiency
3.2. The Overexpression of AtPSP1 Coordinates Sulfur Assimilation, Starch Synthesis, and Carbon Metabolism under Sulfur Deficiency
4. Materials and Methods
4.1. Duckweed Culture and Sulfur-Deficiency Treatment
4.2. Vector Construction and Acquisition of Transgenic Duckweed
4.3. Measurement of the PSP Enzyme Activity
4.4. Biomass and Relative Growth Rate Determination
4.5. Determination of Photosynthetic Pigments and Chlorophyll Fluorescence Parameters
4.6. Starch, Total Sugar, and Soluble Protein Determination
4.7. DNA and RNA Extraction and qRT-PCR
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Les, D.H.; Crawford, D.J.; Landolt, E.; Gabel, J.D.; Rettig, J.H. Phylogeny and Systematics of Lemnaceae, the Duckweed Family. Syst. Bot. 2002, 27, 221–240. [Google Scholar] [CrossRef]
- Stomp, A.M. The duckweeds: A valuable plant for biomanufacturing. Biotechnol. Annu. Rev. 2005, 11, 69–99. [Google Scholar] [CrossRef]
- Bog, M.; Appenroth, K.J.; Sree, K.S. Key to the determination of taxa of Lemnaceae: An update. Nord. J. Bot. 2020, 38. [Google Scholar] [CrossRef]
- Chen, G.; Zhao, K.; Li, W.; Yan, B.; Yu, Y.; Li, J.; Zhang, Y.; Xia, S.; Cheng, Z.; Lin, F.; et al. A review on bioenergy production from duckweed. Biomass Bioenergy 2022, 161, 1064–1068. [Google Scholar] [CrossRef]
- Cui, W.; Cheng, J.J.; Appenroth, K. Growing duckweed for biofuel production: A review. Plant Biol. 2015, 17, 16–23. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, X.; Gao, X.; Sun, J.; Ji, X.; Feng, G.; Shen, G.; Xiang, B.; Wang, Y. Molecular mechanism underlying the effect of maleic hydrazide treatment on starch accumulation in S. polyrrhiza 7498 fronds. Biotechnol. Biofuels 2021, 14, 99. [Google Scholar] [CrossRef]
- Wang, X.; Cui, W.; Hu, W.; Feng, C. Abscisic acid-enhanced starch accumulation of bioenergy crop duckweed (Spirodela polyrrhiza). RSC Adv. 2020, 10, 10394–10401. [Google Scholar] [CrossRef]
- Faizal, A.; Putra, R. Uniconazole increases starch content in duckweed (Lemna aequinoctialis Welw). 3bio J. Biol. Sci. Technol. Manag. 2019, 1, 1. [Google Scholar] [CrossRef]
- Kurepa, J.; Shull, T.E.; Smalle, J.A. Cytokinin-induced growth in the duckweeds Lemna gibba and Spirodela polyrhiza. Plant Growth Regul. 2018, 86, 477–486. [Google Scholar] [CrossRef]
- de Morais, M.B.; Barbosa-Neto, A.G.; Willadino, L.; Ulisses, C.; Calsa Junior, T. Salt Stress Induces Increase in Starch Accumulation in Duckweed (Lemna aequinoctialis, Lemnaceae): Biochemical and Physiological Aspects. J. Plant Growth Regul. 2019, 38, 683–700. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, H.; Wang, Q.; Shao, M.; Li, X.; Chen, D.; Zeng, R.; Song, Y. Intraspecific variations in cadmium tolerance and phytoaccumulation in giant duckweed (Spirodela polyrhiza). J. Hazard. Mater. 2020, 395, 122672. [Google Scholar] [CrossRef]
- Shao, J.; Liu, Z.; Ding, Y.; Wang, J.; Li, X.; Yang, Y. Biosynthesis of the starch is improved by the supplement of nickel (Ni2+) in duckweed (Landoltia punctata). J. Plant Res. 2020, 133, 587–596. [Google Scholar] [CrossRef]
- Yu, C.; Xiaowen, Z.; Qi, G.; Bai, Z.; Wang, Y.; Wang, S.; Ma, Y.; Liu, Q.; Hu, R.; Zhou, G. Integrated analysis of transcriptome and metabolites reveals an essential role of metabolic flux in starch accumulation under nitrogen starvation in duckweed. Biotechnol. Biofuels 2017, 10, 167. [Google Scholar] [CrossRef]
- Guo, L.; Jin, Y.; Xiao, Y.; Tan, L.; Xueping, T.; Ding, Y.; He, K.-Z.; Du, A.; Li, J.; Yi, Z.; et al. Energy-efficient and environmentally friendly production of starch-rich duckweed biomass using nitrogen-limited cultivation. J. Clean. Prod. 2019, 251, 119726. [Google Scholar] [CrossRef]
- Wang, L.; Li, S.; Sun, L.; Tong, Y.; Yang, L.; Zhu, Y.; Wang, Y. Over-Expression of Phosphoserine Aminotransferase-Encoding Gene (AtPSAT1) Prompts Starch Accumulation in L. turionifera under Nitrogen Starvation. Int. J. Mol. Sci. 2022, 23, 11563. [Google Scholar] [CrossRef]
- Huang, M.; Fang, Y.; Liu, Y.; Jin, Y.; Sun, J.; Tao, X.; Ma, X.; He, K.; Zhao, H. Using proteomic analysis to investigate uniconazole-induced phytohormone variation and starch accumulation in duckweed (Landoltia punctata). BMC Biotechnol. 2015, 15, 81. [Google Scholar] [CrossRef]
- Sree, K.S.; Adelmann, K.; Garcia, C.; Lam, E.; Appenroth, K.-J. Natural variance in salt tolerance and induction of starch accumulation in duckweeds. Planta 2015, 241, 1395–1404. [Google Scholar] [CrossRef]
- Wang, W.; Messing, J. Analysis of ADP-glucose pyrophosphorylase expression during turion formation induced by abscisic acid in Spirodela polyrhiza (greater duckweed). BMC Plant Biol. 2012, 12, 5. [Google Scholar] [CrossRef]
- Guo, L.; Ding, Y.; Xu, Y.; Li, Z.; Jin, Y.; He, K.; Fang, Y.; Zhao, H. Responses of Landoltia punctata to cobalt and nickel: Removal, growth, photosynthesis, antioxidant system and starch metabolism. Aquat. Toxicol. 2017, 190, 87–93. [Google Scholar] [CrossRef]
- Sun, Z.; Guo, W.; Zhao, X.; Chen, Y.; Yang, J.; Xu, S.; Hou, H. Sulfur limitation boosts more starch accumulation than nitrogen or phosphorus limitation in duckweed (Spirodela polyrhiza). Ind. Crops Prod. 2022, 185, 115098. [Google Scholar] [CrossRef]
- Bauwe, H.; Hagemann, M.; Fernie, A. Photorespiration: Players, partners and origin. Trends Plant Sci. 2010, 15, 330–336. [Google Scholar] [CrossRef]
- Douce, R.; Bourguignon, J.; Neuburger, M.; Rébeillé, F. The glycine decarboxylase system: A fascinating complex. Trends Plant Sci. 2001, 6, 167–176. [Google Scholar] [CrossRef]
- Kleczkowski, L.; Givan, C. Serine Formation in Leaves by Mechanisms other than the Glycolate Pathway. J. Plant Physiol. 1988, 132, 641–652. [Google Scholar] [CrossRef]
- Ros, R.; Muñoz-Bertomeu, J.; Krueger, S. Serine in plants: Biosynthesis, metabolism, and functions. Trends Plant Sci. 2014, 19, 564–569. [Google Scholar] [CrossRef]
- Benstein, R.; Ludewig, K.; Wulfert, S.; Wittek, S.; Gigolashvili, T.; Frerigmann, H.; Gierth, M.; Flügge, U.-I.; Krueger, S. Arabidopsis Phosphoglycerate Dehydrogenase1 of the Phosphoserine Pathway Is Essential for Development and Required for Ammonium Assimilation and Tryptophan Biosynthesis. Plant Cell 2013, 25, 5011–5029. [Google Scholar] [CrossRef]
- Flores-Tornero, M.; Anoman, A.; Rosa Téllez, S.; Ros, R. Lack of phosphoserine phosphatase activity alters pollen and tapetum development in Arabidopsis thaliana. Plant Sci. 2015, 235, 81–88. [Google Scholar] [CrossRef]
- Han, X.; Peng, K.; Wu, H.; Song, S.; Zhu, Y.; Bai, Y.; Wang, Y. Overexpression of PSP1 enhances growth of transgenic Arabidopsis plants under ambient air conditions. Plant Mol. Biol. 2017, 94, 419–431. [Google Scholar] [CrossRef]
- Ros, R.; Cascales-Miñana, B.; Segura, J.; Anoman, A.; Toujani, W.; Flores-Tornero, M.; Rosa Téllez, S.; Muñoz-Bertomeu, J. Serine biosynthesis by photorespiratory and non-photorespiratory pathways: An interesting interplay with unknown regulatory networks. Plant Biol. 2012, 15, 707–712. [Google Scholar] [CrossRef]
- Takahashi, H.; Kopriva, S.; Giordano, M.; Saito, K.; Hell, R. Sulfur Assimilation in Photosynthetic Organisms: Molecular Functions and Regulations of Transporters and Assimilatory Enzymes. Annu. Rev. Plant Biol. 2010, 62, 157–184. [Google Scholar] [CrossRef]
- Rennenberg, H.; Herschbach, P.D.C. A detailed view on sulphur metabolism at the cellular and whole-plant level illustrates challenges in metabolite flux analyses. J. Exp. Bot. 2014, 65, 5711–5724. [Google Scholar] [CrossRef]
- Kopriva, S.; Rennenberg, H. Control of sulphate assimilation and glutathione synthesis: Interaction with N and C metabolism. J. Exp. Bot. 2004, 55, 1831–1842. [Google Scholar] [CrossRef]
- Smith, A.M.; Zeeman, S.C. Starch: A Flexible, Adaptable Carbon Store Coupled to Plant Growth. Annu. Rev. Plant Biol. 2020, 71, 217–245. [Google Scholar] [CrossRef]
- Streb, S.; Zeeman, S. Starch Metabolism in Arabidopsis. Arab. Book Am. Soc. Plant Biol. 2012, 10, e0160. [Google Scholar] [CrossRef]
- Smith, A.M. Starch in the Arabidopsis plant. Starch—Stärke 2012, 64, 421–434. [Google Scholar] [CrossRef]
- Sun, Z.; Guo, W.; Zhao, X.; Yang, J.; Duan, P.; Xu, S.; Hou, H. Sulfur limitation increases duckweed starch accumulation without compromising growth. bioRxiv 2021, 185, 115098. [Google Scholar] [CrossRef]
- Tao, X.; Fang, Y.; Xiao, Y.; Jin, Y.-L.; Ma, X.-R.; Zhao, Y.; He, K.-Z.; Zhao, H.; Wang, H.-Y. Comparative transcriptome analysis to investigate the high starch accumulation of duckweed (Landoltia punctata) under nutrient starvation. Biotechnol. Biofuels 2013, 6, 72. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Fang, Y.; Huang, M.; Chen, X.; Zhang, Y.; Zhao, H. The effects of photoperiod and nutrition on duckweed (Landoltia punctata) growth and starch accumulation. Ind. Crops Prod. 2018, 115, 243–249. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A. Metabolic changes sustain the plant life in low-sulfur environments. Curr. Opin. Plant Biol. 2017, 39, 144–151. [Google Scholar] [CrossRef]
- Samuilov, S.; Rademacher, N.; Brilhaus, D.; Kurz, S.; Arab, L.; Kopriva, S.; Weber, A.; Mettler-Altmann, T.; Rennenberg, H. Knock-Down of the Phosphoserine Phosphatase Gene Effects Rather N- Than S-Metabolism in Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 1830. [Google Scholar] [CrossRef]
- Niehaus, T.D. Phosphatases are involved in modulating the TCA cycle in plants. Mol. Plant 2021, 14, 1036–1037. [Google Scholar] [CrossRef]
- Lehmann, M.; Schwarzländer, M.; Obata, T.; Sirikantaramas, S.; Burow, M.; Olsen, C.E.; Tohge, T.; Fricker, M.D.; Møller, B.L.; Fernie, A.R.; et al. The Metabolic Response of Arabidopsis Roots to Oxidative Stress is Distinct from that of Heterotrophic Cells in Culture and Highlights a Complex Relationship between the Levels of Transcripts, Metabolites, and Flux. Mol. Plant 2009, 2, 390–406. [Google Scholar] [CrossRef]
- Yang, L.; Han, Y.; Wu, D.; Yong, W.; Liu, M.; Wang, S.; Liu, W.; Lu, M.; Wei, Y.; Sun, J. Salt and cadmium stress tolerance caused by overexpression of the Glycine Max Na+/H+ Antiporter (GmNHX1) gene in duckweed (Lemna turionifera 5511). Aquat. Toxicol. 2017, 192, 127–135. [Google Scholar] [CrossRef]
- Yang, L.; Han, H.; Liu, M.; Zuo, Z.; Zhou, K.; Lü, J.; Zhu, Y.; Bai, Y.; Wang, Y. Overexpression of the Arabidopsis photorespiratory pathway gene, serine: Glyoxylate aminotransferase (AtAGT1), leads to salt stress tolerance in transgenic duckweed (Lemna minor). Plant Cell Tissue Organ Cult. (PCTOC) 2013, 113, 407–416. [Google Scholar] [CrossRef]
- Su, C.; Jiang, Y.; Yang, Y.; Zhang, W.; Xu, Q. Responses of duckweed (Lemna minor L.) to aluminum stress: Physiological and proteomics analyses. Ecotoxicol. Environ. Saf. 2019, 170, 127–140. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Kuang, Y.; Zheng, S.; Tong, Y.; Zhu, Y.; Wang, Y. Overexpression of the Phosphoserine Phosphatase-Encoding Gene (AtPSP1) Promotes Starch Accumulation in Lemna turionifera 5511 under Sulfur Deficiency. Plants 2023, 12, 1012. https://doi.org/10.3390/plants12051012
Wang L, Kuang Y, Zheng S, Tong Y, Zhu Y, Wang Y. Overexpression of the Phosphoserine Phosphatase-Encoding Gene (AtPSP1) Promotes Starch Accumulation in Lemna turionifera 5511 under Sulfur Deficiency. Plants. 2023; 12(5):1012. https://doi.org/10.3390/plants12051012
Chicago/Turabian StyleWang, Lei, Yingying Kuang, Siyu Zheng, Yana Tong, Yerong Zhu, and Yong Wang. 2023. "Overexpression of the Phosphoserine Phosphatase-Encoding Gene (AtPSP1) Promotes Starch Accumulation in Lemna turionifera 5511 under Sulfur Deficiency" Plants 12, no. 5: 1012. https://doi.org/10.3390/plants12051012
APA StyleWang, L., Kuang, Y., Zheng, S., Tong, Y., Zhu, Y., & Wang, Y. (2023). Overexpression of the Phosphoserine Phosphatase-Encoding Gene (AtPSP1) Promotes Starch Accumulation in Lemna turionifera 5511 under Sulfur Deficiency. Plants, 12(5), 1012. https://doi.org/10.3390/plants12051012








