Chronic Ionizing Radiation of Plants: An Evolutionary Factor from Direct Damage to Non-Target Effects
Abstract
:1. Introduction
2. Natural Sources of Ionizing Radiation
3. Anthropogenic IR Impact on the Environment and Plant Biota
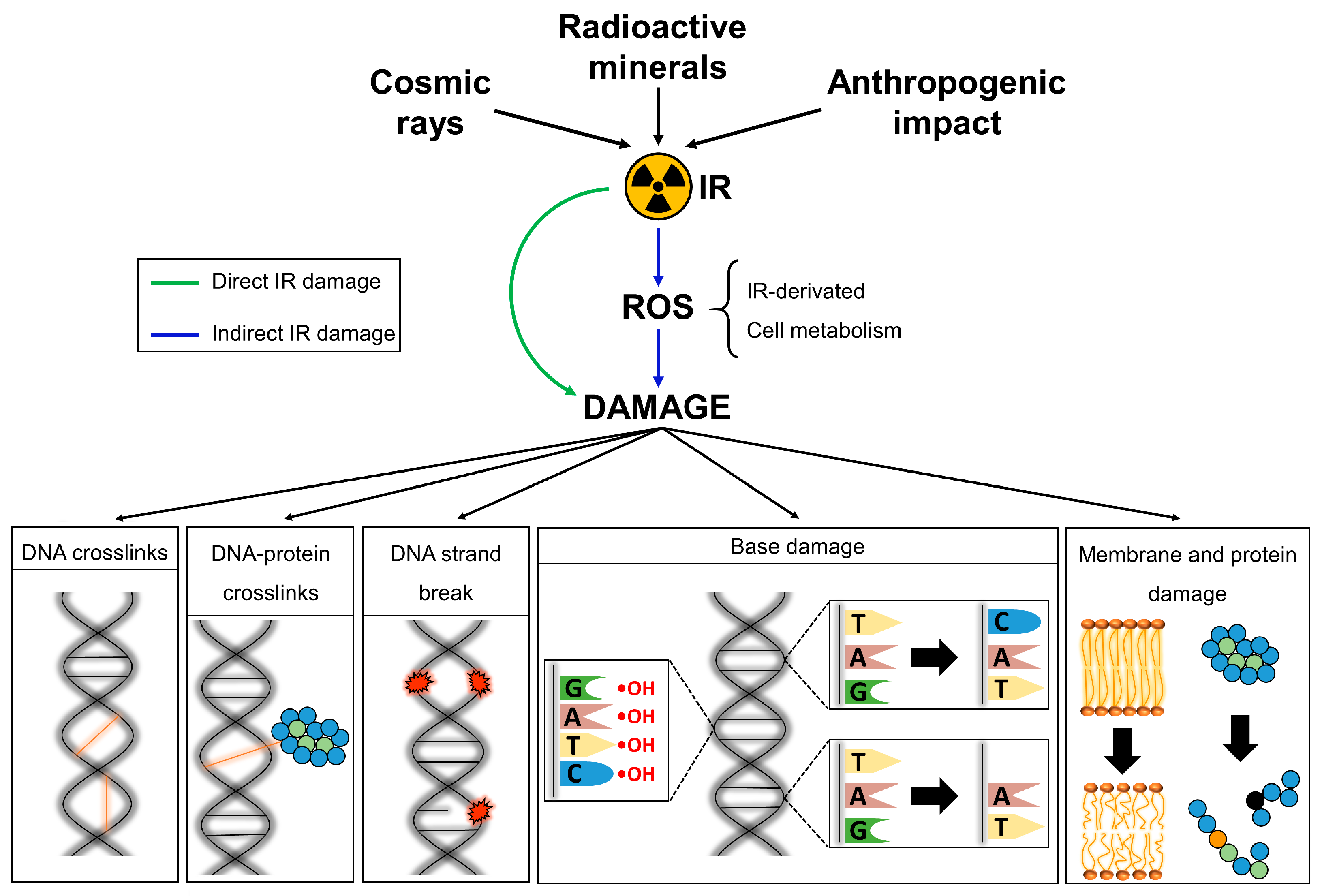
4. Cellular Effects of IR Exposure
5. IR Defence System
5.1. DNA Repair Mechanisms
5.2. Antioxidant Mechanisms
6. From Acute High-Dose to Chronic Low-Dose IR Exposure: Non-Target Effects in Plants
7. DNA Damage Response to Chronic IR under an Evolutionary Perspective
7.1. Early Earth Conditions: A Chronically Radioactive Environment
7.2. Plants Diversification and Terrestrialization: Was Chronic IR Exposure a Limiting Factor?
8. Perspectives
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Kobayashi, K.; Kaneko, T.; Saito, T.; Oshima, T. Amino acid formation in gas mixtures by high energy particle irradiation. Orig. Life Evol. Biosph. 1998, 28, 155–165. [Google Scholar] [CrossRef]
- Kobayashi, K.; Kaneko, T.; Saito, T. Characterization of complex organic compounds formed in simulated planetary atmospheres by the action of high energy particles. Adv. Space Res. 1999, 24, 461–464. [Google Scholar] [CrossRef]
- Geras’kin, S.A.; Fesenko, S.V.; Alexakhin, R.M. Effects of non-human species irradiation after the Chernobyl NPP accident. Environ. Int. 2008, 34, 880–897. [Google Scholar] [CrossRef]
- ICRP. Environmental Protection—The Concept and Use of Reference Animals and Plants; Annals of the ICRP; ICRP Publication: Ottawa, ON, Canada, 2008; Volume 38. [Google Scholar]
- Shu, Q.Y.; Forster, B.P.; Nakagawa, H.; Nakagawa, H. Plant Mutation Breeding and Biotechnology; FAO/IAEA: Vienna, Austria; CABI: Wallingford, UK, 2012. [Google Scholar]
- Sparrow, R.C.; Sparrow, A.H. Relative Radiosensitivities of Woody and Herbaceous Spermatophytes. Science 1965, 147, 1449–1451. [Google Scholar] [CrossRef] [PubMed]
- Capella, J.A.; Conger, A.D. Radiosensitivity and interphase chromosome volume in the gymnosperms. Radiat. Bot. 1967, 7, 137–149. [Google Scholar] [CrossRef]
- Yushkova, E. Involvement of DNA Repair Genes and System of Radiation-Induced Activation of Transposons in Formation of Transgenerational Effects. Front. Genet. 2020, 11, 596947. [Google Scholar] [CrossRef]
- Desouky, O.; Ding, N.; Zhou, G. Targeted and non-targeted effects of ionizing radiation. J. Radiat. Res. Appl. Sci. 2015, 8, 247–254. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, J.L.; Jordan, R.; Sun, J.; Ma, H.; Hsieb, A.W. Dose-dependent changes in the spectrum of mutations induced by ionizing radiation. Radiat. Res. 2000, 153, 312–317. [Google Scholar] [CrossRef]
- Dartnell, L.R. Ionizing radiation and life. Astrobiology 2011, 11, 551–582. [Google Scholar] [CrossRef] [Green Version]
- Joint Research Centre European Commission. European Atlas of Natural Radiation; Cinelli, G., De Cort, M., Tollefsen, T., Achatz, M., Ajtić, J., Ballabio, C., Barnet, I., Bochicchio, F., Borelli, P., Bossew, P., et al., Eds.; Publications Office of the European Union: Luxembourg, 2019. [Google Scholar]
- Ferrari, F.; Szuszkiewicz, E. Cosmic rays: A review for astrobiologists. Astrobiology 2009, 9, 413–436. [Google Scholar] [CrossRef] [PubMed]
- Turekian, K.K.; Graustein, W.C. 4.10—Natural Radionuclides in the Atmosphere. In Treatise on Geochemistry; Holland, H.D., Turekian, K.K., Eds.; Pergamon: Oxford, UK, 2003; pp. 261–279. [Google Scholar]
- Doglioni, C.; Pignatti, J.; Coleman, M. Why did life develop on the surface of the Earth in the Cambrian? Geosci. Front. 2016, 7, 865–873. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, K.; Tsuji, T. Abiotic Synthesis of Uracil from Carbon Monoxide, Nitrogen and Water by Proton Irradiation. Chem. Lett. 1997, 26, 903–904. [Google Scholar] [CrossRef]
- Bonilla, I.; Garcia-González, M.; Mateo, P. Boron requirement in cyanobacteria: Its possible role in the early evolution of photosynthetic organisms. Plant Physiol. 1990, 94, 1554–1560. [Google Scholar] [CrossRef] [Green Version]
- Bolaños, L.; Lukaszewski, K.; Bonilla, I.; Blevins, D. Why boron? Plant Physiol. Biochem. 2004, 42, 907–912. [Google Scholar] [CrossRef]
- Yoshinari, A.; Takano, J. Insights into the Mechanisms Underlying Boron Homeostasis in Plants. Front. Plant Sci. 2017, 8, 1951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, R.J.; Hayes, J.E.; Post, A.; Stangoulis, J.C.R.; Graham, R.D. A critical analysis of the causes of boron toxicity in plants. Plant Cell Environ. 2004, 27, 1405–1414. [Google Scholar] [CrossRef]
- Bassil, E.; Hu, H.; Brown, P.H. Use of Phenylboronic Acids to Investigate Boron Function in Plants. Possible Role of Boron in Transvacuolar Cytoplasmic Strands and Cell-to-Wall Adhesion. Plant Physiol. 2004, 136, 3383–3395. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, T.; Inui, Y.T.; Uraguchi, S.; Yoshizumi, T.; Matsunaga, S.; Mastui, M.; Umeda, M.; Fukui, K.; Fujiwara, T. Condensin II Alleviates DNA Damage and Is Essential for Tolerance of Boron Overload Stress in Arabidopsis. Plant Cell 2011, 23, 3533–3546. [Google Scholar] [CrossRef] [Green Version]
- Voxeur, A.; Fry, S.C. Glycosylinositol phosphorylceramides from Rosa cell cultures are boron-bridged in the plasma membrane and form complexes with rhamnogalacturonan II. Plant J. 2014, 79, 139–149. [Google Scholar] [CrossRef] [Green Version]
- Funakawa, H.; Miwa, K. Synthesis of borate cross-linked rhamnogalacturonan II. Front. Plant Sci. 2015, 6, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricardo, A.; Carrigan, M.A.; Olcott, A.N.; Benner, S.A. Borate Minerals Stabilize Ribose. Science 2004, 303, 196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grew, E.S.; Bada, J.L.; Hazen, R.M. Borate minerals and origin of the RNA world. Orig. Life Evol. Biosph. 2011, 41, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, M. The state-of-the-art on worldwide studies in some environments with elevated naturally occurring radioactive materials (NORM). Appl. Radiat. Isot. 1998, 49, 169–188. [Google Scholar] [CrossRef] [PubMed]
- Ghiassi-Nejad, M.; Mortazavi, S.M.; Cameron, J.R.; Niroomand-rad, A.; Karam, P.A. Very high background radiation areas of Ramsar, Iran: Preliminary biological studies. Health Phys. 2002, 82, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Ghiassi-Nejad, M.; Beitollahi, M.M.; Asefi, M.; Reza-Nejad, F. Exposure to (226)Ra from consumption of vegetables in the high level natural radiation area of Ramsar-Iran. J. Environ. Radioact. 2003, 66, 215–225. [Google Scholar] [CrossRef]
- Saghirzadeh, M.; Gharaati, M.R.; Mohammadi, S.; Ghiassi-Nejad, M. Evaluation of DNA damage in the root cells of Allium cepa seeds growing in soil of high background radiation areas of Ramsar-Iran. J. Environ. Radioact. 2008, 99, 1698–1702. [Google Scholar] [CrossRef]
- Boisson, F.; Miquel, J.C.; Cotret, O.; Fowler, S.W. 210Po and 210Pb cycling in a hydrothermal vent zone in the coastal Aegean Sea. Sci. Total Environ. 2001, 281, 111–119. [Google Scholar] [CrossRef]
- Charmasson, S.; Sarradin, P.-M.; Le Faouder, A.; Agarande, M.; Loyen, J.; Desbruyères, D. High levels of natural radioactivity in biota from deep-sea hydrothermal vents: A preliminary communication. J. Environ. Radioact. 2009, 100, 522–526. [Google Scholar] [CrossRef]
- Ardyna, M.; Lacour, L.; Sergi, S.; d’Ovidio, F.; Sallée, J.-B.; Rembauville, M.; Blain, S.; Tagliabue, A.; Schlitzer, R.; Jeandel, C.; et al. Hydrothermal vents trigger massive phytoplankton blooms in the Southern Ocean. Nat. Commun. 2019, 10, 2451. [Google Scholar] [CrossRef] [Green Version]
- Beresford, N.A.; Horemans, N.; Copplestone, D.; Raines, K.E.; Orizaola, G.; Wood, M.D.; Laanen, P.; Whitehead, H.C.; Burrows, J.E.; Tinsley, M.C.; et al. Towards solving a scientific controversy—The effects of ionising radiation on the environment. J. Environ. Radioact. 2020, 211, 106033. [Google Scholar] [CrossRef]
- Gibon, T.; Hertwich, E.G.; Arvesen, A.; Singh, B.; Verones, F. Health benefits, ecological threats of low-carbon electricity. Environ. Res. Lett. 2017, 12, 034023. [Google Scholar] [CrossRef]
- Kim, J.H.; Ryu, T.H.; Lee, S.S.; Lee, S.; Chung, B.Y. Ionizing radiation manifesting DNA damage response in plants: An overview of DNA damage signaling and repair mechanisms in plants. Plant Sci. 2019, 278, 44–53. [Google Scholar] [CrossRef]
- Van Harten, A.M. Mutation Breeding: Theory and Practical Applications; Cambridge University Press: Cambridge, UK, 1998. [Google Scholar]
- Weiss, W.; Larsson, C.M.; McKenney, C.; Minon, J.P.; Mobbs, S.; Schneider, T.; Umeki, H.; Hilden, W.; Pescatore, C.; Vesterlind, M. ICRP Publication 122: Radiological protection in geological disposal of long-lived solid radioactive waste. Ann. ICRP 2013, 42, 1–57. [Google Scholar] [CrossRef]
- UNSCEAR. Sources and Effects of Ionizing Radiation, United Nations Scientific Committee on the Effects of Atomic Radiation (UNSCEAR) 2008 Report, Volume I; United Nations: San Francisco, CA, USA, 2010. [Google Scholar]
- United Nations. End Nuclear Tests Day—History|United Nations. Available online: https://www.un.org/en/observances/end-nuclear-tests-day/history (accessed on 11 October 2022).
- Prăvălie, R. Nuclear weapons tests and environmental consequences: A global perspective. AMBIO 2014, 43, 729–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, E.W.; Molina, M.R.; Abella, M.K.I.L.; Nikolić-Hughes, I.; Ruderman, M.A. Radiation maps of ocean sediment from the Castle Bravo crater. Proc. Natl. Acad. Sci. USA 2019, 116, 15420–15424. [Google Scholar] [CrossRef] [Green Version]
- Fesenko, S. Review of radiation effects in non-human species in areas affected by the Kyshtym accident. J. Radiol. Prot. 2019, 39, R1–R17. [Google Scholar] [CrossRef]
- Beresford, N.A.; Fesenko, S.; Konoplev, A.; Skuterud, L.; Smith, J.T.; Voigt, G. Thirty years after the Chernobyl accident: What lessons have we learnt? J. Environ. Radioact. 2016, 157, 77–89. [Google Scholar] [CrossRef] [Green Version]
- Ludovici, G.M.; Chierici, A.; de Souza, S.O.; d’Errico, F.; Iannotti, A.; Malizia, A. Effects of Ionizing Radiation on Flora Ten Years after the Fukushima Dai-ichi Disaster. Plants 2022, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- IAEA. Effects of Ionizing Radiation on Plants and Animals at Levels Implied by Current Radiation Protection Standards; International Atomic Energy Agency: Vienna, Austria, 1992. [Google Scholar]
- Yoschenko, V.; Nanba, K.; Yoshida, S.; Watanabe, Y.; Takase, T.; Sato, N.; Keitoku, K. Morphological abnormalities in Japanese red pine (Pinus densiflora) at the territories contaminated as a result of the accident at Fukushima Dai-Ichi Nuclear Power Plant. J. Environ. Radioact. 2016, 165, 60–67. [Google Scholar] [CrossRef]
- Geras’kin, S.; Yoschenko, V.; Bitarishvili, S.; Makarenko, E.; Vasiliev, D.; Prazyan, A.; Lychenkova, M.; Nanba, K. Multifaceted effects of chronic radiation exposure in Japanese red pines from Fukushima prefecture. Sci. Total Environ. 2021, 763, 142946. [Google Scholar] [CrossRef]
- Watanabe, Y.; Ichikawa, S.E.; Kubota, M.; Hoshino, J.; Kubota, Y.; Maruyama, K.; Fuma, S.; Kawaguchi, I.; Yoschenko, V.I.; Yoshida, S. Morphological defects in native Japanese fir trees around the Fukushima Daiichi Nuclear Power Plant. Sci. Rep. 2015, 5, 13232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinhauser, G.; Brandl, A.; Johnson, T.E. Comparison of the Chernobyl and Fukushima nuclear accidents: A review of the environmental impacts. Sci. Total Environ. 2014, 470–471, 800–817. [Google Scholar] [CrossRef] [PubMed]
- Caplin, N.; Willey, N. Ionizing Radiation, Higher Plants, and Radioprotection: From Acute High Doses to Chronic Low Doses. Front. Plant Sci. 2018, 9, 847. [Google Scholar] [CrossRef]
- ICRP. The 2007 Recommendations of the International Commission on Radiological Protection. ICRP publication 103. Ann. ICRP 2007, 37, 1–332. [Google Scholar] [CrossRef]
- Harrison, F.L.; Anderson, S.L. Taxonomic and developmental aspects of radiosensitivity. In Proceedings of the Conference: International Symposium on Ionising Radiation, Stockholm, Sweden, 20–24 May 1996; Lawrence Livermore National Laboratory: Livermore, CA, USA, 1996. 33p. [Google Scholar]
- Adam-Guillermin, C.; Hertal-Aas, T.; Oughton, D.; Blanchard, L.; Alonzo, F.; Armant, O.; Horemans, N. Radiosensitivity and transgenerational effects in non-human species. Ann. ICRP 2018, 47, 327–341. [Google Scholar] [CrossRef]
- Kam, W.W.-Y.; Banati, R.B. Effects of ionizing radiation on mitochondria. Free Radic. Biol. Med. 2013, 65, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Meessen, J.; Backhaus, T.; Brandt, A.; Raguse, M.; Bottger, U.; de Vera, J.P.; de la Torre, R. The Effect of High-Dose Ionizing Radiation on the Isolated Photobiont of the Astrobiological Model Lichen Circinaria gyrosa. Astrobiology 2017, 17, 154–162. [Google Scholar] [CrossRef]
- Maremonti, E.; Brede, D.A.; Olsen, A.-K.; Eide, D.M.; Berg, E.S. Ionizing radiation, genotoxic stress, and mitochondrial DNA copy-number variation in Caenorhabditis elegans: Droplet digital PCR analysis. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2020, 858–860, 503277. [Google Scholar] [CrossRef]
- Gill, S.S.; Anjum, N.A.; Gill, R.; Jha, M.; Tuteja, N. DNA Damage and Repair in Plants under Ultraviolet and Ionizing Radiations. Sci. World J. 2015, 2015, 250158. [Google Scholar] [CrossRef] [Green Version]
- Nakano, T.; Xu, X.; Salem, A.M.H.; Shoulkamy, M.I.; Ide, H. Radiation-induced DNA-protein cross-links: Mechanisms and biological significance. Free Radic. Biol. Med. 2017, 107, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Manova, V.; Gruszka, D. DNA damage and repair in plants—From models to crops. Front. Plant Sci. 2015, 6, 885. [Google Scholar] [CrossRef] [Green Version]
- Cadet, J.; Wagner, J.R. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol. 2013, 5, a012559. [Google Scholar] [CrossRef] [PubMed]
- Poetsch, A.R. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput. Struct. Biotechnol. J. 2020, 18, 207–219. [Google Scholar] [CrossRef]
- Saha, G.B. Physics and Radiobiology of Nuclear Medicine, 4th ed.; Springer: New York, NY, USA, 2013. [Google Scholar]
- Le Caër, S. Water Radiolysis: Influence of Oxide Surfaces on H2 Production under Ionizing Radiation. Water 2011, 3, 235–253. [Google Scholar] [CrossRef] [Green Version]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, A.; Kumar, A.; Kaur, N. ROS and oxidative burst: Roots in plant development. Plant Divers. 2020, 42, 33–43. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef] [Green Version]
- Enderle, J.; Dorn, A.; Puchta, H. DNA- and DNA-Protein-Crosslink Repair in Plants. Int. J. Mol. Sci. 2019, 20, 4304. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.-Q. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, X.; He, Z.; Chen, N.; Tang, Z.; Wang, Q.; Cai, Y. The Roles of Environmental Factors in Regulation of Oxidative Stress in Plant. Biomed. Res. Int. 2019, 2019, 9732325. [Google Scholar] [CrossRef] [PubMed]
- Dextraze, M.-E.; Gantchev, T.; Girouard, S.; Hunting, D. DNA interstrand cross-links induced by ionizing radiation: An unsung lesion. Mutat. Res. Rev. Mutat. Res. 2010, 704, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N.; Ahmad, P.; Panda, B.B.; Tuteja, R. Genotoxic stress in plants: Shedding light on DNA damage, repair and DNA repair helicases. Mutat. Res. Rev. Mutat. Res. 2009, 681, 134–149. [Google Scholar] [CrossRef] [PubMed]
- Nisa, M.U.; Huang, Y.; Benhamed, M.; Raynaud, C. The Plant DNA Damage Response: Signaling Pathways Leading to Growth Inhibition and Putative Role in Response to Stress Conditions. Front. Plant Sci. 2019, 10, 653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raina, A.; Sahu, P.K.; Laskar, R.A.; Rajora, N.; Sao, R.; Khan, S.; Ganai, R.A. Mechanisms of Genome Maintenance in Plants: Playing It Safe With Breaks and Bumps. Front. Genet. 2021, 12, 675686. [Google Scholar] [CrossRef]
- de Medeiros, N.M.C.; de Medeiros, A.L.M.; Silva, H.C.; Scortecci, K.C. Recent Advances in Plant DNA Repair. In Advances in DNA Repair; Chen, C.C., Ed.; IntechOpen: London, UK, 2015. [Google Scholar]
- Grogan, D.W. Photoreactivation in an archaeon from geothermal environments. Microbiology 1997, 143, 1071–1076. [Google Scholar] [CrossRef] [Green Version]
- Rohner, N. Evolution: A Dark Past. Curr. Biol. 2018, 28, R1190–R1192. [Google Scholar] [CrossRef] [Green Version]
- Adachi, S.; Minamisawa, K.; Okushima, Y.; Inagaki, S.; Yoshiyama, K.; Kondou, Y.; Kaminuma, E.; Kawashima, M.; Toyoda, T.; Matsui, M.; et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 10004–10009. [Google Scholar] [CrossRef] [Green Version]
- Volkova, P.Y.; Duarte, G.T.; Soubigou-Taconnat, L.; Kazakova, E.A.; Pateyron, S.; Bondarenko, V.S.; Bitarishvili, S.V.; Makarenko, E.S.; Churyukin, R.S.; Lychenkova, M.A.; et al. Early response of barley embryos to low- and high-dose gamma irradiation of seeds triggers changes in the transcriptional profile and an increase in hydrogen peroxide content in seedlings. J. Agron. Crop Sci. 2020, 206, 277–295. [Google Scholar] [CrossRef]
- Rea, G.; Esposito, D.; Damasso, M.; Serafini, A.; Margonelli, A.; Faraloni, C.; Torzillo, G.; Zanini, A.; Bertalan, I.; Johanningmeier, U.; et al. Ionizing radiation impacts photochemical quantum yield and oxygen evolution activity of Photosystem II in photosynthetic microorganisms. Int. J. Radiat. Biol. 2008, 84, 867–877. [Google Scholar] [CrossRef]
- Saenen, E.; Horemans, N.; Vanhoudt, N.; Vandenhove, H.; Biermans, G.; Van Hees, M.; Wannijn, J.; Vangronsveld, J.; Cuypers, A. Effects of pH on uranium uptake and oxidative stress responses induced in Arabidopsis thaliana. Environ. Toxicol. Chem. 2013, 32, 2125–2133. [Google Scholar] [CrossRef]
- Vanhoudt, N.; Horemans, N.; Biermans, G.; Saenen, E.; Wannijn, J.; Nauts, R.; Van Hees, M.; Vandenhove, H. Uranium affects photosynthetic parameters in Arabidopsis thaliana. Environ. Exp. Bot. 2014, 97, 22–29. [Google Scholar] [CrossRef]
- Vanhoudt, N.; Horemans, N.; Wannijn, J.; Nauts, R.; Van Hees, M.; Vandenhove, H. Primary stress responses in Arabidopsis thaliana exposed to gamma radiation. J. Environ. Radioact. 2014, 129, 1–6. [Google Scholar] [CrossRef]
- Babina, D.; Podobed, M.; Bondarenko, E.; Kazakova, E.; Bitarishvili, S.; Podlutskii, M.; Mitsenyk, A.; Prazyan, A.; Gorbatova, I.; Shesterikova, E.; et al. Seed Gamma Irradiation of Arabidopsis thaliana ABA-Mutant Lines Alters Germination and Does Not Inhibit the Photosynthetic Efficiency of Juvenile Plants. Dose-Response 2020, 18, 1559325820979249. [Google Scholar] [CrossRef]
- Volkova, P.Y.; Duarte, G.T.; Kazakova, E.A.; Makarenko, E.S.; Bitarishvili, S.V.; Bondarenko, V.S.; Perevolotskii, A.N.; Geras’kin, S.A.; Garbaruk, D.K.; Turchin, L.M. Radiosensitivity of herbaceous plants to chronic radiation exposure: Field study in the Chernobyl exclusion zone. Sci. Total Environ. 2021, 777, 146206. [Google Scholar] [CrossRef]
- Richter, C.; Park, J.W.; Ames, B.N. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl. Acad. Sci. USA 1988, 85, 6465–6467. [Google Scholar] [CrossRef] [Green Version]
- May, A.; Bohr, V.A. Gene-Specific Repair of γ-Ray-Induced DNA Strand Breaks in Colon Cancer Cells: No Coupling to Transcription and No Removal from the Mitochondrial Genome. Biochem. Biophys. Res. Commun. 2000, 269, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Goto, S.; Kawakatsu, M.; Urata, Y.; Li, T.-S. Mitochondrial dysfunction, a probable cause of persistent oxidative stress after exposure to ionizing radiation. Free Radic. Res. 2012, 46, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Larsen, N.B.; Rasmussen, M.; Rasmussen, L.J. Nuclear and mitochondrial DNA repair: Similar pathways? Mitochondrion 2005, 5, 89–108. [Google Scholar] [CrossRef]
- Boesch, P.; Weber-Lotfi, F.; Ibrahim, N.; Tarasenko, V.; Cosset, A.; Paulus, F.; Lightowlers, R.N.; Dietrich, A. DNA repair in organelles: Pathways, organization, regulation, relevance in disease and aging. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2011, 1813, 186–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexeyev, M.; Shokolenko, I.; Wilson, G.; LeDoux, S. The maintenance of mitochondrial DNA integrity—Critical analysis and update. Cold Spring Harb. Perspect. Biol. 2013, 5, a012641. [Google Scholar] [CrossRef] [Green Version]
- Nair, K.A.S.; Netrawali, M.S. Sensitivity of Light-grown and Dark-grown Euglena Cells to Gamma-irradiation. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1979, 36, 223–231. [Google Scholar] [CrossRef]
- Hayashi, H.; Narumi, I.; Wada, S.; Kikuchi, M.; Furuta, M.; Uehara, K.; Watanabe, H. Light dependency of resistance to ionizing radiation in Euglena gracilis. J. Plant Physiol. 2004, 161, 1101–1106. [Google Scholar] [CrossRef]
- Martin, L.M.; Marples, B.; Coffey, M.; Lawler, M.; Lynch, T.H.; Hollywood, D.; Marignol, L. DNA mismatch repair and the DNA damage response to ionizing radiation: Making sense of apparently conflicting data. Cancer Treat. Rev. 2010, 36, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S. Base excision repair: A critical player in many games. DNA Repair 2014, 19, 14–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masaoka, A.; Horton, J.K.; Beard, W.A.; Wilson, S.H. DNA polymerase β and PARP activities in base excision repair in living cells. DNA Repair 2009, 8, 1290–1299. [Google Scholar] [CrossRef] [Green Version]
- Roldán-Arjona, T.; Ariza, R.R.; Córdoba-Cañero, D. DNA Base Excision Repair in Plants: An Unfolding Story With Familiar and Novel Characters. Front. Plant Sci. 2019, 10, 1055. [Google Scholar] [CrossRef] [Green Version]
- Amiard, S.; Charbonnel, C.; Allain, E.; Depeiges, A.; White, C.I.; Gallego, M.E. Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell 2010, 22, 3020–3033. [Google Scholar] [CrossRef] [Green Version]
- Saldivar, J.C.; Cortez, D.; Cimprich, K.A. The essential kinase ATR: Ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017, 18, 622–636. [Google Scholar] [CrossRef] [Green Version]
- Ogita, N.; Okushima, Y.; Tokizawa, M.; Yamamoto, Y.Y.; Tanaka, M.; Seki, M.; Makita, Y.; Matsui, M.; Okamoto-Yoshiyama, K.; Sakamoto, T.; et al. Identifying the target genes of SUPPRESSOR OF GAMMA RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 2018, 94, 439–453. [Google Scholar] [CrossRef] [Green Version]
- Meschichi, A.; Zhao, L.; Reeck, S.; White, C.; Da Ines, O.; Sicard, A.; Pontvianne, F.; Rosa, S. The plant-specific DDR factor SOG1 increases chromatin mobility in response to DNA damage. EMBO Rep. 2022, 23, e54736. [Google Scholar] [CrossRef] [PubMed]
- Biedermann, S.; Harashima, H.; Chen, P.; Heese, M.; Bouyer, D.; Sofroni, K.; Schnittger, A. The retinoblastoma homolog RBR1 mediates localization of the repair protein RAD51 to DNA lesions in Arabidopsis. EMBO J. 2017, 36, 1279–1297. [Google Scholar] [CrossRef] [PubMed]
- Horvath, B.M.; Kourova, H.; Nagy, S.; Nemeth, E.; Magyar, Z.; Papdi, C.; Ahmad, Z.; Sanchez-Perez, G.F.; Perilli, S.; Blilou, I.; et al. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J. 2017, 36, 1261–1278. [Google Scholar] [CrossRef]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Danielsen, J.M.R.; Yang, Y.-G.; Qi, Y. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Wei, W.; Li, M.-M.; Wu, Y.-S.; Ba, Z.; Jin, K.-X.; Li, M.-M.; Liao, Y.-Q.; Adhikari, S.; Chong, Z.; et al. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 2014, 24, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Weimer, A.K.; Biedermann, S.; Harashima, H.; Roodbarkelari, F.; Takahashi, N.; Foreman, J.; Guan, Y.; Pochon, G.; Heese, M.; Van Damme, D.; et al. The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 2016, 35, 2068–2086. [Google Scholar] [CrossRef]
- Enderle, J.; Dorn, A.; Beying, N.; Trapp, O.; Puchta, H. The Protease WSS1A, the Endonuclease MUS81, and the Phosphodiesterase TDP1 Are Involved in Independent Pathways of DNA-protein Crosslink Repair in Plants. Plant Cell 2019, 31, 775–790. [Google Scholar] [CrossRef]
- Hanada, K.; Budzowska, M.; Modesti, M.; Maas, A.; Wyman, C.; Essers, J.; Kanaar, R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006, 25, 4921–4932. [Google Scholar] [CrossRef] [Green Version]
- Geuting, V.; Kobbe, D.; Hartung, F.; Dürr, J.; Focke, M.; Puchta, H. Two Distinct MUS81-EME1 Complexes from Arabidopsis Process Holliday Junctions. Plant Physiol. 2009, 150, 1062–1071. [Google Scholar] [CrossRef] [Green Version]
- Barker, S.; Weinfeld, M.; Zheng, J.; Li, L.; Murray, D. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J. Biol. Chem. 2005, 280, 33826–33838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Block-Schmidt, A.S.; Dukowic-Schulze, S.; Wanieck, K.; Reidt, W.; Puchta, H. BRCC36A is epistatic to BRCA1 in DNA crosslink repair and homologous recombination in Arabidopsis thaliana. Nucleic Acids Res. 2010, 39, 146–154. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Sofo, A.; Scopa, A.; Nuzzaci, M.; Vitti, A. Ascorbate Peroxidase and Catalase Activities and Their Genetic Regulation in Plants Subjected to Drought and Salinity Stresses. Int. J. Mol. Sci. 2015, 16, 13561–13578. [Google Scholar] [CrossRef] [Green Version]
- Bela, K.; Riyazuddin, R.; Csiszár, J. Plant Glutathione Peroxidases: Non-Heme Peroxidases with Large Functional Flexibility as a Core Component of ROS-Processing Mechanisms and Signalling. Antioxidants 2022, 11, 1624. [Google Scholar] [CrossRef]
- Wituszyńska, W.; Karpiński, S. Programmed cell death as a response to high light, UV and drought stress in plants. In Abiotic Stress: Plant Responses and Applications in Agriculture; Books on Demand: Norderstedt, Germany, 2013; pp. 207–246. [Google Scholar] [CrossRef] [Green Version]
- Sadiq, M.; Akram, N.A.; Ashraf, M.; Al-Qurainy, F.; Ahmad, P. Alpha-Tocopherol-Induced Regulation of Growth and Metabolism in Plants Under Non-stress and Stress Conditions. J. Plant Growth Regul. 2019 38, 1325–1340. [CrossRef]
- Falk, J.; Munné-Bosch, S. Tocochromanol functions in plants: Antioxidation and beyond. J. Exp. Bot. 2010, 61, 1549–1566. [Google Scholar] [CrossRef] [Green Version]
- Kavi Kishor, P.B.; Suravajhala, P.; Rathnagiri, P.; Sreenivasulu, N. Intriguing Role of Proline in Redox Potential Conferring High Temperature Stress Tolerance. Front. Plant Sci. 2022, 13, 867531. [Google Scholar] [CrossRef] [PubMed]
- Kocsy, G.; Laurie, R.; Szalai, G.; Szilágyi, V.; Simon-Sarkadi, L.; Galiba, G.; De Ronde, J.A. Genetic manipulation of proline levels affects antioxidants in soybean subjected to simultaneous drought and heat stresses. Physiol. Plant. 2005, 124, 227–235. [Google Scholar] [CrossRef]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Mahmud, J.A.; Fujita, M. Regulation of Ascorbate-Glutathione Pathway in Mitigating Oxidative Damage in Plants under Abiotic Stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef] [Green Version]
- Zaka, R.; Vandecasteele, C.M.; Misset, M.T. Effects of low chronic doses of ionizing radiation on antioxidant enzymes and G6PDH activities in Stipa capillata (Poaceae). J. Exp. Bot. 2002, 53, 1979–1987. [Google Scholar] [CrossRef]
- Al-Rumaih, M.M.; Al-Rumaih, M.M. Influence of Ionizing Radiation on Antioxidant Enzymes in Three Species of Trigonella. Am. J. Environ. Sci. 2008, 4, 151–156. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.S.; Kim, J.-B.; Goh, E.J.; Kim, W.-J.; Kim, S.H.; Seo, Y.W.; Jang, C.S.; Kang, S.-Y. Antioxidant response of Arabidopsis plants to gamma irradiation: Genome-wide expression profiling of the ROS scavenging and signal transduction pathways. J. Plant Physiol. 2011, 168, 1960–1971. [Google Scholar] [CrossRef]
- Van Hoeck, A.; Horemans, N.; Van Hees, M.; Nauts, R.; Knapen, D.; Vandenhove, H.; Blust, R. β-Radiation Stress Responses on Growth and Antioxidative Defense System in Plants: A Study with Strontium-90 in Lemna minor. Int. J. Mol. Sci. 2015, 16, 15309–15327. [Google Scholar] [CrossRef] [Green Version]
- Volkova, P.Y.; Geras’kin, S.A.; Kazakova, E.A. Radiation exposure in the remote period after the Chernobyl accident caused oxidative stress and genetic effects in Scots pine populations. Sci. Rep. 2017, 7, 43009. [Google Scholar] [CrossRef] [Green Version]
- Hong, M.J.; Kim, D.Y.; Ahn, J.W.; Kang, S.Y.; Seo, Y.W.; Kim, J.B. Comparison of radiosensitivity response to acute and chronic gamma irradiation in colored wheat. Genet. Mol. Biol. 2018, 41, 611–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kariuki, J.; Horemans, N.; Saenen, E.; Van Hees, M.; Verhoeven, M.; Nauts, R.; Van Gompel, A.; Wannijn, J.; Cuypers, A. The responses and recovery after gamma irradiation are highly dependent on leaf age at the time of exposure in rice (Oryza sativa L.). Environ. Exp. Bot. 2019, 162, 157–167. [Google Scholar] [CrossRef]
- Pan, J.; Zhang, L.; Chen, M.; Ruan, Y.; Li, P.; Guo, Z.; Liu, B.; Ruan, Y.; Xiao, M.; Huang, Y. Identification and charactering of APX genes provide new insights in abiotic stresses response in Brassica napus. PeerJ 2022, 10, e13166. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.W.; Doolittle, R.F. A comparison of evolutionary rates of the two major kinds of superoxide dismutase. J. Mol. Evol. 1992, 34, 175–184. [Google Scholar] [CrossRef]
- Bannister, W.H.; Bannister, J.V.; Barra, D.; Bond, J.; Bossa, F. Evolutionary aspects of superoxide dismutase: The copper/zinc enzyme. Free Radic. Res. Commun. 1991, 12–13 Pt 1, 349–361. [Google Scholar] [CrossRef]
- Geigenberger, P.; Thormählen, I.; Daloso, D.M.; Fernie, A.R. The Unprecedented Versatility of the Plant Thioredoxin System. Trends Plant Sci. 2017, 22, 249–262. [Google Scholar] [CrossRef]
- Reichheld, J.P.; Khafif, M.; Riondet, C.; Droux, M.; Bonnard, G.; Meyer, Y. Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 2007, 19, 1851–1865. [Google Scholar] [CrossRef] [Green Version]
- Talla, S.; Riazunnisa, K.; Padmavathi, L.; Sunil, B.; Rajsheel, P.; Raghavendra, A.S. Ascorbic acid is a key participant during the interactions between chloroplasts and mitochondria to optimize photosynthesis and protect against photoinhibition. J. Biosci. 2011, 36, 163–173. [Google Scholar] [CrossRef]
- Xiao, M.; Li, Z.; Zhu, L.; Wang, J.; Zhang, B.; Zheng, F.; Zhao, B.; Zhang, H.; Wang, Y.; Zhang, Z. The Multiple Roles of Ascorbate in the Abiotic Stress Response of Plants: Antioxidant, Cofactor, and Regulator. Front. Plant Sci. 2021, 12, 598173. [Google Scholar] [CrossRef]
- Horemans, N.; Foyer, C.H.; Asard, H. Transport and action of ascorbate at the plant plasma membrane. Trends Plant Sci. 2000, 5, 263–267. [Google Scholar] [CrossRef]
- Ma, L.; Wang, Y.; Liu, W.; Liu, Z. Overexpression of an alfalfa GDP-mannose 3, 5-epimerase gene enhances acid, drought and salt tolerance in transgenic Arabidopsis by increasing ascorbate accumulation. Biotechnol. Lett. 2014, 36, 2331–2341. [Google Scholar] [CrossRef] [PubMed]
- Gaafar, A.A.; Ali, S.I.; El-Shawadfy, M.A.; Salama, Z.A.; Sękara, A.; Ulrichs, C.; Abdelhamid, M.T. Ascorbic Acid Induces the Increase of Secondary Metabolites, Antioxidant Activity, Growth, and Productivity of the Common Bean under Water Stress Conditions. Plants 2020, 9, 627. [Google Scholar] [CrossRef] [PubMed]
- Broad, R.C.; Bonneau, J.P.; Beasley, J.T.; Roden, S.; Sadowski, P.; Jewell, N.; Brien, C.; Berger, B.; Tako, E.; Glahn, R.P.; et al. Effect of Rice GDP-L-Galactose Phosphorylase Constitutive Overexpression on Ascorbate Concentration, Stress Tolerance, and Iron Bioavailability in Rice. Front. Plant Sci. 2020, 11, 595439. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xiao, Y.; Chen, W.; Tang, K.; Zhang, L. Increased vitamin C content accompanied by an enhanced recycling pathway confers oxidative stress tolerance in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Conger, B.V. Radioprotective effects of ascorbic acid in barley seeds. Radiat. Bot. 1975, 15, 39–48. [Google Scholar] [CrossRef]
- Lohr, M. Chapter 21—Carotenoids. In The Chlamydomonas Sourcebook, 2nd ed.; Harris, E.H., Stern, D.B., Witman, G.B., Eds.; Academic Press: London, UK, 2009; pp. 799–817. [Google Scholar]
- Han, H.L.; Liu, J.; Feng, X.J.; Zhang, M.; Lin, Q.F.; Wang, T.; Qi, S.L.; Xu, T.; Hua, X.J. SSR1 is involved in maintaining the function of mitochondria electron transport chain and iron homeostasis upon proline treatment in Arabidopsis. J. Plant Physiol. 2021, 256, 153325. [Google Scholar] [CrossRef]
- Averbeck, D. Non-targeted effects as a paradigm breaking evidence. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2010, 687, 7–12. [Google Scholar] [CrossRef]
- Zemp, F.; Kovalchuk, I. Inter-plant Communication of Genome Instability in Radiation Exposed Arabidopsis. In Radiobiology and Environmental Security; NATO Science for Peace and Security Series C: Environmental Security; Mothersill, C., Korogodina, V., Seymour, C., Eds.; Springer: Dordrecht, Germany, 2012; pp. 87–97. [Google Scholar]
- Li, F.; Liu, P.; Wang, T.; Bian, P.; Wu, Y.; Wu, L.; Yu, Z. The induction of bystander mutagenic effects in vivo by alpha-particle irradiation in whole Arabidopsis thaliana plants. Radiat. Res. 2010, 174, 228–237. [Google Scholar] [CrossRef]
- Wang, T.; Li, F.; Xu, S.; Bian, P.; Wu, Y.; Wu, L.; Yu, Z. The time course of long-distance signaling in radiation-induced bystander effect in vivo in Arabidopsis thaliana demonstrated using root micro-grafting. Radiat. Res. 2011, 176, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wang, T.; Xu, S.; Xu, S.; Wu, L.; Wu, Y.; Bian, P. Radiation-induced epigenetic bystander effects demonstrated in Arabidopsis thaliana. Radiat. Res. 2015, 183, 511–524. [Google Scholar] [CrossRef]
- Wang, T.; Sun, Q.; Xu, W.; Li, F.; Li, H.; Lu, J.; Wu, L.; Wu, Y.; Liu, M.; Bian, P. Modulation of modeled microgravity on radiation-induced bystander effects in Arabidopsis thaliana. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2015, 773, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xu, W.; Deng, C.; Xu, S.; Li, F.; Wu, Y.; Wu, L.; Bian, P. A pivotal role of the jasmonic acid signal pathway in mediating radiation-induced bystander effects in Arabidopsis thaliana. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2016, 791–792, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Volkova, P.Y.; Bondarenko, E.V.; Kazakova, E.A. Radiation hormesis in plants. Curr. Opin. Toxicol. 2022, 30, 100334. [Google Scholar] [CrossRef]
- Wiegant, F.A.; de Poot, S.A.; Boers-Trilles, V.E.; Schreij, A.M. Hormesis and Cellular Quality Control: A Possible Explanation for the Molecular Mechanisms that Underlie the Benefits of Mild Stress. Dose Response 2012, 11, 413–430. [Google Scholar] [CrossRef] [Green Version]
- Tang, F.R.; Loke, W.K. Molecular mechanisms of low dose ionizing radiation-induced hormesis, adaptive responses, radioresistance, bystander effects, and genomic instability. Int. J. Radiat. Biol. 2015, 91, 13–27. [Google Scholar] [CrossRef]
- Bitarishvili, S.V.; Volkova, P.Y.; Geras’kin, S.A. γ-Irradiation of Barley Seeds and Its Effect on the Phytohormonal Status of Seedlings. Russ. J. Plant Physiol. 2018, 65, 446–454. [Google Scholar] [CrossRef]
- Volkova, P.Y.; Clement, G.; Makarenko, E.S.; Kazakova, E.A.; Bitarishvili, S.V.; Lychenkova, M.A. Metabolic Profiling of γ-Irradiated Barley Plants Identifies Reallocation of Nitrogen Metabolism and Metabolic Stress Response. Dose Response 2020, 18, 1559325820914186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duarte-Sierra, A.; Tiznado-Hernández, M.E.; Jha, D.K.; Janmeja, N.; Arul, J. Abiotic stress hormesis: An approach to maintain quality, extend storability, and enhance phytochemicals on fresh produce during postharvest. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3659–3682. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, S.; Rashydov, N. The Transcriptional Response of Arabidopsis thaliana L. Genes AtKu70, AtRAD51 and AtRad1 to X-Ray Radiation. J. Agric. Sci. Technol. 2017, 7, 52–60. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Ma, R.; Cui, D.; Cao, Q.; Shan, Z.; Jiao, Z. Physio-biochemical and molecular mechanism underlying the enhanced heavy metal tolerance in highland barley seedlings pre-treated with low-dose gamma irradiation. Sci. Rep. 2017, 7, 14233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezk, A.A.; Al-Khayri, J.M.; Al-Bahrany, A.M.; El-Beltagi, H.S.; Mohamed, H.I. X-ray irradiation changes germination and biochemical analysis of two genotypes of okra (Hibiscus esculentus L.). J. Radiat. Res. Appl. Sci. 2019, 12, 393–402. [Google Scholar] [CrossRef] [Green Version]
- Yasmin, K.; Arulbalachandran, D.; Soundarya, V.; Vanmathi, S. Effects of gamma radiation (γ) on biochemical and antioxidant properties in black gram (Vigna mungo L. Hepper). Int. J. Radiat. Biol. 2019, 95, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Van Hoeck, A.; Horemans, N.; Nauts, R.; Van Hees, M.; Vandenhove, H.; Blust, R. Lemna minor plants chronically exposed to ionising radiation: RNA-seq analysis indicates a dose rate dependent shift from acclimation to survival strategies. Plant Sci. 2017, 257, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Agathokleous, E.; Feng, Z.; Peñuelas, J. Chlorophyll hormesis: Are chlorophylls major components of stress biology in higher plants? Sci. Total Environ. 2020, 726, 138637. [Google Scholar] [CrossRef]
- Song, K.E.; Lee, S.H.; Jung, J.G.; Choi, J.E.; Jun, W.; Chung, J.-W.; Hong, S.H.; Shim, S. Hormesis effects of gamma radiation on growth of quinoa (Chenopodium quinoa). Int. J. Radiat. Biol. 2021, 97, 906–915. [Google Scholar] [CrossRef]
- Oliveira, N.M.; Medeiros, A.D.D.; Nogueira, M.D.L.; Arthur, V.; Mastrangelo, T.D.A.; Barboza da Silva, C. Hormetic effects of low-dose gamma rays in soybean seeds and seedlings: A detection technique using optical sensors. Comput. Electron. Agric. 2021, 187, 106251. [Google Scholar] [CrossRef]
- Yokota, Y.; Funayama, T.; Hase, Y.; Hamada, N.; Kobayashi, Y.; Tanaka, A.; Narumi, I. Enhanced micronucleus formation in the descendants of gamma-ray-irradiated tobacco cells: Evidence for radiation-induced genomic instability in plant cells. Mutat. Res. Mol. Mech. Mutagen. 2010, 691, 41–46. [Google Scholar] [CrossRef]
- Geras’kin, S.; Volkova, P.; Vasiliyev, D.; Dikareva, N.; Oudalova, A.; Kazakova, E.; Makarenko, E.; Duarte, G.; Kuzmenkov, A. Scots pine as a promising indicator organism for biomonitoring of the polluted environment: A case study on chronically irradiated populations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2019, 842, 3–13. [Google Scholar] [CrossRef]
- Kovalchuk, I.; Molinier, J.; Yao, Y.; Arkhipov, A.; Kovalchuk, O. Transcriptome analysis reveals fundamental differences in plant response to acute and chronic exposure to ionizing radiation. Mutat. Res. Mol. Mech. Mutagen. 2007, 624, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Duarte, G.T.; Volkova, P.Y.; Geras’kin, S.A. The response profile to chronic radiation exposure based on the transcriptome analysis of Scots pine from Chernobyl affected zone. Environ. Pollut. 2019, 250, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Podlutskii, M.; Babina, D.; Podobed, M.; Bondarenko, E.; Bitarishvili, S.; Blinova, Y.; Shesterikova, E.; Prazyan, A.; Turchin, L.; Garbaruk, D.; et al. Arabidopsis thaliana Accessions from the Chernobyl Exclusion Zone Show Decreased Sensitivity to Additional Acute Irradiation. Plants 2022, 11, 3142. [Google Scholar] [CrossRef]
- Weng, M.-L.; Becker, C.; Hildebrandt, J.; Neumann, M.; Rutter, M.T.; Shaw, R.G.; Weigel, D.; Fenster, C.B. Fine-Grained Analysis of Spontaneous Mutation Spectrum and Frequency in Arabidopsis thaliana. Genetics 2019, 211, 703–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisen, J.A.; Hanawalt, P.C. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. DNA Repair 1999, 435, 171–213. [Google Scholar] [CrossRef] [Green Version]
- DiRuggiero, J.; Robb, F.T. Early Evolution of DNA Repair Mechanisms. In The Genetic Code and the Origin of Life; Ribas de Pouplana, L., Ed.; Springer: Boston, MA, USA, 2004; pp. 169–182. [Google Scholar]
- Britt, A. Repair of damaged bases. Arab. Book 2002, 1, e0005. [Google Scholar] [CrossRef]
- Merino, N.; Aronson, H.S.; Bojanova, D.P.; Feyhl-Buska, J.; Wong, M.L.; Zhang, S.; Giovannelli, D. Living at the Extremes: Extremophiles and the Limits of Life in a Planetary Context. Front. Microbiol. 2019, 10, 780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nouspikel, T.; Hanawalt, P.C. Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Mol. Cell. Biol. 2000, 20, 1562–1570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saint-Ruf, C.; Pesut, J.; Sopta, M.; Matic, I. Causes and consequences of DNA repair activity modulation during stationary phase in Escherichia coli. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 259–270. [Google Scholar] [CrossRef]
- Torres-Barceló, C.; Cabot, G.; Oliver, A.; Buckling, A.; MacLean, R.C. A trade-off between oxidative stress resistance and DNA repair plays a role in the evolution of elevated mutation rates in bacteria. Proc. R. Soc. B Biol. Sci. 2013, 280, 20130007. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, S.; Santosh, M. Models on Snowball Earth and Cambrian explosion: A synopsis. Gondwana Res. 2008, 14, 22–32. [Google Scholar] [CrossRef]
- Mothersill, C.; Rusin, A.; Elliott, A.; Seymour, C. Chapter 34—Radiation and chemical induced genomic instability as a driver for environmental evolution. In Genome Stability, 2nd ed.; Kovalchuk, I., Kovalchuk, O., Eds.; Academic Press: Boston, MA, USA, 2021; Volume 26, pp. 639–658. [Google Scholar]
- Maruyama, S.; Ikoma, M.; Genda, H.; Hirose, K.; Yokoyama, T.; Santosh, M. The naked planet Earth: Most essential pre-requisite for the origin and evolution of life. Geosci. Front. 2013, 4, 141–165. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Lazcano, A. Historical development of origins research. Cold Spring Harb. Perspect. Biol. 2010, 2, a002089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cech, T.R. The RNA Worlds in Context. Cold Spring Harb. Perspect. Biol. 2012, 4, a006742. [Google Scholar] [CrossRef] [Green Version]
- Becker, S.; Thoma, I.; Deutsch, A.; Gehrke, T.; Mayer, P.; Zipse, H.; Carell, T. A high-yielding, strictly regioselective prebiotic purine nucleoside formation pathway. Science 2016, 352, 833–836. [Google Scholar] [CrossRef] [PubMed]
- Ikehara, K. Chapter 9—Under what kind of life system could space life emerge? In Model Ecosystems in Extreme Environments; Seckbach, J., Rampelotto, P., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 189–208. [Google Scholar]
- Gough, D.O. Solar interior structure and luminosity variations. Sol. Phys. 1981, 74, 21–34. [Google Scholar] [CrossRef]
- Sagan, C.; Chyba, C. The Early Faint Sun Paradox: Organic Shielding of Ultraviolet-Labile Greenhouse Gases. Science 1997, 276, 1217–1221. [Google Scholar] [CrossRef]
- Karam, P.A.; Leslie, S.A. Changes in terrestrial natural radiation levels over the history of life. In Radioactivity in the Environment; Elsevier: Amsterdam, The Netherlands, 2005; Volume 7, pp. 107–117. [Google Scholar]
- Ribas, I.; Guinan, E.F.; Güdel, M.; Audard, M. Evolution of the Solar Activity over Time and Effects on Planetary Atmospheres. I. High-Energy Irradiances (1–1700 Å). Astrophys. J. 2005, 622, 680. [Google Scholar] [CrossRef] [Green Version]
- Ribas, I.; Porto De Mello, G.F.; Ferreira, L.D.; Hébrard, E.; Selsis, F.; Catalán, S.; Garcés, A.; Do Nascimento, J.D., Jr.; De Medeiros, J.R. Evolution of the solar activity over time and effects on planetary atmospheres. II. κ1 Ceti, an analog of the sun when life arose on earth. Astrophys. J. 2010, 714, 384–395. [Google Scholar] [CrossRef] [Green Version]
- Karam, P.A. Inconstant sun: How solar evolution has affected cosmic and ultraviolet radiation exposure over the history of life on Earth. Health Phys. 2003, 84, 322–333. [Google Scholar] [CrossRef]
- Svensmark, H. Cosmic rays and the biosphere over 4 billion years. Astron. Nachr. 2006, 327, 871–875. [Google Scholar] [CrossRef]
- Kataoka, R.; Ebisuzaki, T.; Miyahara, H.; Nimura, T.; Tomida, T.; Sato, T.; Maruyama, S. The Nebula Winter: The united view of the snowball Earth, mass extinctions, and explosive evolution in the late Neoproterozoic and Cambrian periods. Gondwana Res. 2014, 25, 1153–1163. [Google Scholar] [CrossRef]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Phylogeny and Molecular Evolution of the Green Algae. Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef] [Green Version]
- Umen, J.G. Green algae and the origins of multicellularity in the plant kingdom. Cold Spring Harb. Perspect. Biol. 2014, 6, a016170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strother, P.K.; Battison, L.; Brasier, M.D.; Wellman, C.H. Earth’s earliest non-marine eukaryotes. Nature 2011, 473, 505–509. [Google Scholar] [CrossRef]
- Fang, L.; Leliaert, F.; Zhang, Z.-H.; Penny, D.; Zhong, B.-J. Evolution of the Chlorophyta: Insights from chloroplast phylogenomic analyses. J. Syst. Evol. 2017, 55, 322–332. [Google Scholar] [CrossRef] [Green Version]
- Tang, Q.; Pang, K.; Yuan, X.; Xiao, S. A one-billion-year-old multicellular chlorophyte. Nat. Ecol. Evol. 2020, 4, 543–549. [Google Scholar] [CrossRef]
- Steemans, P.; Hérissé, A.L.; Melvin, J.; Miller, M.A.; Paris, F.; Verniers, J.; Wellman, C.H. Origin and Radiation of the Earliest Vascular Land Plants. Science 2009, 324, 353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jill Harrison, C. Development and genetics in the evolution of land plant body plans. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20150490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, D.; Cherns, L.; Raven, J.A. Could land-based early photosynthesizing ecosystems have bioengineered the planet in mid-Palaeozoic times? Palaeontology 2015, 58, 803–837. [Google Scholar] [CrossRef]
- Lenton, T.M.; Crouch, M.; Johnson, M.; Pires, N.; Dolan, L. First plants cooled the Ordovician. Nat. Geosci. 2012, 5, 86–89. [Google Scholar] [CrossRef]
- Algeo, T.J.; Scheckler, S.E.; Maynard, J.B. 12. Effects of the Middle to Late Devonian Spread of Vascular Land Plants on Weathering Regimes, Marine Biotas, and Global Climate. In Plants Invade the Land; Columbia University Press: New York, NY, USA, 2001; pp. 213–236. [Google Scholar]
- Whitfield, J. Plants detonated Cambrian explosion. Nature 2003. [Google Scholar] [CrossRef]
- Knauth, L.P.; Kennedy, M.J. The late Precambrian greening of the Earth. Nature 2009, 460, 728–732. [Google Scholar] [CrossRef]
- Karol, K.G.; McCourt, R.M.; Cimino, M.T.; Delwiche, C.F. The Closest Living Relatives of Land Plants. Science 2001, 294, 2351–2353. [Google Scholar] [CrossRef]
- Qiu, Y.-L.; Li, L.; Wang, B.; Chen, Z.; Knoop, V.; Groth-Malonek, M.; Dombrovska, O.; Lee, J.; Kent, L.; Rest, J.; et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc. Natl. Acad. Sci. USA 2006, 103, 15511–15516. [Google Scholar] [CrossRef] [Green Version]
- Donoghue, P.C.J.; Harrison, C.J.; Paps, J.; Schneider, H. The evolutionary emergence of land plants. Curr. Biol. 2021, 31, R1281–R1298. [Google Scholar] [CrossRef]
- Morris, J.L.; Puttick, M.N.; Clark, J.W.; Edwards, D.; Kenrick, P.; Pressel, S.; Wellman, C.H.; Yang, Z.; Schneider, H.; Donoghue, P.C.J. The timescale of early land plant evolution. Proc. Natl. Acad. Sci. USA 2018, 115, E2274–E2283. [Google Scholar] [CrossRef] [Green Version]
- Bowman, J.L. Walkabout on the long branches of plant evolution. Curr. Opin. Plant Biol. 2013, 16, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Maruyama, F.; Fujisawa, T.; Togashi, T.; Yamamoto, N.; Seo, M.; Sato, S.; Yamada, T.; Mori, H.; Tajima, N.; et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 2014, 5, 3978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harholt, J.; Moestrup, Ø.; Ulvskov, P. Why Plants Were Terrestrial from the Beginning. Trends Plant Sci. 2016, 21, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Lutzoni, F.; Nowak, M.D.; Alfaro, M.E.; Reeb, V.; Miadlikowska, J.; Krug, M.; Arnold, A.E.; Lewis, L.A.; Swofford, D.L.; Hibbett, D.; et al. Contemporaneous radiations of fungi and plants linked to symbiosis. Nat. Commun. 2018, 9, 5451. [Google Scholar] [CrossRef] [Green Version]
- Naranjo-Ortiz, M.A.; Gabaldón, T. Fungal evolution: Major ecological adaptations and evolutionary transitions. Biol. Rev. 2019, 94, 1443–1476. [Google Scholar] [CrossRef] [Green Version]
- Strother, P.K. Thalloid carbonaceous incrustations and the asynchronous evolution of embryophyte characters during the Early Paleozoic. Int. J. Coal Geol. 2010, 83, 154–161. [Google Scholar] [CrossRef]
- Rubinstein, C.V.; Gerrienne, P.; de la Puente, G.S.; Astini, R.A.; Steemans, P. Early Middle Ordovician evidence for land plants in Argentina (eastern Gondwana). New Phytol. 2010, 188, 365–369. [Google Scholar] [CrossRef]
- Griesemer, J.R. Niche: Historical perspectives. In Keywords in Evolutionary Biology; Keller, E.F.L., Elisabeth, A., Eds.; Harvard University Press: Cambridge, MA, USA, 1999. [Google Scholar]
- Brockhurst, M.A.; Colegrave, N.; Hodgson, D.J.; Buckling, A. Niche Occupation Limits Adaptive Radiation in Experimental Microcosms. PLoS ONE 2007, 2, e193. [Google Scholar] [CrossRef] [Green Version]
- Biggin, A.J.; Strik, G.H.M.A.; Langereis, C.G. The intensity of the geomagnetic field in the late-Archaean: New measurements and an analysis of the updated IAGA palaeointensity database. Earth Planets Space 2009, 61, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Miki, M.; Taniguchi, A.; Yokoyama, M.; Gouzu, C.; Hyodo, H.; Uno, K.; Zaman, H.; Otofuji, Y.I. Palaeomagnetism and geochronology of the proterozoic dolerite dyke from southwest greenland: Indication of low palaeointensity. Geophys. J. Int. 2009, 179, 18–34. [Google Scholar] [CrossRef] [Green Version]
- Tarduno, J.A.; Cottrell, R.D.; Watkeys, M.K.; Hofmann, A.; Doubrovine, P.V.; Mamajek, E.E.; Liu, D.; Sibeck, D.G.; Neukirch, L.P.; Usui, Y. Geodynamo, Solar Wind, and Magnetopause 3.4 to 3.45 Billion Years Ago. Science 2010, 327, 1238–1240. [Google Scholar] [CrossRef] [Green Version]
- Schreider, A.; Schreider, A.A.; Varga, P.; Denis, C. Variations of the Earth’s magnetic field in the Phanerozoic. Oceanology 2011, 51, 506–510. [Google Scholar] [CrossRef]
- Cockell, C.S. Ultraviolet radiation and the photobiology of earth’s early oceans. Orig. Life Evol. Biosph. 2000, 30, 467–500. [Google Scholar] [CrossRef]
- Seki, K.; Elphic, R.C.; Hirahara, M.; Terasawa, T.; Mukai, T. On atmospheric loss of oxygen ions from earth through magnetospheric processes. Science 2001, 291, 1939–1941. [Google Scholar] [CrossRef] [Green Version]
- Meert, J.G.; Levashova, N.M.; Bazhenov, M.L.; Landing, E. Rapid changes of magnetic Field polarity in the late Ediacaran: Linking the Cambrian evolutionary radiation and increased UV-B radiation. Gondwana Res. 2016, 34, 149–157. [Google Scholar] [CrossRef] [Green Version]
- Cnossen, I.; Sanz-Forcada, J.; Favata, F.; Witasse, O.; Zegers, T.; Arnold, N.F. Habitat of early life: Solar X-ray and UV radiation at Earth’s surface 4–3.5 billion years ago. J. Geophys. Res. Planets 2007, 112. [Google Scholar] [CrossRef]
- Cockell, C.S.; Raven, J.A. Ozone and life on the Archaean Earth. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2007, 365, 1889–1901. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.C.; Amon, A. Gene copy-number alterations: A cost-benefit analysis. Cell 2013, 152, 394–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suryawanshi, V.; Talke, I.N.; Weber, M.; Eils, R.; Brors, B.; Clemens, S.; Krämer, U. Between-species differences in gene copy number are enriched among functions critical for adaptive evolution in Arabidopsis halleri. BMC Genom. 2016, 17, 1034. [Google Scholar] [CrossRef] [Green Version]
- El-Brolosy, M.A.; Stainier, D.Y.R. Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet. 2017, 13, e1006780. [Google Scholar] [CrossRef] [Green Version]
- Schopf, J.W.; Kitajima, K.; Spicuzza, M.J.; Kudryavtsev, A.B.; Valley, J.W. SIMS analyses of the oldest known assemblage of microfossils document their taxon-correlated carbon isotope compositions. Proc. Natl. Acad. Sci. USA 2018, 115, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Meng, F.; Kong, F.; Niu, C. Microfossils from the Paleoproterozoic Hutuo Group, Shanxi, North China: Early evidence for eukaryotic metabolism. Precambrian Res. 2020, 342, 105650. [Google Scholar] [CrossRef]
- Miao, L.; Moczydłowska, M.; Zhu, S.; Zhu, M. New record of organic-walled, morphologically distinct microfossils from the late Paleoproterozoic Changcheng Group in the Yanshan Range, North China. Precambrian Res. 2019, 321, 172–198. [Google Scholar] [CrossRef]
- Lyons, T.W.; Reinhard, C.T.; Planavsky, N.J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 2014, 506, 307–315. [Google Scholar] [CrossRef]
- Catling, D.C.; Zahnle, K.J. The Archean atmosphere. Sci. Adv. 2020, 6, eaax1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acosta, K.; Appenroth, K.J.; Borisjuk, L.; Edelman, M.; Heinig, U.; Jansen, M.A.K.; Oyama, T.; Pasaribu, B.; Schubert, I.; Sorrels, S.; et al. Return of the Lemnaceae: Duckweed as a model plant system in the genomics and postgenomics era. Plant Cell 2021, 33, 3207–3234. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duarte, G.T.; Volkova, P.Y.; Fiengo Perez, F.; Horemans, N. Chronic Ionizing Radiation of Plants: An Evolutionary Factor from Direct Damage to Non-Target Effects. Plants 2023, 12, 1178. https://doi.org/10.3390/plants12051178
Duarte GT, Volkova PY, Fiengo Perez F, Horemans N. Chronic Ionizing Radiation of Plants: An Evolutionary Factor from Direct Damage to Non-Target Effects. Plants. 2023; 12(5):1178. https://doi.org/10.3390/plants12051178
Chicago/Turabian StyleDuarte, Gustavo Turqueto, Polina Yu. Volkova, Fabricio Fiengo Perez, and Nele Horemans. 2023. "Chronic Ionizing Radiation of Plants: An Evolutionary Factor from Direct Damage to Non-Target Effects" Plants 12, no. 5: 1178. https://doi.org/10.3390/plants12051178
APA StyleDuarte, G. T., Volkova, P. Y., Fiengo Perez, F., & Horemans, N. (2023). Chronic Ionizing Radiation of Plants: An Evolutionary Factor from Direct Damage to Non-Target Effects. Plants, 12(5), 1178. https://doi.org/10.3390/plants12051178







