Transgene Bioconfinement: Don’t Flow There
Abstract
1. Introduction
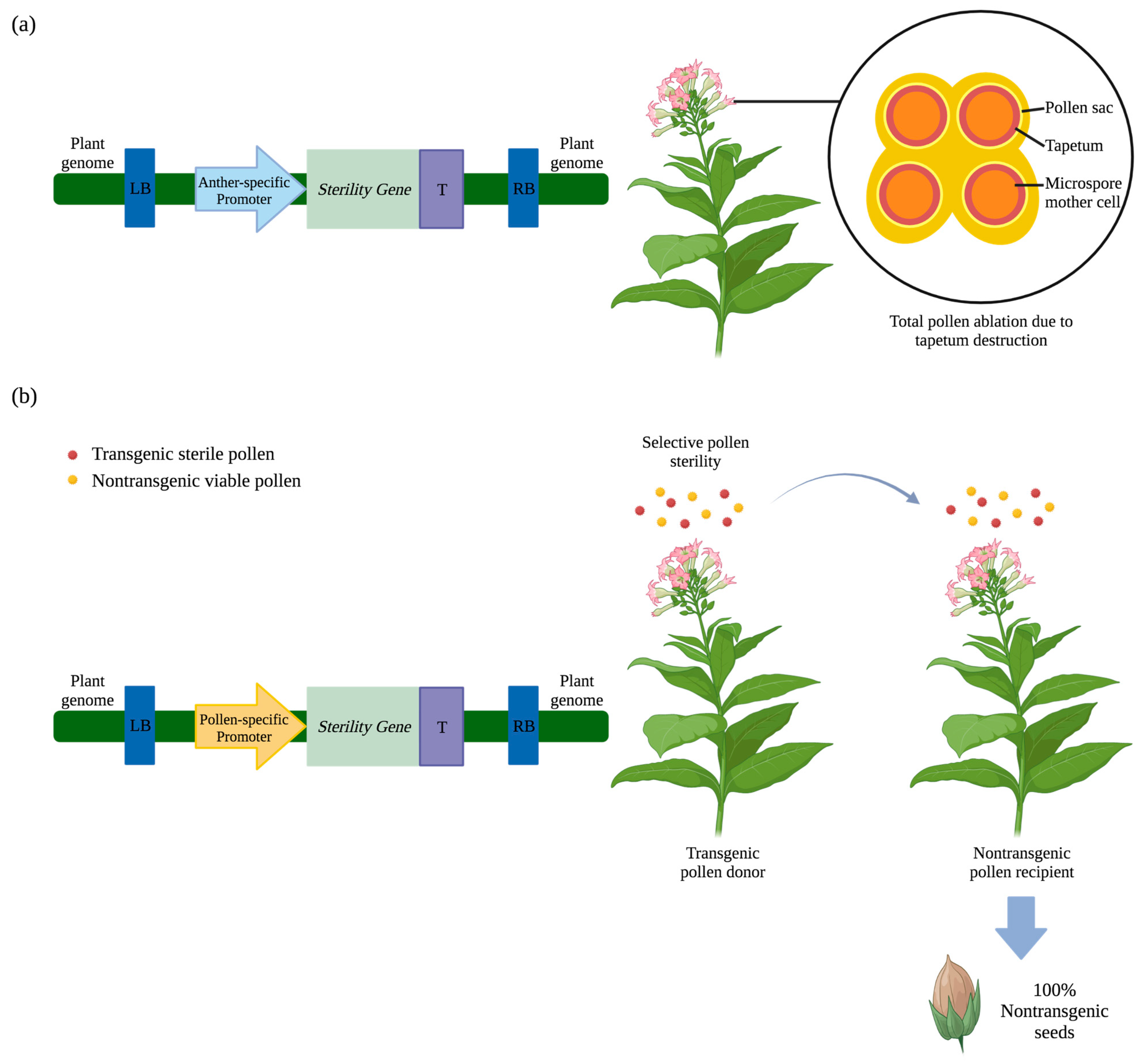
2. Engineered Reproductive Sterility
2.1. Male Sterility
2.1.1. Barnase-Induced Male Sterility
2.1.2. Diphtheria Toxin A-Chain-Induced Male Sterility
2.1.3. EcoRI Endonuclease-Induced Male Sterility
2.2. Conditional Seed Sterility
3. Transgene Excision
3.1. Transgene Excision via Cre/loxP Recombinase
3.2. Transgene Excision from Pollen via CinH-RS2
3.3. Transgene Excision from Pollen and/or Seeds via Fused loxP-FRT Recombinase System
3.4. CRISPR/Cas9-Mediated Transgene Excision
4. Delayed Flowering
Overexpression of miR156 Delayed Flowering
5. Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ISAAA. Global Status of Commercialized Biotech/GM Crops in 2019: Biotech Crops Drive SocioEconomic Development and Sustainable Environment in the New Frontier; ISAAA Brief No. 55; ISAAA: Ithaca, NY, USA, 2019. [Google Scholar]
- Chandler, S.; Dunwell, J.M. Gene flow, risk assessment and the environmental release of transgenic plants. Crit. Rev. Plant Sci. 2008, 27, 25–49. [Google Scholar] [CrossRef]
- Ellstrand, N.C. Current knowledge of gene flow in plants: Implications for transgene flow. Philos. Trans. Biol. Sci. 2003, 358, 1163–1170. [Google Scholar] [CrossRef]
- Rizwan, M.; Hussain, M.; Shimelis, H.; Hameed, M.U.; Atif, R.M.; Azhar, M.T.; Asif, M. Gene flow from major genetically modified crops and strategies for containment and mitigation of transgene escape: A review. Appl. Ecol. Environ. Res. 2019, 17, 11191–11208. [Google Scholar] [CrossRef]
- Lu, B.R. Transgene escape from GM crops and potential biosafety consequences: An environmental perspective. Collect. Biosaf. Rev. 2008, 4, 66–141. [Google Scholar]
- Chang, H.; Snow, A.A.; Mutegi, E.; Lewis, E.M.; Heaton, E.A.; Miriti, M.N. Extent of pollen-mediated gene flow and seed longevity in switchgrass (Panicum virgatum L.): Implications for biosafety procedures. Biomass Bioenergy 2018, 109, 114–124. [Google Scholar] [CrossRef]
- Kesoju, S.R.; Kramer, M.; Brunet, J.; Greene, S.L.; Jordan, A.; Martin, R.C. Gene flow in commercial alfalfa (Medicago sativa subsp. sativa L.) seed production fields: Distance is the primary but not the sole influence on adventitious presence. PLoS ONE 2021, 16, e0248746. [Google Scholar] [CrossRef]
- Millwood, R.; Nageswara-Rao, M.; Ye, R.; Terry-Emert, E.; Johnson, C.R.; Hanson, M.; Burris, J.N.; Kwit, C.; Stewart, C.N., Jr. Pollen-mediated gene flow from transgenic to non-transgenic switchgrass (Panicum virgatum L.) in the field. BMC Biotechnol. 2017, 17, 40. [Google Scholar] [CrossRef] [PubMed]
- Arriola, P.E.; Ellstrand, N.C. Crop-to-weed gene flow in the genus Sorghum (Poaceae): Spontaneous interspecific hybridization between johnsongrass, Sorghum halepense, and crop sorghum, S. bicolor. Am. J. Bot. 1996, 83, 1153–1159. [Google Scholar] [CrossRef]
- Cao, Q.J.; Xia, H.; Yang, X.; Lu, B.R. Performance of hybrids between weedy rice and insect-resistant transgenic rice under field experiments: Implication for environmental biosafety assessment. J. Integr. Plant Biol. 2009, 51, 1138–1148. [Google Scholar] [CrossRef] [PubMed]
- Ellstrand, N.C. “Born to run”? Not necessarily: Species and trait bias in persistent free-living transgenic plants. Front. Bioeng. Biotechnol. 2018, 6, 88. [Google Scholar] [CrossRef]
- Bauer-Panskus, A.; Miyazaki, J.; Kawall, K.; Then, C. Risk assessment of genetically engineered plants that can persist and propagate in the environment. Environ. Sci. Eur. 2020, 32, 32. [Google Scholar] [CrossRef]
- Zapiola, M.L.; Campbell, C.K.; Butler, M.D.; Mallory-Smith, C.A. Escape and establishment of transgenic glyphosate-resistant creeping bentgrass Agrostis stolonifera in Oregon, USA: A 4-year study. J. Appl. Ecol. 2007, 45, 486–494. [Google Scholar] [CrossRef]
- Greene, S.L.; Kesoju, S.R.; Martin, R.C.; Kramer, M. Occurrence of transgenic feral alfalfa (Medicago sativa subsp. sativa L.) in alfalfa seed production areas in the United States. PLoS ONE 2015, 10, e0143296. [Google Scholar] [CrossRef]
- Reichman, J.R.; Watrud, L.S.; Lee, E.H.; Burdick, C.A.; Bollman, M.A.; Storm, M.J.; King, G.A.; Mallory-Smith, C. Establishment of transgenic herbicide-resistant creeping bentgrass (Agrostis stolonifera L.) in nonagronomic habitats. Mol. Ecol. 2006, 15, 4243–4255. [Google Scholar] [CrossRef] [PubMed]
- Schafer, M.G.; Ross, A.A.; Londo, J.P.; Burdick, C.A.; Lee, E.H.; Travers, S.E.; Van de Water, P.K.; Sagers, C.L. The establishment of genetically engineered canola populations in the US. PLoS ONE 2011, 6, e25736. [Google Scholar] [CrossRef] [PubMed]
- NASEM. Genetically Engineered Crops: Experiences and Prospects; NASEM: Washington, DC, USA, 2016. [Google Scholar]
- Ellstrand, N.C.; Rieseberg, L.H. When gene flow really matters: Gene flow in applied evolutionary biology. Evol. Appl. 2016, 9, 833–836. [Google Scholar] [CrossRef]
- Stewart, C.N., Jr. Controlling transgene flow from engineered crops to unintended hosts by molecular approaches. In Gene Flow: Monitoring, Modeling and Mitigation; Wei, W., Stewart, C.N., Eds.; CABI: Wallingford, UK, 2021; pp. 118–124. [Google Scholar]
- Warwick, S.I.; Beckie, H.J.; Hall, L.M. Gene flow, invasiveness, and ecological impact of genetically modified crops. Annu. N. Y. Acad. Sci. 2009, 1168, 72–99. [Google Scholar] [CrossRef]
- Ding, J.; Duan, H.; Deng, Z.; Zhao, D.; Yi, G.; McAvoy, R.; Li, Y. Molecular strategies for addressing gene flow problems and their potential applications in abiotic stress tolerant transgenic plants. Crit. Rev. Plant Sci. 2014, 33, 190–204. [Google Scholar] [CrossRef]
- Pilson, D.; Prendeville, H.R. Ecological effects of transgenic crops and the escape of transgenes into wild populations. Annu. Rev. Ecol. Evol. Syst. 2004, 35, 149–174. [Google Scholar] [CrossRef]
- Haygood, R.; Ives, A.R.; Andow, D.A. Consequences of recurrent gene flow from crops to wild relatives. Proc. R. Soc. London. B Biol. Sci. 2003, 270, 1879–1886. [Google Scholar] [CrossRef]
- Kausch, A.P.; Hague, J.; Oliver, M.J.; Li, Y.; Daniell, H.; Mascia, P.; Watrud, L.S.; Stewart, C.N., Jr. Transgenic perennial biofuel feedstocks and strategies for bioconfinement. Biofuels 2010, 1, 163–176. [Google Scholar] [CrossRef]
- Daniell, H. Molecular strategies for gene containment in transgenic crops. Nat. Biotechnol. 2002, 20, 581–586. [Google Scholar] [CrossRef]
- Sandhu, S.; Blount, A.R.; Quesenberry, K.H.; Altpeter, F. Apomixis and ploidy barrier suppress pollen-mediated gene flow in field grown transgenic turf and forage grass (Paspalum notatum Flüggé). Theor. Appl. Genet. 2010, 121, 919–929. [Google Scholar] [CrossRef]
- Faisal, S.; Guo, Y.; Zang, S.; Cao, B.; Qu, G.; Hu, S. Morphological and genetic analysis of a cleistogamous mutant in rapeseed (Brassica napus L.). Genet. Resour. Crop Evol. 2018, 65, 397–403. [Google Scholar] [CrossRef]
- Fargue, A.; Colbach, N.; Pierre, J.; Picault, H.; Renard, M.; Meynard, J.M. Predictive study of the advantages of cleistogamy in oilseed rape in limiting unwanted gene flow. Euphytica 2006, 151, 1–13. [Google Scholar] [CrossRef]
- Klocko, A.L. Strategies to facilitate containment of genetically engineered crops. CABI Rev. 2020, 15, 1–11. [Google Scholar] [CrossRef]
- Mariani, C.; Beuckeleer, M.D.; Truettner, J.; Leemans, J.; Goldberg, R.B. Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature 1990, 347, 737–741. [Google Scholar] [CrossRef]
- Echlin, P. The role of the tapetum during microsporogenesis of angiosperms. Pollen 1971, 41–61. [Google Scholar] [CrossRef]
- Goldberg, R.B.; Beals, T.P.; Sanders, P.M. Anther development: Basic principles and practical applications. Plant Cell 1993, 5, 1217–1229. [Google Scholar] [CrossRef] [PubMed]
- Hartley, R.W. Barnase and barstar: Expression of its cloned inhibitor permits expression of a cloned ribonuclease. J. Mol. Biol. 1988, 202, 913–915. [Google Scholar] [CrossRef]
- Mariani, C.; Gossele, V.; Beuckeleer, M.D.; Block, M.D.; Goldberg, R.B.; Greef, W.D.; Leemans, J. A chimaeric ribonuclease-inhibitor gene restores fertility to male sterile plants. Nature 1992, 357, 384–387. [Google Scholar] [CrossRef]
- Block, M.D.; Debrouwer, D.; Moens, T. The development of a nuclear male sterility system in wheat. Expression of the barnase gene under the control of tapetum-specific promoters. Theor. Appl. Genet. 1997, 95, 125–131. [Google Scholar] [CrossRef]
- Luo, H.; Kausch, A.P.; Hu, Q.; Nelson, K.; Wipff, J.K.; Fricker, C.C.R.; Owen, T.P.; Moreno, M.A.; Lee, J.Y.; Hodges, T.K. Controlling transgene escape in GM creeping bentgrass. Mol. Breed. 2005, 16, 185–188. [Google Scholar] [CrossRef]
- Hague, J.P.; Dellaporta, S.L.; Moreno, M.A.; Longo, C.; Nelson, K.; Kausch, A.P. Pollen sterility—A promising approach to gene confinement and breeding for genetically modified bioenergy crops. Agriculture 2012, 2, 295–315. [Google Scholar] [CrossRef]
- Zhang, C.; Norris-Caneda, K.H.; Rottmann, W.H.; Gulledge, J.E.; Chang, S.; Kwan, B.Y.H.; Thomas, A.M.; Mandel, L.C.; Kothera, R.T.; Victor, A.D.; et al. Control of pollen-mediated gene flow in transgenic trees. Plant Physiol. 2012, 159, 1319–1334. [Google Scholar] [CrossRef]
- Elorriaga, E.; Meilan, R.; Ma, C.; Skinner, J.S.; Etherington, E.; Brunner, A.; Strauss, S.H. A tapetal ablation transgene induces stable male sterility and slows field growth in Populus. Tree Genet. Genomes 2014, 10, 1583–1593. [Google Scholar] [CrossRef]
- Pappenheimer, A.M., Jr. Diphtheria toxin. Annu. Rev. Biochem. 1977, 46, 69–94. [Google Scholar] [CrossRef]
- Twell, D. Diphtheria toxin-mediated cell ablation in developing pollen: Vegetative cell ablation blocks generative cell migration. Protoplasma 1995, 187, 144–154. [Google Scholar] [CrossRef]
- Koltunow, A.M.; Truettner, J.; Cox, K.H.; Wallroth, M.; Goldberg, R.B. Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 1990, 2, 1201–1224. [Google Scholar] [CrossRef]
- Fourel, G.; Phalipon, A.; Kaczorek, M. Evidence for direct regulation of diphtheria toxin gene transcription by an Fe2+-dependent DNA-binding repressor, DtoxR, in Corynebacterium diphtheriae. Infect. Immun. 1989, 57, 3221–3225. [Google Scholar] [CrossRef]
- Yamaizumi, M.; Mekada, E.; Uchida, T.; Okada, Y. One molecule of diphtheria toxin fragment A introduced into a cell can kill the cell. Cell 1978, 15, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Millwood, R.; Moon, H.S.; Poovaiah, C.R.; Muthukumar, B.; Rice, J.H.; Abercrombie, J.M.; Abercrombie, L.L.; Green, W.D.; Stewart, C.N., Jr. Engineered selective plant male sterility through pollen-specific expression of the EcoRI restriction endonuclease. Plant Biotechnol. J. 2016, 14, 1281–1290. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.G. Type II restriction—Modification systems. Trends Genet. 1988, 4, 314–318. [Google Scholar] [CrossRef]
- Oliver, M.J.; Quisenberry, J.E.; Trolinder, N.; Keim, D.L. Control of Plant Gene Expression. U.S. Patent No. 5,723,765, 3 March 1998. [Google Scholar]
- Oliver, M.J.; Quisenberry, J.E.; Trolinder, N.; Keim, D.L. Control of Gene Expression. U.S. Patent No. 5,925,808, 20 July 1999. [Google Scholar]
- Oliver, M.J.; Quisenberry, J.E.; Trolinder, N.; Keim, D.L. Control of Gene Expression. U.S. Patent No. 5,977,441, 2 November 1999. [Google Scholar]
- Sang, Y.; Millwood, R.; Stewart, C.N., Jr. Gene use restriction technologies for transgenic plant bioconfinement. Plant Biotechnol. J. 2013, 11, 649–658. [Google Scholar] [CrossRef] [PubMed]
- FAO. Potential Impacts of Genetic Use Restriction Technologies (GURTs) on Agricultural Biodiversity and Agricultural Production Systems; FAO: Rome, Italy, 2001. [Google Scholar]
- Lombardo, L. Genetic use restriction technologies: A review. Plant Biotechnol. J. 2014, 12, 995–1005. [Google Scholar] [CrossRef]
- Oliver, M.J.; Hake, K. Seed-based containment strategies. In Plant Gene Containment; Oliver, M.J., Li, Y., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 113–124. [Google Scholar]
- Ferreras, J.; Barbieri, L.; Girbés, T.; Battelli, M.G.; Rojo, M.A.; Arias, F.J.; Rocher, M.A.; Soriano, F.; Mendéz, E.; Stirpe, F. Distribution and properties of major ribosome-inactivating proteins (28 S rRNA N-glycosidases) of the plant Saponaria officinalis L. (Caryophyllaceae). Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 1993, 1216, 31–42. [Google Scholar] [CrossRef]
- Oliver, M.J.; Luo, H.; Kausch, A.P.; Collins, H. Seed-based strategies for transgene containment. In Proceedings of the 8th International Symposium on the Biosafety of Genetically Modified Organisms, Montpellier, France, 26–30 September 2004; pp. 154–161. [Google Scholar]
- Luo, K.; Duan, H.; Zhao, D.; Zheng, X.; Deng, W.; Chen, Y.; Stewart, C.N., Jr.; McAvoy, R.; Jiang, X.; Wu, Y.; et al. ‘GM-gene-deletor’: Fused loxP-FRT recognition sequences dramatically improve the efficiency of FLP or CRE recombinase on transgene excision from pollen and seed of tobacco plants. Plant Biotechnol. J. 2007, 5, 263–374. [Google Scholar] [CrossRef]
- Liu, W.; Stewart, C.N., Jr. Plant synthetic promoters and transcription factors. Curr. Opin. Biotechnol. 2016, 37, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Kosentka, P.Z.; Liu, W. Synthetic biology approaches in regulation of targeted gene expression. Curr. Opin. Plant Biol. 2021, 63, 102036. [Google Scholar] [CrossRef]
- Yang, Y.; Chaffin, T.A.; Ahkami, A.H.; Blumwald, E.; Stewart, C.N., Jr. Plant synthetic biology innovations for biofuels and bioproducts. Trends Biotechnol. 2022, 40, 1454–1468. [Google Scholar] [CrossRef]
- Ali, S.; Kim, W.C. A fruitful decade using synthetic promoters in the improvement of transgenic plants. Front. Plant Sci. 2019, 10, 1433. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.S.; Abercrombie, L.L.; Eda, S.; Blanvillain, R.; Thomson, J.G.; Ow, D.W.; Stewart, C.N., Jr. Transgene excision in pollen using a codon optimized serine resolvase CinH-RS2 site-specific recombination system. Plant Mol. Biol. 2011, 75, 621–631. [Google Scholar] [CrossRef]
- Yau, Y.Y.; Stewart, C.N., Jr. Less is more: Strategies to remove marker genes from transgenic plants. BMC Biotechnol. 2013, 13, 36. [Google Scholar] [CrossRef]
- Hoess, R.H.; Abremski, K. The Cre-lox recombination system. In Nucleic Acids and Molecular Biology; Eckstein, F., Lilley, D.M.J., Eds.; Springer: Berlin/Heidelberg, Germany, 1990; Volume 4, pp. 99–109. [Google Scholar]
- Senecoff, J.F.; Bruckner, R.C.; Cox, M.M. The FLP recombinase of the yeast 2-micron plasmid: Characterization of its recombination site. Proc. Natl. Acad. Sci. USA 1985, 82, 7270–7274. [Google Scholar] [CrossRef]
- Araki, H.; Jearnpipatkul, A.; Tatsumi, H.; Sakurai, T.; Ushio, K.; Muta, T.; Oshima, Y. Molecular and functional organization of yeast plasmid pSR1. J. Mol. Biol. 1985, 182, 191–203. [Google Scholar] [CrossRef]
- Gidoni, D.; Srivastava, V.; Carmi, N. Site-specific excisional recombination strategies for elimination of undesirable transgenes from crop plants. Vitr. Cell. Dev. Biol.- Plant 2008, 44, 457–467. [Google Scholar] [CrossRef]
- Mlynárová, L.; Conner, A.J.; Nap, J.P. Directed microspore-specific recombination of transgenic alleles to prevent pollen-mediated transmission of transgenes. Plant Biotechnol. J. 2006, 4, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Hamzeh, S.; Motallebi, M.; Zamani, M.R. Efficient seed-specifically regulated autoexcision of marker gene (nptII) with inducible expression of interest gene in transgenic Nicotiana tabacum. Turk. J. Biol. 2016, 40, 1–11. [Google Scholar] [CrossRef]
- Kopertekh, L.; Schulze, K.; Frolov, A.; Strack, D.; Broer, I.; Schiemann, J. Cre-mediated seed-specific transgene excision in tobacco. Plant Mol. Biol. 2010, 72, 597–605. [Google Scholar] [CrossRef]
- Polóniová, Z.; Jopčík, M.; Matušíková, I.; Libantová, J.; Moravčíková, J. The pollen- and embryo- specific Arabidopsis DLL promoter bears good potential for application in marker-free Cre/loxP self-excision strategy. Plant Cell Rep. 2015, 34, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Vaghchhipawala, Z.; Williams, E.J.; Fu, C.; Liu, J.; Lu, F.; Hall, E.L.; Guo, S.X.; Frank, L.; Gilbertson, L.A. Cre-mediated autoexcision of selectable marker genes in soybean, cotton, canola and maize transgenic plants. Plant Cell Rep. 2022, 42, 45–55. [Google Scholar] [CrossRef]
- Liu, W.; Yuan, J.S.; Stewart, C.N., Jr. Advanced genetic tools for plant biotechnology. Nat. Rev. Genet. 2013, 14, 781–793. [Google Scholar] [CrossRef]
- Sobecky, P.A.; Easter, C.L.; Bear, P.D.; Helinski, D.R. Characterization of the stable maintenance properties of the par region of broad-host-range plasmid RK2. J. Bacteriol. 1996, 178, 2086–2093. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kholodii, G.; Mindlin, S.; Gorlenko, Z.; Petrova, M.; Hobman, J.; Nikiforov, V. Translocation of transposition-deficient (TndPKLH2-like) transposons in the natural environment: Mechanistic insights from the study of adjacent DNA sequences. Microbiology 2004, 150, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Blechl, A.; Thomson, J.G. Small serine recombination systems ParA-MRS and CinH-RS2 perform precise excision of plastid DNA. Plant Biotechnol. J. 2017, 15, 1577–1589. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.G.; Ow, D.W. Site-specific recombination systems for the genetic manipulation of eukaryotic genomes. Genesis 2006, 44, 465–476. [Google Scholar] [CrossRef]
- He, Y.; Mudgett, M.; Zhao, Y. Advances in gene editing without residual transgenes in plants. Plant Physiol. 2022, 188, 1757–1768. [Google Scholar] [CrossRef]
- Srivastava, V.; Underwood, J.L.; Zhao, S. Dual-targeting by CRISPR/Cas9 for precise excision of transgenes from rice genome. Plant Cell Tissue Organ Cult. 2017, 129, 153–160. [Google Scholar] [CrossRef]
- Tan, J.; Wang, Y.; Chen, S.; Lin, Z.; Zhao, Y.; Xue, Y.; Luo, Y.; Liu, Y.G.; Zhu, Q. An efficient marker gene excision strategy based on CRISPR/Cas9-mediated homology-directed repair in rice. Int. J. Mol. Sci. 2022, 23, 1588. [Google Scholar] [CrossRef]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasmussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread translational inhibition by plant miRNAs and siRNAs. Science 2008, 320, 1185–1190. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Brookes, G.; Barfoot, P.; Melé, E.; Messeguer, J.; Bénétrix, F.; Bloc, D.; Foueillassar, X.; Fabié, A.; Poeydomenge, C. Genetically Modified Maize: Pollen Movement and Crop Co-Existence; PG Economics Ltd.: Dorchester, UK, 2004; pp. 3–20. [Google Scholar]
- Ortega-Molina, J. Results of the studies into the coexistence of genetically modified and conventional maize. In Proceedings of the COPA-COGECA Colloquy on the Co-Existence and Thresholds of Adventitious Presence on GMOs in Conventional Seeds, Brussels, Belgium, 2004. [Google Scholar]
- Luo, Y.; Guo, Z.; Li, L. Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev. Biol. 2013, 380, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Sunkar, R.; Zhou, C.; Shen, H.; Zhang, J.Y.; Matts, J.; Wolf, J.; Mann, D.G.J.; Stewart, C.N., Jr.; Tang, Y.; et al. Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol. J. 2012, 10, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.R.; Millwood, R.; Tang, Y.; Gou, J.; Sykes, R.W.; Turner, G.B.; Davis, M.F.; Sang, Y.; Wang, Z.Y.; Stewart, C.N., Jr. Field-grown miR156 transgenic switchgrass reproduction, yield, global gene expression analysis, and bioconfinement. Biotechnol. Biofuels 2017, 10, 255. [Google Scholar] [CrossRef] [PubMed]
- Trumbo, J.L.; Zhang, B.; Stewart, C.N., Jr. Manipulating microRNAs for improved biomass and biofuels from plant feedstocks. Plant Biotechnol. J. 2015, 13, 337–354. [Google Scholar] [CrossRef] [PubMed]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stockdale, J.N.; Millwood, R.J. Transgene Bioconfinement: Don’t Flow There. Plants 2023, 12, 1099. https://doi.org/10.3390/plants12051099
Stockdale JN, Millwood RJ. Transgene Bioconfinement: Don’t Flow There. Plants. 2023; 12(5):1099. https://doi.org/10.3390/plants12051099
Chicago/Turabian StyleStockdale, Jessica N., and Reginald J. Millwood. 2023. "Transgene Bioconfinement: Don’t Flow There" Plants 12, no. 5: 1099. https://doi.org/10.3390/plants12051099
APA StyleStockdale, J. N., & Millwood, R. J. (2023). Transgene Bioconfinement: Don’t Flow There. Plants, 12(5), 1099. https://doi.org/10.3390/plants12051099





