Moonlight Is Perceived as a Signal Promoting Genome Reorganization, Changes in Protein and Metabolite Profiles and Plant Growth
Abstract
:1. Introduction
2. Results
2.1. Exposure to FML Induces Epigenetic Variation
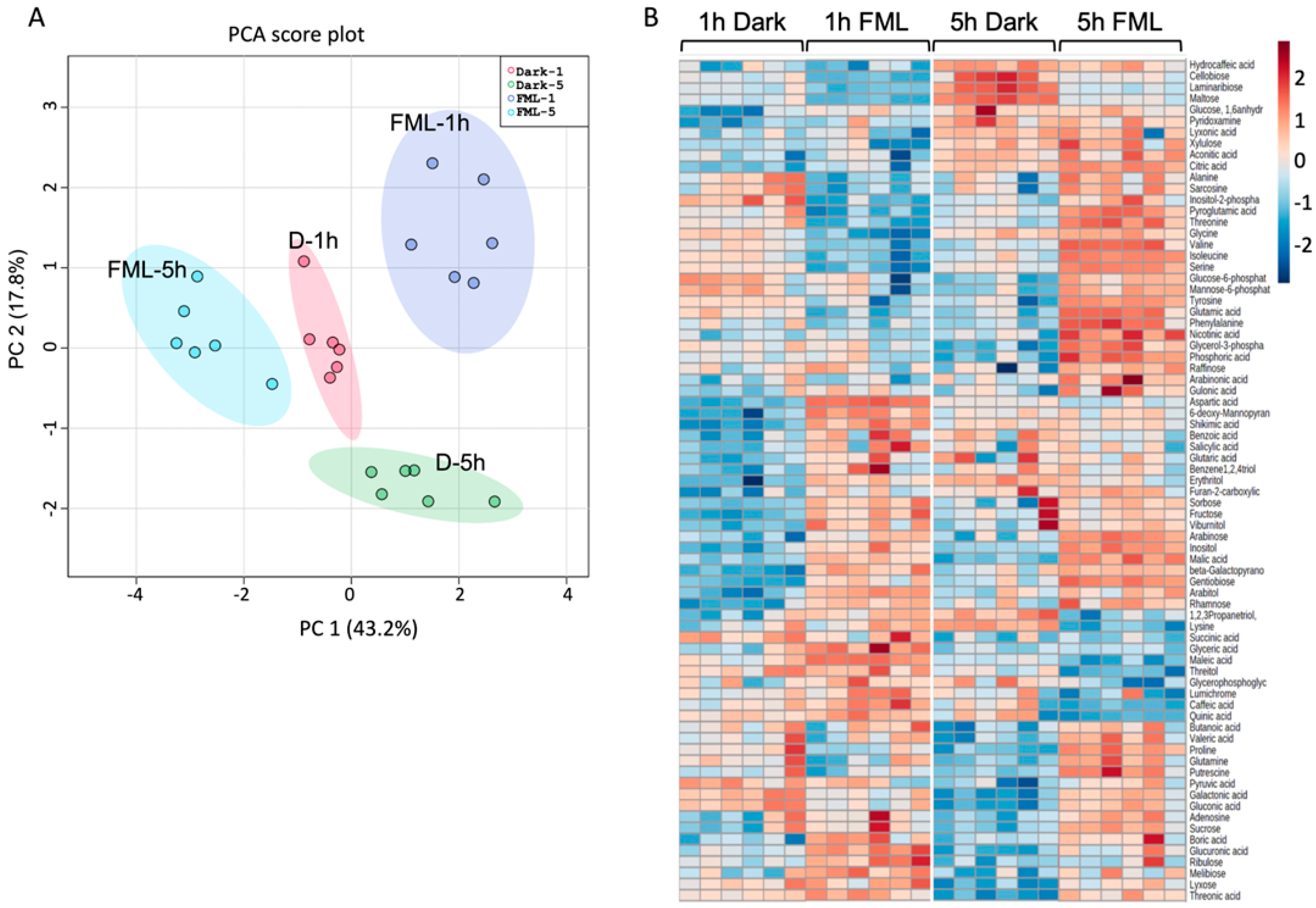
2.2. Exposure of Tobacco Plants to FML Induces Changes in Metabolic Profile
2.3. Exposure to FML Induces Changes in Proteome Profile: Upregulation of Photoreceptors and Stress Proteins
2.4. FML Enhances Growth of Indian Mustard Seedlings
3. Discussion
3.1. FML Is Perceived as a Signal Promoting Activation of Stress-Associated Substances
3.2. Upregulation of Photoreceptor by FML
3.3. Epigenetic Variation Induced by FML
3.4. Conclusions
4. Materials and Methods
4.1. Plant Growth Conditions and Exposure to Moonlight
4.2. Nuclei Isolation and Confocal Microscope Inspection
4.3. Acid Extraction of Proteins and Immunoblotting
4.4. DNA Extraction and Methylation Analysis
4.5. Metabolite Analysis
4.6. Proteome Analysis
4.7. RNA Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raven, J.A.; Cockell, C.S. Influence on photosynthesis of starlight, moonlight, planetlight, and light pollution (reflections on photosynthetically active radiation in the universe). Astrobiology 2006, 6, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Kronfeld-Schor, N.; Dominoni, D.; de la Iglesia, H.; Levy, O.; Herzog, E.D.; Dayan, T.; Helfrich-Forster, C. Chronobiology by moonlight. Proc. Biol. Sci. 2013, 280, 20123088. [Google Scholar] [CrossRef] [Green Version]
- Gorbunov, M.; Falkowski, P.G. Photoreceptors in Cnidarian hosts allow symbiotic corals to sense the light of the blue moon. Limnol. Oceanogr. 2002, 47, 309–315. [Google Scholar] [CrossRef]
- Phillips, S. An Encyclopaedia of Plants in Myth, Legend, Magic and Lore; The Crowood Press Ltd.: Marlborough, UK, 2013. [Google Scholar]
- Mayoral, O.; Solbes, J.; Cantó, J.; Pina, T. What Has Been Thought and Taught on the Lunar Influence on Plants in Agriculture? Perspective from Physics and Biology. Agronomy 2020, 10, 955. [Google Scholar] [CrossRef]
- Zürcher, E. Plants and the moon-traditions and phenomena. HerbalEGram 2011, 8, 1–14. [Google Scholar]
- Kolisko, L. The moon and the growth of plants. In Bray-on-Thames; Anthroposophical Agricultural Foundation: London, UK, 1936. [Google Scholar]
- Semmens, E. Effect of moonlight on the germination of seeds. Nature 1923, 111, 49–50. [Google Scholar] [CrossRef]
- Brown, F.A.; Chow, C.S. Lunar-correlated variations in water uptake by bean seeds. Biol. Bull. 1973, 145, 265–278. [Google Scholar] [CrossRef]
- Spruyt, E.; Verbelen, J.-P.; de Greef, J.A. Expression of circaseptan and circannual rhythmicity in the imbibition of dry stored bean seeds. Plant Physiol. 1987, 84, 707–710. [Google Scholar] [CrossRef] [Green Version]
- Graviou, E. Analogies between rhythms in plant material, in atmospheric pressure, and solar lunar periodicities. Int. J. Biometeorol. 1978, 22, 103–111. [Google Scholar] [CrossRef]
- Restrepo, J. La Luna: El sol Nocturno en los Trópicos y su Influencia en la Agricultura; (No. 630.2233 R436.); Servicio de Información Mesoamericano Sobre Agricultura Sostenible: Managua, Nicaragua, 2004. [Google Scholar]
- Ben-Attia, M.; Reinberg, A.; Smolensky, M.H.; Gadacha, W.; Khedaier, A.; Sani, M.; Touitou, Y.; Boughamni, N.G. Blooming rhythms of cactus Cereus peruvianus with nocturnal peak at full moon during seasons of prolonged daytime photoperiod. Chronobiol. Int. 2016, 33, 419–430. [Google Scholar] [CrossRef]
- Breitler, J.C.; Djerrab, D.; Leran, S.; Toniutti, L.; Guittin, C.; Severac, D.; Pratlong, M.; Dereeper, A.; Etienne, H.; Bertrand, B. Full moonlight-induced circadian clock entrainment in Coffea arabica. BMC Plant Biol. 2020, 20, 24. [Google Scholar] [CrossRef] [PubMed]
- Bourbousse, C.; Mestiri, I.; Zabulon, G.; Bourge, M.; Formiggini, F.; Koini, M.A.; Brown, S.C.; Fransz, P.; Bowler, C.; Barneche, F. Light signaling controls nuclear architecture reorganization during seedling establishment. Proc. Natl. Acad. Sci. USA 2015, 112, E2836–E2844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovarík, A.; Koukalová, B.; Lim, K.Y.; Matyásek, R.; Lichtenstein, C.P.; Leitch, A.R.; Bezdek, M. Comparative analysis of DNA methylation in tobacco heterochromatic sequences. Chromosome Res. 2000, 8, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Nagaki, K.; Shibata, F.; Suzuki, G.; Kanatani, A.; Ozaki, S.; Hironaka, A.; Kashihara, K.; Murata, M. Coexistence of NtCENH3 and two retrotransposons in tobacco centromeres. Chromosome Res. 2011, 19, 591–605. [Google Scholar] [CrossRef]
- Batista-Silva, W.; Heinemann, B.; Rugen, N.; Nunes-Nesi, A.; Araújo, W.L.; Braun, H.P.; Hildebrandt, T.M. The role of amino acid metabolism during abiotic stress release. Plant Cell Environ. 2019, 42, 1630–1644. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Zhang, Y.; Liu, Y.; Li, X.; Hao, G.; Han, Q.; Dirk, L.M.A.; Downie, A.B.; Ruan, Y.L.; Wang, J.; et al. Raffinose synthase enhances drought tolerance through raffinose synthesis or galactinol hydrolysis in maize and Arabidopsis plants. J. Biol. Chem. 2020, 295, 8064–8077. [Google Scholar] [CrossRef]
- Rai, V. Role of Amino Acids in Plant Responses to Stresses. Biol. Plant. 2002, 45, 481–487. [Google Scholar] [CrossRef]
- Janowitz, T.; Kneifel, H.; Piotrowski, M. Identification and characterization of plant agmatine iminohydrolase, the last missing link in polyamine biosynthesis of plants. FEBS Lett. 2003, 544, 258–261. [Google Scholar] [CrossRef] [Green Version]
- Adams, D.O.; Yang, S.F. Ethylene biosynthesis: Identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. USA 1979, 76, 170–174. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.I.; Fatma, M.; Per, T.S.; Anjum, N.A.; Khan, N.A. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 2015, 6, 462. [Google Scholar] [CrossRef] [Green Version]
- Johnson, P.R.; Ecker, J.R. The ethylene gas signal transduction pathway: A molecular perspective. Annu. Rev. Genet. 1998, 32, 227–254. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Wang, W.; Wu, H.; Gong, X.; Moriguchi, T. Polyamines function in stress tolerance: From synthesis to regulation. Front. Plant Sci. 2015, 6, 827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, U.K.; Islam, M.N.; Siddiqui, M.N.; Cao, X.; Khan, M.A.R. Proline, a multifaceted signalling molecule in plant responses to abiotic stress: Understanding the physiological mechanisms. Plant Biol. Stuttg. 2022, 24, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Kan, C.C.; Chung, T.Y.; Juo, Y.A.; Hsieh, M.H. Glutamine rapidly induces the expression of key transcription factor genes involved in nitrogen and stress responses in rice roots. BMC Genom. 2015, 16, 731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rausenberger, J.; Hussong, A.; Kircher, S.; Kirchenbauer, D.; Timmer, J.; Nagy, F.; Schäfer, E.; Fleck, C. An integrative model for phytochrome B mediated photomorphogenesis: From protein dynamics to physiology. PLoS ONE 2010, 5, e10721. [Google Scholar] [CrossRef]
- Pham, V.N.; Kathare, P.K.; Huq, E. Phytochromes and phytochrome interacting factors. Plant Physiol. 2018, 176, 1025–1038. [Google Scholar] [CrossRef] [Green Version]
- Fichman, Y.; Xiong, H.; Sengupta, S.; Morrow, J.; Loog, H.; Azad, R.K.; Hibberd, J.M.; Liscum, E.; Mittler, R. Phytochrome B regulates reactive oxygen signaling during abiotic and biotic stress in plants. New Phytol. 2023, 237, 1711–1727. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, J.H.; Park, C.M. A multifaceted action of phytochrome B in plant environmental adaptation. Front. Plant Sci. 2021, 12, 659712. [Google Scholar] [CrossRef]
- Klose, C.; Venezia, F.; Hussong, A.; Kircher, S.; Schäfer, E.; Fleck, C. Systematic analysis of how phytochrome B dimerization determines its specificity. Nat. Plants 2015, 1, 15090. [Google Scholar] [CrossRef]
- Chen, M.; Chory, J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011, 21, 664–671. [Google Scholar] [CrossRef] [Green Version]
- Khanna, R.; Shen, Y.; Marion, C.M.; Tsuchisaka, A.; Theologis, A.; Schäfer, E.; Quail, P.H. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 2007, 19, 3915–3929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labuz, J.; Sztatelman, O.; Banaś, A.K.; Gabryś, H. The expression of phototropins in Arabidopsis leaves: Developmental and light regulation. J. Exp. Bot. 2012, 63, 1763–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medzihradszky, M.; Bindics, J.; Ádám, É.; Viczián, A.; Klement, É.; Lorrain, S.; Gyula, P.; Mérai, Z.; Fankhauser, C.; Medzihradszky, K.F.; et al. Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell 2013, 25, 535–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phee, B.K.; Kim, J.I.; Shin, D.H.; Yoo, J.; Park, K.J.; Han, Y.J.; Kwon, Y.K.; Cho, M.H.; Jeon, J.S.; Bhoo, S.H.; et al. A novel protein phosphatase indirectly regulates phytochrome-interacting factor 3 via phytochrome. Biochem. J. 2008, 415, 247–255. [Google Scholar] [CrossRef]
- Kinoshita, T.; Doi, M.; Suetsugu, N.; Kagawa, T.; Wada, M.; Shimazaki, K. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 2001, 414, 656–660. [Google Scholar] [CrossRef]
- Inoue, S.; Matsushita, T.; Tomokiyo, Y.; Matsumoto, M.; Nakayama, K.I.; Kinoshita, T.; Shimazaki, K. Functional analyses of the activation loop of phototropin2 in Arabidopsis. Plant Physiol. 2011, 156, 117–128. [Google Scholar] [CrossRef] [Green Version]
- Inoue, S.; Kinoshita, T.; Matsumoto, M.; Nakayama, K.I.; Doi, M.; Shimazaki, K. Blue light-induced autophosphorylation of phototropin is a primary step for signaling. Proc. Natl. Acad. Sci. USA 2008, 105, 5626–5631. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, S.; Thomson, C.E.; Lamont, D.J.; Jones, M.A.; Christie, J.M. In vivo phosphorylation site mapping and functional characterization of Arabidopsis phototropin 1. Mol. Plant 2008, 1, 178–194. [Google Scholar] [CrossRef]
- Jones, M.A.; Christie, J.M. Phototropin receptor kinase activation by blue light. Plant Signal. Behav. 2008, 3, 44–46. [Google Scholar] [CrossRef] [Green Version]
- Takemiya, A.; Inoue, S.; Doi, M.; Kinoshita, T.; Shimazaki, K. Phototropins promote plant growth in response to blue light in low light environments. Plant Cell 2005, 17, 1120–1127. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Morozova, N.; Williams, L.; Libs, L.; Avivi, Y.; Grafi, G. Two phases of chromatin decondensation during cellular dedifferentiation of plant cells: Distinction between competence for cell-fate switch and a commitment for S phase. J. Biol. Chem. 2001, 276, 22772–22778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tessadori, F.; Chupeau, M.C.; Chupeau, Y.; Knip, M.; Germann, S.; van Driel, R.; Fransz, P.; Gaudin, V. Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J. Cell Sci. 2007, 120, 1200–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damri, M.; Granot, G.; Ben-Meir, H.; Avivi, Y.; Plaschkes, I.; Chalifa-Caspi, V.; Wolfson, M.; Fraifeld, V.; Grafi, G. Senescing cells share common features with dedifferentiating cells. Rejuvenation Res. 2009, 12, 435–443. [Google Scholar] [CrossRef] [PubMed]
- van Zanten, M.; Tessadori, F.; Bossen, L.; Peeters, A.J.M.; Fransz, P. Large-scale chromatin de-compaction induced by low light is not accompanied by nucleosomal displacement. Plant Signal. Behav. 2010, 5, 1677–1678. [Google Scholar] [CrossRef] [Green Version]
- Florentin, A.; Damri, M.; Grafi, G. Stress induces plant somatic cells to acquire some features of stem cells accompanied by selective chromatin reorganization. Dev. Dyn. 2013, 242, 1121–1133. [Google Scholar] [CrossRef]
- Givaty-Rapp, Y.; Yadav, N.S.; Khan, A.; Grafi, G. S1-type endonuclease 2 in dedifferentiating Arabidopsis protoplasts: Translocation to the nucleus in senescing protoplasts is associated with de-glycosylation. PLoS ONE 2017, 12, e0170067. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Freeman, L.; Kurumizaka, H.; Wolffe, A.P. Functional domains for assembly of histones H3 and H4 into the chromatin of Xenopus embryos. Proc. Natl. Acad. Sci. USA 1996, 93, 12780–12785. [Google Scholar] [CrossRef] [Green Version]
- Bourbousse, C.; Barneche, F.; Laloi, C. Plant Chromatin Catches the Sun. Front Plant Sci. 2020, 10, 1728. [Google Scholar] [CrossRef] [Green Version]
- Tessadori, F.; van Zanten, M.; Pavlova, P.; Clifton, R.; Pontvianne, F.; Snoek, L.B.; Millenaar, F.F.; Schulkes, R.K.; van Driel, R.; Voesenek, L.A.; et al. Phytochrome B and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet. 2009, 5, e1000638. [Google Scholar] [CrossRef] [Green Version]
- van Zanten, M.; Tessadori, F.; McLoughlin, F.; Smith, R.; Millenaar, F.F.; van Driel, R.; Voesenek, L.A.; Peeters, A.J.; Fransz, P. Photoreceptors CRYPTOCHROME2 and phytochrome B control chromatin compaction in Arabidopsis. Plant Physiol. 2010, 154, 1686–1696. [Google Scholar] [CrossRef] [Green Version]
- van Zanten, M.; Tessadori, F.; Peeters, A.J.; Fransz, P. Shedding light on large-scale chromatin reorganization in Arabidopsis thaliana. Mol. Plant 2012, 5, 583–590. [Google Scholar] [CrossRef] [Green Version]
- Trovato, M.; Funck, D.; Forlani, G.; Okumoto, S.; Amir, R. Editorial: Amino acids in plants: Regulation and functions in development and stress defense. Front. Plant Sci. 2021, 12, 772810. [Google Scholar] [CrossRef]
- Yan, S.; Liu, Q.; Li, W.; Yan, J.; Fernie, A.R. Raffinose family oligosaccharides: Crucial regulators of plant development and stress responses. Crit. Rev. Plant Sci. 2022, 41, 286–303. [Google Scholar] [CrossRef]
- Han, S.H.; Park, Y.J.; Park, C.M. Light priming of thermotolerance development in plants. Plant Signal. Behav. 2019, 14, 1554469. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Able, A.J.; Able, J.A. Priming crops for the future: Rewiring stress memory. Trends Plant Sci. 2022, 27, 699–716. [Google Scholar] [CrossRef]
- Saxena, P.K.; Fowke, L.C.; King, J. An efficient procedure for isolation of nuclei from plant protoplasts. Protoplasma 1985, 128, 184–189. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version, I.I. Plant Mol. Biol. Report. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef]
- Reshef, N.; Fait, A.; Agam, N. Grape berry position affects the diurnal dynamics of its metabolic profile. Plant Cell Environ. 2019, 42, 1897–1912. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinform. 2016, 55, 14.10.1–14.10.91. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| 1 h Exposure to FML vs. Dark | 5 h Exposure to FML vs. Dark | ||||
|---|---|---|---|---|---|
| Metabolite | FC (FML/D) | Up/Down | Metabolite | FC (FML/D) | Up/Down |
| Amino Acids | Amino Acids | ||||
| Alanine | 0.46 | Down | Alanine | 1.70 | Up |
| Aspartic acid | 2.49 | Up | Glutamic acid | 7.44 | Up |
| Glutamic acid | 0.32 | Down | Glutamine | 6.14 | Up |
| Glutamine | 0.58 | Down | Isoleucine | 2.11 | Up |
| Glycine | 0.32 | Down | Lysine | 0.70 | Down |
| Isoleucine | 0.50 | Down | Phenylalanine | 3.54 | Up |
| Phenylalanine | 0.70 | Down | Proline | 3.93 | Up |
| Proline | 0.56 | Down | Serine | 2.04 | Up |
| Serine | 0.41 | Down | Threonine | 1.80 | Up |
| Threonine | 0.62 | Down | Tyrosine | 5.58 | Up |
| Tyrosine | 0.43 | Down | Valine | 2.03 | Up |
| Valine | 0.61 | Down | |||
| Sugars | Sugars | ||||
| 6-deoxy-Mannopyranose | 1.41 | Up | Cellobiose | 0.73 | Down |
| β-Galactopyranosyl-1,3-arab. abinose | 1.62 | Up | Gentiobiose | 1.41 | Up |
| Fructose | 1.32 | Up | Glucose-6-phosphate | 1.34 | Up |
| Gentiobiose | 1.39 | Up | Glycerol-3-phosphate | 1.37 | Up |
| Inositol-2-phosphate | 0.75 | Down | Laminaribiose | 0.61 | Down |
| Ribulose | 1.50 | Up | Maltose | 0.67 | Down |
| Sorbose | 1.31 | Up | Mannose-6-phosphate | 1.42 | Up |
| Viburnitol | 1.31 | Up | Raffinose | 2.09 | Up |
| Other metabolites | Other metabolites | ||||
| Boric acid | 1.64 | Up | Boric acid | 1.40 | Up |
| Caffeic acid | 1.45 | Up | Butyric acid | 2.04 | Up |
| Quinic acid | 1.48 | Up | Phosphoric acid | 1.39 | Up |
| Sarcosine | 0.48 | Down | Putrescine | 1.52 | Up |
| Shikimic acid | 1.33 | Up | Quinic acid | 0.25 | Down |
| Sarcosine | 1.70 | Up | |||
| Gene Names | Protein Names | Avg (FML) | Avg (Dark) | FC (FML/Dark) | Adj p Value | Cell Comp. |
|---|---|---|---|---|---|---|
| LOC107767359 | Phytochrome B | 9.0 × 108 | 2.9 × 107 | 30.627 | 0.00614 | Nuc; Mem |
| LOC107777858 | Glutamine synthetase | 9.1 × 108 | 1.1 × 108 | 8.152 | 0.00618 | Cyt. |
| LOC107775921 | Heat shock 70 kDa protein | 8.3 × 109 | 1.4 × 109 | 5.927 | 0.00535 | Mem; Mit |
| LOC107828946 | Peroxidase | 7.1 × 108 | 1.3 × 108 | 5.664 | 0.00906 | CW |
| LOC107790714 | Heat shock 70 kDa protein 15-like | 8.5 × 109 | 1.6 × 109 | 5.298 | 0.00535 | Nuc; Mem; Cyt |
| LOC107769513 | PSI subunit V | 7.0 × 109 | 1.5 × 109 | 4.719 | 0.00822 | Chl |
| LOC107781119 | LHCP translocation defect-like | 5.9 × 109 | 1.3 × 109 | 4.467 | 0.00454 | Chl |
| LOC107769068 | Heat shock protein 90 | 9.2 × 108 | 2.1 × 108 | 4.418 | 0.04268 | nd |
| LOC107763119 | 23 kDa subunit of OES of photosystem II | 7.0 × 109 | 1.8 × 109 | 3.818 | 0.03765 | Chl |
| LOC107770030 | 60S ribosomal protein L30 | 1.1 × 108 | 3.0 × 107 | 3.558 | 0.03079 | Cyt |
| LOC107793292 | Phototropin-2 | 7.7 × 108 | 2.2 × 108 | 3.531 | 0.00657 | Nuc; Cyt |
| LOC107810219 | Chaperonin 60 subunit beta 2 | 7.7 × 1010 | 2.5 × 1010 | 3.046 | 0.00736 | Chl |
| LOC107806960 | Superoxide dismutase | 8.1 × 109 | 2.9 × 109 | 2.784 | 0.00535 | Chl |
| LOC107778847 | Malic enzyme | 7.8 × 109 | 2.9 × 109 | 2.657 | 0.00951 | Mem; Chl |
| LOC107820445 | Chaperone protein dnaJ A6 | 6.0 × 108 | 2.3 × 108 | 2.596 | 0.02283 | Cyt |
| LOC107825601 | Aconitate hydratase | 9.9 × 109 | 4.1 × 109 | 2.425 | 0.00005 | Cyt |
| LOC107768788 | 60S ribosomal protein L3-2-like | 1.1 × 108 | 5.1 × 107 | 2.196 | 0.02283 | Cyt |
| LOC107785603 | F-box protein PP2-B11-like | 2.5 × 108 | 5.2 × 108 | −2.08 | 0.00454 | nd |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singiri, J.R.; Priyanka, G.; Trishla, V.S.; Adler-Agmon, Z.; Grafi, G. Moonlight Is Perceived as a Signal Promoting Genome Reorganization, Changes in Protein and Metabolite Profiles and Plant Growth. Plants 2023, 12, 1121. https://doi.org/10.3390/plants12051121
Singiri JR, Priyanka G, Trishla VS, Adler-Agmon Z, Grafi G. Moonlight Is Perceived as a Signal Promoting Genome Reorganization, Changes in Protein and Metabolite Profiles and Plant Growth. Plants. 2023; 12(5):1121. https://doi.org/10.3390/plants12051121
Chicago/Turabian StyleSingiri, Jeevan R., Govindegowda Priyanka, Vikas S. Trishla, Zachor Adler-Agmon, and Gideon Grafi. 2023. "Moonlight Is Perceived as a Signal Promoting Genome Reorganization, Changes in Protein and Metabolite Profiles and Plant Growth" Plants 12, no. 5: 1121. https://doi.org/10.3390/plants12051121
APA StyleSingiri, J. R., Priyanka, G., Trishla, V. S., Adler-Agmon, Z., & Grafi, G. (2023). Moonlight Is Perceived as a Signal Promoting Genome Reorganization, Changes in Protein and Metabolite Profiles and Plant Growth. Plants, 12(5), 1121. https://doi.org/10.3390/plants12051121








