New Biological and Chemical Insights into Optimization of Chamomile Extracts by Using Artificial Neural Network (ANN) Model
Abstract
1. Introduction
2. Results and Discussion
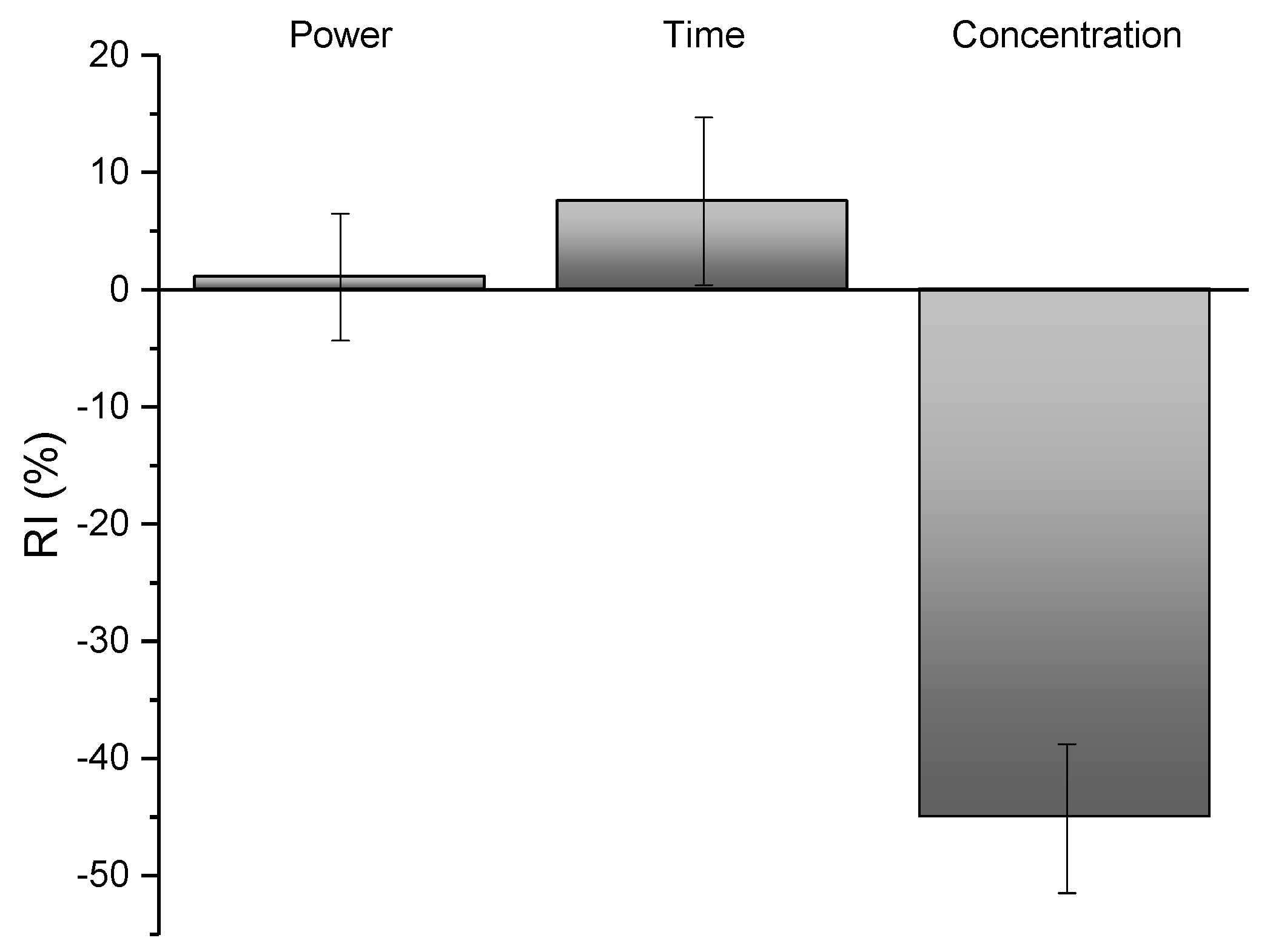
2.1. Extraction and Optimization of the Extraction via ANN
2.2. Chemical Profiling
2.3. Biological Activity
2.3.1. Antioxidant Activity
2.3.2. Enzyme-Inhibitory Activity
2.4. Evaluation of Potential of MAE for the Growth of La. rhamnosus ATCC 7469
3. Material and Methods
3.1. Chemicals
3.2. Plant Material
3.3. Extraction, Experimental Design, and ANN Optimization
3.4. Chemical Analysis of Extracts
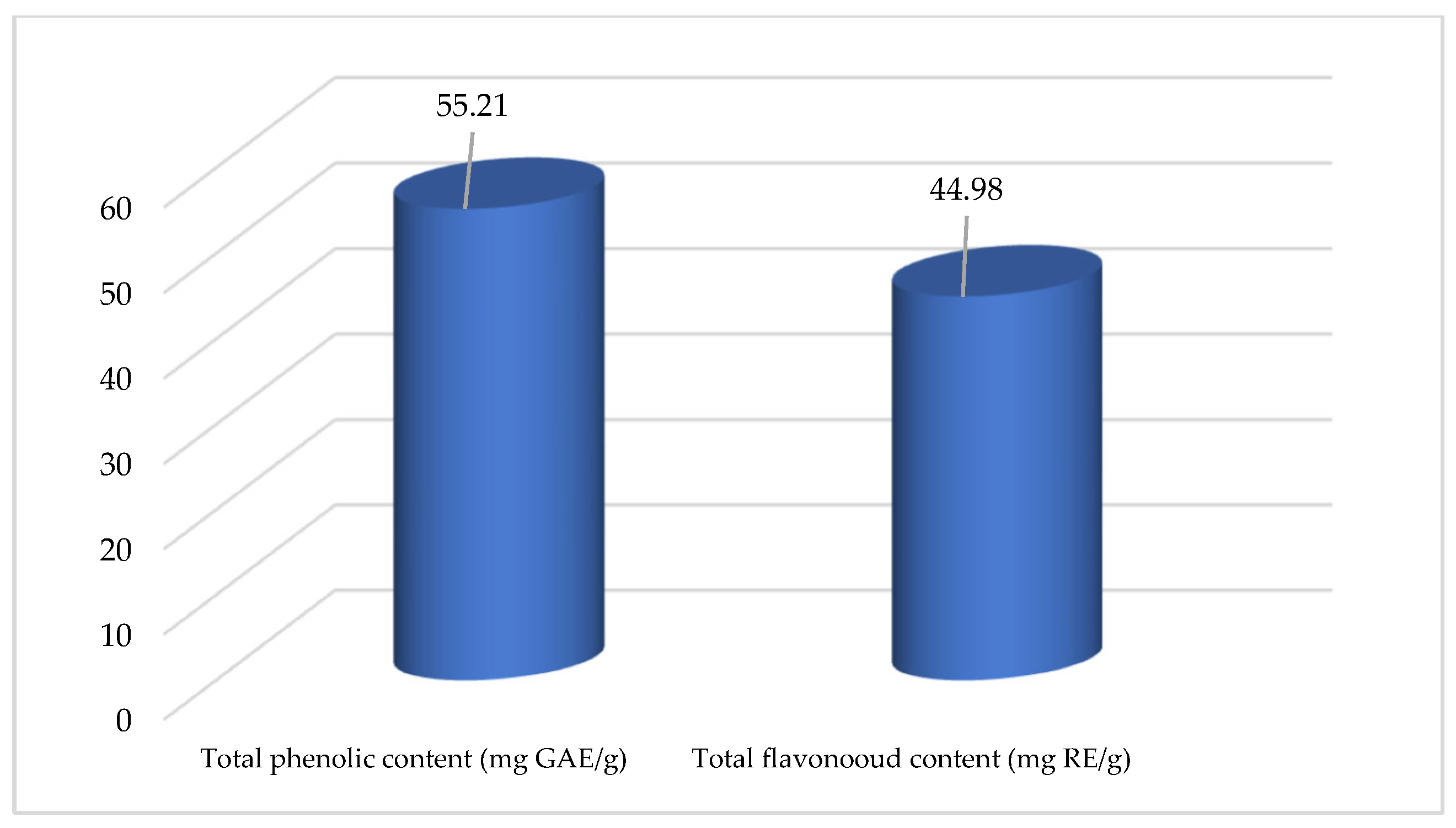
3.4.1. Assays for Total Phenolic and Flavonoid Content
3.4.2. UHPLC-LTQ OrbiTrap MS Analysis of Polyphenolic Compounds
3.5. Determination of Biological Activity of Extracts
3.6. Evaluation of Chamomile Potential for the Growth of La. rhamnosus ATCC 7469
3.6.1. Preparation of Growth Media and Inoculum, and Bacteria Growth Conditions
3.6.2. Determination of Reducing Sugars Concentration and Viability of La. rhamnosus Cells
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cvetanović, A. Optimization of Novel Extraction Techniques for Apigenin Isolation from Chamomile Flowers (Chamomilla recutita L.) and Characterization of Biological Activity of Obtained Extracts. Ph.D. Thesis, University of Novi Sad, Faculty of Technology Novi Sad, Novi Sad, Serbia, 2016. [Google Scholar]
- Newall, C.A.; Anderson, L.A.; Phillipson, J.D. Herbal Medicines: A Guide for Health-Care Professionals; Pharmaceutical Press: London, UK, 1996. [Google Scholar]
- Srivastava, J.K.; Shankar, E.; Gupta, S. Chamomile: A herbal medicine of the past with a bright future. Mol. Med. Rep. 2010, 3, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekhar, V.M.; Halagali, K.S.; Nidavani, R.B.; Shalavadi, M.H.; Biradar, B.S.; Biswas, D.; Muchchandi, I.S. Anti-allergic activity of German chamomile (Matricaria recutita L.) in mast cell mediated allergy model. J. Ethnopharmacol. 2011, 137, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Lis-Balchin, M.; Deans, S.G.; Eaglesham, E. Relationship between bioactivity and chemical composition of commercial essential oils. Flavour Fragr. J. 1998, 13, 98–104. [Google Scholar] [CrossRef]
- Franke, R.; Schilcher, H. Chamomile: Industrial Profiles, Series: Medicinal and Aromatic Plants: Industrial Profiles; CRC Press/Taylor & Francis Group: New York, NY, USA, 2005. [Google Scholar]
- McKay, D.L.; Blumberg, J.B. A Review of the bioactivity and potential health benefits of chamomile tea (Matricaria recutita L.). Phyther. Res. 2006, 20, 519–530. [Google Scholar] [CrossRef]
- Sahebkar, A.; Emami, S.A. Medicinal plants for the treatment of uterus inflammation: Implications from iranian folk medicine. J. Acupunct. Meridian Stud. 2013, 6, 1. [Google Scholar] [CrossRef]
- Cvetanović, A.; Švarc-Gajić, J.; Zeković, Z.; Savić, S.; Vulić, J.; Mašković, P.; Ćetković, G. Comparative analysis of antioxidant, antimicrobiological and cytotoxic activities of native and fermented chamomile ligulate flower extracts. Planta 2015, 242, 721–732. [Google Scholar] [CrossRef]
- Cvetanović, A.; Švarc-Gajić, J.; Mašković, P.; Savić, S.; Nikolić, L. Antioxidant and biological activity of chamomile extracts obtained by different techniques: Perspective of using superheated water for isolation of biologically active compounds. Ind. Crops Prod. 2015, 65, 582–591. [Google Scholar] [CrossRef]
- Ameer, K.; Chun, B.-S.; Kwon, J.-H. Optimization of Supercritical Fluid Extraction of Steviol Glycosides and Total Phenolic Content from Stevia Rebaudiana (Bertoni) Leaves Using Response Surface Methodology and Artificial Neural Network Modeling. Ind. Crops Prod. 2017, 109, 672–685. [Google Scholar] [CrossRef]
- Sinha, K.; Chowdhury, S.; Saha, P.D.; Datta, S. Modeling of Microwave-Assisted Extraction of Natural Dye from Seeds of Bixa Orellana (Annatto) Using Response Surface Methodology (RSM) and Artificial Neural Network (ANN). Ind. Crops Prod. 2013, 41, 165–171. [Google Scholar] [CrossRef]
- Thakker, M.R.; Parikh, J.K.; Desai, M.A. Microwave Assisted Extraction of Essential Oil from the Leaves of Palmarosa: Multi-Response Optimization and Predictive Modelling. Ind. Crops Prod. 2016, 86, 311–319. [Google Scholar] [CrossRef]
- Kennedy, J.; Eberhart, R. Particle Swarm Optimization. In Proceedings of the ICNN’95—International Conference on Neural Networks, Perth, WA, Australia, 27 November—1 December 1995; Volume 4, pp. 1942–1948. [Google Scholar] [CrossRef]
- Mezura-Montes, E.; Coello, C.A.C. Constraint-Handling in Nature-Inspired Numerical Optimization: Past, Present and Future. Swarm Evol. Comput. 2011, 1, 173–194. [Google Scholar] [CrossRef]
- Font, N.; Hernández, F.; Hogendoorn, E.; Baumann, R.; van Zoonen, P. Microwave-assisted solvent extraction and reversed-phase liquid chromatography—UV detection for screening soils for sulfonylurea herbicides. J. Chromatogr. A. 1998, 798, 179–186. [Google Scholar] [CrossRef]
- Brachet, A.; Christen, P.; Veuthey, J.L. Focused microwave-assisted extraction of cocaine and benzoylecgonine from coca leaves. Phytochem. Anal. 2002, 13, 162–169. [Google Scholar] [CrossRef]
- Blekić, M.; Jambrak, A.R.; Chemat, F. Mikrovalna ekstrakcija bioaktivnih spojeva. Croat. J. Food Sci. Technol. 2011, 3, 32–47. [Google Scholar]
- Pavlić, A.; Kaplan, M.; Bera, O.; Olgun, E.E.; Canli, O.; Milosavljević, N.; Antić, B.; Zeković, Z. Microwave-assisted extraction of peppermint polyphenols—Artificial neural networks approach. Food Bioprod. Process. 2019, 118, 258–269. [Google Scholar] [CrossRef]
- Lin, L.Z.; Harnly, J.M. LC-PDA-ESI/MS identification of the phenolic components of three compositae spices: Chamomile, tarragon, and Mexican arnica. Nat. Prod. Commun. 2012, 7, 749–752. [Google Scholar] [CrossRef]
- Švehlíková, V.; Bennett, R.N.; Mellon, F.A.; Needs, P.W.; Piacente, S.; Kroon, P.A.; Bao, Y. Isolation, identification and stability of acylated derivatives of apigenin 7-O-glucoside from chamomile (Chamomilla recutita [L.] Rauschert). Phytochemistry 2004, 65, 2323–2332. [Google Scholar] [CrossRef]
- Franco, E.P.D.; Contesini, F.J.; Lima da Silva, B.; Alves de Piloto Fernandes, A.M.; Wielewski Leme, C.; Gonçalves Cirino, J.P.; Bueno Campos, P.R.; de Oliveira Carvalho, P. Enzyme-assisted modification of flavonoids from Matricaria chamomilla: Antioxidant activity and inhibitory effect on digestive enzymes. J. Enzym. Inhib. Med. Ch. 2020, 35, 42–49. [Google Scholar] [CrossRef]
- Dondi, F.; Kahie, Y.D.; Lodi, G.; Reschiglian, P.; Pietrogrande, C.; Bighi, C.; Cartoni, G.P. Comparison of the Sequential Simplex Method and Linear Solvent Strength Theory in HPLC Gradient Elution Optimization of Multicomponent Flavonoid Mixtures. Chromatographia 1987, 23, 844–849. [Google Scholar] [CrossRef]
- Petroianu, G.; Szoke, E.; Kalász, H.; Szegi, P.; Laufer, R.; Benko, B.; Darvas, F.; Tekes, K. Monitoring by HPLC of Chamomile Flavonoids Exposed to Rat Liver Microsomal Metabolism. Open J. Med. Chem. 2009, 3, 1–7. [Google Scholar] [CrossRef]
- Yang, D.; Wang, T.; Long, M.; Li, P. Quercetin: Its Main Pharmacological Activity and Potential Application in Clinical Medicine. Oxid. Med. Cell. Longev. 2020, 2020, 8825387. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Rauf, A.; Shah, Z.A.; Saeed, F.; Imran, A.; Arshad, M.U.; Ahmad, B.; Bawazeer, S.; Peters, D.; Mubarak, M.S. Chemo-preventive and therapeutic effect of the dietary flavonoid kaempferol: A comprehensive review. Phytother. Res. 2019, 33, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.K.; Chin, K.Y.; Ima-Nirwana, S. The osteoprotective effects of kaempferol: The evidence from in vivo and in vitro studies. Drug Des. Devel. Ther. 2019, 13, 3497–3514. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Lu, Y.; Qian, Y.; Chen, B.; Wu, T.; Ji, G. Recent progress regarding kaempferol for the treatment of various diseases. Exp. Ther. Med. 2019, 18, 2759–2776. [Google Scholar] [CrossRef]
- Zengin, G.; Cvetanović, A.; Gašić, U.; Dragičević, D.; Stupar, A.; Uysal, A.; Senkardes, I.; Ibrahime Sinan, K.; Picot-Allain, M.C.N.; Ak, G.; et al. UHPLC-LTQ OrbiTrap MS analysis and biological properties of Origanum vulgare subsp. viridulum obtained by different extraction methods. Ind. Crops Prod. 2020, 146, 112747. [Google Scholar] [CrossRef]
- Zengin, G.; Cvetanović, A.; Gašić, U.; Stupar, A.; Bulut, G.; Şenkardes, I.; Dogan, A.; Ibrahime Sinan, K.; Uysal, S.; Aumeeruddy-Elalfi, Z.; et al. Modern and traditional extraction techniques affect chemical composition and bioactivity of Tanacetum parthenium (L.) Sch.Bip. Ind. Crops Prod. 2020, 146, 112202. [Google Scholar] [CrossRef]
- Papoutsis, K.; Zhang, J.; Bowyer, M.C.; Brunton, N.; Gibney, E.R.; Lyng, J. Fruit, vegetables, and mushrooms for the preparation of extracts with α-amylase and α-glucosidase inhibition properties: A review. Food Chem. 2021, 338, 128119. [Google Scholar] [CrossRef]
- Karaoglan, E.S.; Koca, M. Tyrosinase and cholinesterase inhibitory activities and molecular docking studies on apigenin and vitexin. J. Pharm. Istanbul Univ. 2020, 50, 268–272. [Google Scholar] [CrossRef]
- Proença, C.; Freitas, M.; Ribeiro, D.; Tomé, S.M.; Oliveira, E.F.; Viegas, M.F.; Araújo, A.N.; Silva, A.M.S.; Fernandes, P.A.; Fernandes, E. Evaluation of a flavonoids library for inhibition of pancreatic α-amylase towards a structure–activity relationship. J. Enzym. Inhib. Med. Ch. 2019, 34, 577–588. [Google Scholar] [CrossRef]
- Agunloye, O.M.; Oboh, G.; Ademiluyi, A.O.; Ademosun, A.O.; Akindahunsi, A.A.; Oyagbemi, A.A.; Omobowale, T.O.; Ajibade, T.O.; Adedapo, A.A. Cardio-protective and antioxidant properties of caffeic acid and chlorogenic acid: Mechanistic role of angiotensin converting enzyme, cholinesterase and arginase activities in cyclosporine induced hypertensive rats. Biomed. Pharmacother. 2019, 109, 450–458. [Google Scholar] [CrossRef]
- Oboh, G.; Agunloye, O.M.; Adefegha, S.A.; Akinyemi, A.J.; Ademiluyi, A.O. Caffeic and chlorogenic acids inhibit key enzymes linked to type 2 diabetes (in vitro): A comparative study. J. Basic Clin. Physiol. Pharmacol. 2015, 26, 165–170. [Google Scholar] [CrossRef]
- Cvetanović, A.; Švarc-Gajić, J.; Zeković, Z.; Gašić, U.; Tešić, Ž.; Zengin, G.; Mašković, P.; Mahomoodally, M.F.; Đurović, S. Subcritical water extraction as a cutting edge technology for the extraction of bioactive compounds from chamomile: Influence of pressure on chemical composition and bioactivity of extracts. Food Chem. 2018, 266, 389–396. [Google Scholar] [CrossRef]
- Cvetanović, A.; Švarc-Gajić, J.; Gašić, U.; Tešić, Ž.; Zengin, G.; Zeković, Z.; Đurović, S. Isolation of apigenin from subcritical water extracts: Optimization of the process. J. Supercrit. Fluids 2017, 120, 32–42. [Google Scholar] [CrossRef]
- Cvetanović, A.; Zeković, Z.; Zengin, G.; Mašković, P.; Petronijević, M.; Radojković, M. Multidirectional approaches on autofermented chamomile ligulate flowers: Antioxidant, antimicrobial, cytotoxic and enzyme inhibitory effects. S. Afr. J. Bot. 2019, 120, 112–118. [Google Scholar] [CrossRef]
- Currò, D.; Ianiro, G.; Pecere, S.; Bibbò, S.; Cammarota, G. Probiotics, fibre and herbal medicinal products for functional and inflammatory bowel disorders. Br. J. Pharmacol. 2017, 174, 1426–1449. [Google Scholar] [CrossRef]
- Cocetta, V.; Catanzaro, D.; Borgonetti, V.; Ragazzi, E.; Giron, M.C.; Governa, P.; Carnevali, I.; Biagi, M.; Montopoli, M. A Fixed Combination of Probiotics and Herbal Extracts Attenuates Intestinal Barrier Dysfunction from Inflammatory Stress. Preprints 2017, 2017, 120199. [Google Scholar] [CrossRef]
- Marhamatizadeh, M.H.; Shahriarpoor, M.S.; Rezazadeh, S. Effects of Chamomile Essence on the Growth of Probiotic Bacteria, Bifidobacterium bifidum and Lactobacillus acidophilus in Milk and Yoghurt. Glob. Vet. 2012, 8, 605–611. [Google Scholar]
- Zarei, K.; Marhamatizadeh, M.H.; Javamardi, F. The Effect of Echium amoenum Hydro-alcoholic Extract on the Growth of Probiotic Bacteria Lactobacillus acidophilus and Bifidobacterium bifidum in Kefir. Alborz Univ. Med. Sci. J. 2020, 9, 219–230. [Google Scholar] [CrossRef]
- Dimofte, A.; Simionescu, N.; Petrovici, A.R.; Spiridon, I. Probiotic Properties of Weissella confusa PP29 on Hibiscus sabdariffa L. Media. Fermentation 2022, 8, 553. [Google Scholar] [CrossRef]
- Martinelli, M.; Ummarino, D.; Giugliano, F.P.; Sciorio, E.; Tortora, C.; Bruzzese, D.; De Giovanni, D.; Rutigliano, I.; Valenti, S.; Romano, C.; et al. Efficacy of a standardized extract of Matricariae chamomilla L., Melissa officinalis L. and tyndallized Lactobacillus acidophilus (HA 122) in infantile colic: An open randomized controlled trial. Neurogastroenterol. Motil. 2017, 29, e13145. [Google Scholar] [CrossRef]
- Hajqasemi, M.; Vaseji, N.; Iranmanesh, M.; Torshizi, K.; Mojgani, N. Synergistic Effects and Viability of Indigenous Probiotic Bacteria in the Presence of Herbal Extracts and Evaluations of their Minimum Inhibitory Concentrations in invitro Conditions. Anim. Sci. J. 2021, 34, 49–64. [Google Scholar] [CrossRef]
- Köberl, M.; Erschen, S.; Etemadi, M.; White, R.A.; El-Arabi, T.F.; Berg, G. Deciphering the microbiome shift during fermentation of medicinal plants. Sci. Rep. 2019, 9, 13461. [Google Scholar] [CrossRef] [PubMed]
- Holkem, A.T.; Silva, M.P.D.; Favaro-Trindade, C.S. Probiotics and plant extracts: A promising synergy and delivery systems. Crit. Rev. Food Sci. Nutr. 2022, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Swales, G., Jr.; Margavio, T.M. A comparison of discriminant analysis versus artificial neural networks. J. Oper. Res. Soc. 1993, 44, 51–60. [Google Scholar] [CrossRef]
- Cvetanović, A.; Zengin, G.; Zeković, Z.; Švarc-Gajić, J.; Ražić, S.; Damjanović, A.; Mašković, P.; Mitić, M. Comparative in vitro studies of the biological potential and chemical composition of stems, leaves and berries Aronia melanocarpa’s extracts obtained by subcritical water extraction. Food Chem. Toxicol. 2018, 121, 458–466. [Google Scholar] [CrossRef]
- Jurič, A.; Gašić, U.; Brčić Karačonji, I.; Jurica, K.; Milojković-Opsenica, D. The phenolic profile of strawberry tree (Arbutus unedo L.) honey. J. Serb. Chem. Soc. 2020, 85, 1011–1019. [Google Scholar] [CrossRef]
- Banjanac, T.; Dragićević, M.; Šiler, B.; Gašić, U.; Bohanec, B.; Nestorović Živković, J.; Trifunović, S.; Mišić, D. Chemodiversity of two closely related tetraploid Centaurium species and their hexaploid hybrid: Metabolomic search for high-resolution taxonomic classifiers. Phytochemistry 2017, 140, 27–44. [Google Scholar] [CrossRef]
- Mulinacci, N.; Romani, A.; Pinelli, P.; Vincieri, F.F.; Prucher, D. Characterization of Matricaria recutita L. Flower extracts by HPLC-MS and HPLC-DAD analysis. Chromatographia 2000, 51, 301–307. [Google Scholar] [CrossRef]
- Viapiana, A.; Struck-Lewicka, W.; Konieczynski, P.; Wesolowski, M.; Kaliszan, R. An Approach Based on HPLC-Fingerprint and Chemometrics to Quality Consistency Evaluation of Matricaria chamomilla L. Commercial Samples. Front. Plant Sci. 2016, 7, 1561. [Google Scholar] [CrossRef]
- Kováčik, J.; Klejdus, B. Dynamics of phenolic acids and lignin accumulation in metal-treated Matricaria chamomilla roots. Plant Cell Rep. 2008, 27, 605–615. [Google Scholar] [CrossRef]
- Nováková, L.; Vildová, A.; Mateus, J.P.; Gonçalves, T.; Solich, P. Development and application of UHPLC–MS/MS method for the determination of phenolic compounds in Chamomile flowers and Chamomile tea extracts. Talanta 2010, 82, 1271–1280. [Google Scholar] [CrossRef]
- Uysal, S.; Zengin, G.; Locatelli, M.; Bahadori, M.B.; Mocan, A.; Bellagamba, G.; De Luca, E.; Mollica, A.; Aktumsek, A. Cytotoxic and enzyme inhibitory potential of two Potentilla species (P. speciosa L. and P. reptans Willd.) and their chemical composition. Front. Pharmacol. 2017, 8, 290. [Google Scholar] [CrossRef]
- Grochowski, D.M.; Uysal, S.; Aktumsek, A.; Granica, S.; Zengin, G.; Ceylan, R.; Locatelli, M.; Tomczyk, M. In vitro enzyme inhibitory properties, antioxidant activities, and phytochemical profile of Potentilla thuringiaca. Phytochem. Lett. 2017, 20, 365–372. [Google Scholar] [CrossRef]
- Secci, D.; Carradori, S.; Bizzarri, B.; Chimenti, P.; De Monte, C.; Mollica, A.; Rivanera, D.; Zicari, A.; Mari, E.; Zengin, G.; et al. Novel 1,3-thiazolidin-4-one derivatives as promising anti-Candida agents endowed with anti-oxidant and chelating properties. Eur. J. Med. Chem. 2016, 117, 144–156. [Google Scholar] [CrossRef]
- Zengin, G.; Nithiyanantham, S.; Locatelli, M.; Ceylan, R.; Uysal, S.; Aktumsek, A.; Selvi, P.K.; Maskovic, P. Screening of in vitro antioxidant and enzyme inhibitory activities of different extracts from two uninvestigated wild plants: Centranthus longiflorus subsp. longiflorus and Cerinthe minor subsp. auriculata. Eur. J. Integr. Med. 2016, 8, 286–292. [Google Scholar] [CrossRef]
- Radosavljević, M.; Pejin, J.; Pribić, M.; Kocić-Tanackov, S.; Romanić, R.; Mladenović, D.; Djukić-Vuković, A.; Mojović, L. Utilization of brewing and malting by-products as carrier and raw materials in l-(+)-lactic acid production and feed application. Appl. Microbiol. Biotechnol. 2019, 103, 3001–3013. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Pejin, J.; Radosavljević, M.; Mojović, L.; Kocić-Tanackov, S.; Djukić-Vuković, A. The influence of calcium-carbonate and yeast extract addition on lactic acid fermentation of brewer’s spent grain hydrolysate. Food Res. Int. 2015, 73, 31–37. [Google Scholar] [CrossRef]
- Miller, G. Use of dinitrosalicylic acid for determining reducing sugars. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]


| No | tR, Min | Compound Name | Molecular Formula, [M − H]– | Calculated Mass, [M − H]– | Exact Mass, [M − H]– | Δ ppm | MS2 Fragments, (% Base Peak) | MS3 Fragments, (% Base Peak) | MS4 Fragments, (% Base Peak) |
|---|---|---|---|---|---|---|---|---|---|
| Phenolic acids and their derivatives | |||||||||
| 1 | 1.95 | Gallic acid hexoside isomer 1 | C13H15O10– | 331.06707 | 331.06680 | 0.82 | 211(15), 193(80), 175(30), 169(100), 151(50) | 151(100) | 110(10), 97(30), 81(100), 53(30) |
| 2 | 2.39 | Gallic acid a | C7H5O5– | 169.01425 | 169.01385 | 2.37 | 125(100) | 107(100) | − |
| 3 | 2.69 | Dihydroxybenzoic acid hexoside isomer 1 | C13H15O9– | 315.07216 | 315.07206 | 0.32 | 153(100), 152(50), 109(15), 108(10) | 109(100) | 123(25), 109(10), 85(10), 81(100) |
| 4 | 3.13 | Gallic acid hexoside isomer 2 | C13H15O10– | 331.06707 | 331.06702 | 0.15 | 313(100), 169(25), 168(90), 151(10), 125(25) | 193(50), 151(100), 125(80) | 123(100), 107(90), 95(65) |
| 5 | 3.82 | Dihydroxybenzoic acid hexoside isomer 2 | C13H15O9– | 315.07216 | 315.07121 | 3.02 | 153(100), 135(10), 109(10) | 135(100), 109(50) | 91(100) |
| 6 | 4.32 | Dihydroxybenzoic acid hexoside isomer 3 | C13H15O9– | 315.07216 | 315.07169 | 1.49 | 153(100), 109(10) | 135(100), 109(50) | 91(100) |
| 7 | 4.51 | Protocatechuic acid a | C7H5O4– | 153.01933 | 153.01872 | 3.99 | 109(100) | 81(60), 80(50), 67(30), 65(100) | − |
| 8 | 4.38 | Caffeoylquinic acid hexoside isomer 1 | C22H27O14– | 515.14008 | 515.13928 | 1.55 | 353(80), 341(5), 323(10), 191(100), 179(5) | 173(65), 127(90), 111(50), 93(55), 85(100) | − |
| 9 | 4.61 | 3-O-Caffeoylquinic acid | C16H17O9– | 353.08781 | 353.08676 | 2.97 | 191(100), 179(35), 135(10) | 173(75), 127(100), 111(40), 93(60), 85(90) | 109(30), 99(40), 85(100) |
| 10 | 4.68 | Caffeic acid hexoside isomer 1 | C15H17O9– | 341.08781 | 341.08716 | 1.91 | 191(10), 179(100), 135(10) | 135(100) | 135(100), 107(50) |
| 11 | 4.88 | Caffeoylquinic acid hexoside isomer 2 | C22H27O14– | 515.14008 | 515.13928 | 1.55 | 353(15), 341(15), 323(100), 191(25), 179(5) | 161(100), 133(5) | 133(100), 117(20) |
| 12 | 5.17 | Caffeic acid hexoside isomer 2 | C15H17O9– | 341.08781 | 341.08731 | 1.47 | 179(100), 135(10) | 135(100) | 107(100), 79(20) |
| 13 | 5.22 | Ferulic acid hexosylhexoside | C22H29O14– | 517.15628 | 517.15466 | 3.13 | 221(25), 193(100), 179(25), 161(10), 149(20) | 149(100) | 134(100) |
| 14 | 5.30 | 4-O-Caffeoylquinic acid | C16H17O9– | 353.08781 | 353.08749 | 0.91 | 223(20), 191(50), 179(60), 173(100), 135(10) | 115(20), 111(50), 93(100), 71(20) | − |
| 15 | 5.32 | Coumaric acid hexoside | C15H17O8– | 325.09289 | 325.09283 | 0.18 | 163(100), 119(10) | 119(100) | − |
| 16 | 5.42 | p-Hydroxybenzoic acid a | C7H5O3– | 137.02442 | 137.02420 | 1.61 | 109(10), 93(100) | 66(100) | − |
| 17 | 5.46 | Gentisic acid a | C7H5O4– | 153.01933 | 153.01895 | 2.48 | 109(100) | 95(10), 81(100), 65(35) | − |
| 18 | 5.73 | Ferulic acid hexoside isomer 1 | C16H19O9– | 355.10346 | 355.10187 | 4.48 | 193(100), 149(25) | 149(100) | 133(100) |
| 19 | 5.78 | 3-O-Feruoylquinic acid | C17H19O9– | 367.10346 | 367.10263 | 2.26 | 193(100), 178(5), 173(5), 134(10) | 178(90), 149(40), 134(100) | 106(100) |
| 20 | 5.82 | Caffeic acid a | C9H7O4– | 179.03498 | 179.03459 | 2.18 | 135(100) | 135(60), 117(15), 107(100), 91(55), 79(15) | − |
| 21 | 5.86 | Hydroxybenzoic acid derivative | C25H27O14– | 551.14063 | 551.13977 | 1.56 | 431(10), 413(100) | 281(5), 179(5), 137(100) | 93(100) |
| 22 | 5.94 | 5-O-p-Coumaroylquinic acid | C16H17O8– | 337.09289 | 337.09268 | 0.62 | 191(100), 179(5), 163(10) | 173(75), 127(100), 111(40), 93(60), 85(90) | 109(30), 99(40), 85(100) |
| 23 | 6.39 | 4-O-Feruoylquinic acid isomer 1 | C17H19O9– | 367.10346 | 367.10275 | 1.93 | 193(5), 173(100), 155(5), 111(5) | 155(15), 111(40), 93(100), 71(10) | − |
| 24 | 6.46 | Ferulic acid acetylhexoside isomer 1 | C18H21O10– | 397.11402 | 397.11346 | 1.41 | 193(100), 149(30), 134(10) | 149(100) | 134(100) |
| 25 | 6.51 | Ferulic acid hexoside isomer 2 | C16H19O9– | 355.10346 | 355.10306 | 1.13 | 193(100), 149(10) | 149(100) | 133(100) |
| 26 | 6.64 | 4-O-Feruoylquinic acid isomer 2 | C17H19O9– | 367.10346 | 367.10294 | 1.42 | 193(5), 179(10), 173(100), 155(5), 111(10) | 155(15), 111(40), 93(100), 71(10) | − |
| 27 | 6.72 | p-Coumaric acid a | C9H7O3– | 163.04007 | 163.04006 | 0.06 | 119(100) | 119(60), 101(20), 93(25), 91(100), 72(10) | − |
| 28 | 6.94 | Dicaffeoylquinic acid isomer 1 | C25H23O12– | 515.11950 | 515.11749 | 3.90 | 353(100), 355(20), 299(10), 191(10), 179(15) | 191(40), 179(70), 173(100), 135(10) | 155(10), 111(50), 93(100), 71(10) |
| 29 | 7.09 | Ferulic acid acetylhexoside isomer 2 | C18H21O10– | 397.11402 | 397.11400 | 0.05 | 193(100), 149(30), 134(10) | 149(100) | 134(100) |
| 30 | 7.16 | Dicaffeoylquinic acid isomer 2 | C25H23O12– | 515.11950 | 515.11780 | 3.30 | 353(100) | 191(100), 179(30), 173(5), 135(10) | 173(65), 127(90), 111(50), 93(55), 85(100) |
| 31 | 7.33 | Dicaffeoylquinic acid isomer 3 | C25H23O12– | 515.11950 | 515.11874 | 1.48 | 353(100), 355(5), 317(5), 299(10), 203(5) | 191(30), 179(60), 173(100), 135(10) | 155(20), 111(60), 93(100), 71(20) |
| 32 | 8.05 | Ferulic acid | C10H9O4– | 193.05063 | 193.04985 | 4.04 | 178(70), 149(100), 134(40) | 134(100) | − |
| Flavonoid aglycones and glycosides | |||||||||
| 33 | 5.62 | Apigenin 6,8-di-C-hexoside | C27H29O15– | 593.15119 | 593.15002 | 1.97 | 533(10), 503(30), 473(100), 383(20), 353(35) | 383(20), 353(100) | 275(20), 265(60), 249(100), 221(35), 173(80) |
| 34 | 6.18 | 6-Hydroxyquercetin 7-O-hexoside | C21H19O13– | 479.08311 | 479.08176 | 2.82 | 318(10), 317(100) | 299(40), 271(100), 167(75), 139(45) | 243(100), 227(30), 215(20), 199(50) |
| 35 | 6.65 | Quercetin 3-O-galactoside a | C21H19O12– | 463.08820 | 463.08753 | 1.45 | 301(100), 300(30) | 273(25), 257(20), 179(100), 151(75) | 151(100) |
| 36 | 6.74 | Luteolin 7-O-hexoside | C21H19O11– | 447.09329 | 447.09262 | 1.50 | 286(10), 285(100) | 257(30), 241(100), 217(75), 199(85), 175(95) | 241(5), 226(15), 213(30), 197(100) |
| 37 | 6.79 | 6-Methoxyquercetin 7-O-hexoside | C22H21O13– | 493.09876 | 493.09772 | 2.11 | 477(20), 373(10), 331(100), 323(30), 316(5) | 316(100), 209(5), 181(5), 166(5) | 287(100), 271(60), 194(40), 166(70), 151(5) |
| 38 | 6.92 | Apigenin 7-O-(6”-rhamnosyl)hexoside | C27H29O14– | 577.15628 | 577.15576 | 0.90 | 270(10), 269(100) | 225(100), 201(20), 197(30), 183(30), 149(25) | 225(5), 210(10), 197(100), 181(50), 169(40) |
| 39 | 7.07 | 6-Methoxyapigenin 7-O-(6”-rhamnosyl)hexoside | C28H31O15– | 607.16684 | 607.16656 | 0.46 | 300(15), 299(100), 284(5) | 284(100) | 284(30), 256(100), 239(5), 227(15), 211(10) |
| 40 | 7.22 | Isorhamnetin 3-O-glucoside a | C22H21O12– | 477.10385 | 477.10278 | 2.24 | 357(20), 315(50), 314(100), 300(5), 299(5) | 300(30), 285(100), 271(75), 257(10), 243(25) | 270(100) |
| 41 | 7.27 | Apigenin 7-O-glucoside a | C21H19O10− | 431.09837 | 431.09811 | 0.60 | 270(10), 269(100) | 225(100), 201(15), 197(20), 183(30), 149(30) | 210(15), 197(80), 183(100), 181(30), 169(30) |
| 42 | 7.30 | Naringenin a | C15H11O5− | 271.06120 | 271.06113 | 0.26 | 177(10), 151(100) | 107(100) | 65(100) |
| 43 | 7.38 | Isorhamnetin 7-O-hexoside | C22H21O12– | 477.10385 | 477.10385 | 0.00 | 462(5), 357(10), 316(10), 315(100), 300(5) | 300(100), 285(5), 151(10) | 283(20), 272(65), 271(70), 227(30), 151(100) |
| 44 | 7.43 | 6-Methoxyapigenin 7-O-hexoside | C22H21O11− | 461.10894 | 461.10834 | 1.30 | 446(80), 341(10), 299(100), 284(20) | 284(100) | 284(30), 256(100), 239(5), 227(15), 211(10) |
| 45 | 7.51 | 6-Methoxyquercetin 7-O-(6”-caffeoyl)hexoside | C31H27O16– | 655.13046 | 655.13043 | 0.05 | 533(15), 505(10), 331(100), 323(20), 316(30) | 316(100), 209(5), 181(5), 166(5) | 287(100), 271(60), 194(40), 166(70), 151(5) |
| 46 | 7.54 | Isorhamnetin 7-O-(6”-acetyl)hexoside | C24H23O13– | 519.11441 | 519.11383 | 1.12 | 357(5), 316(10), 315(100), 300(5), 285(5) | 300(100), 287(5), 272(10) | 272(30), 271(100), 255(50) |
| 47 | 7.81 | Apigenin 7-O-acetylhexoside isomer 1 | C23H21O11– | 473.10894 | 473.10782 | 2.37 | 413(15), 311(10), 270(15), 269(100), 268(60) | 225(100), 201(25), 197(35), 183(30), 149(40) | 210(20), 197(100), 183(30), 181(60), 169(30) |
| 48 | 8.15 | Apigenin 7-O-acetylhexoside isomer 2 | C23H21O11– | 473.10894 | 473.10699 | 4.12 | 413(30), 311(10), 270(10), 269(100), 268(40) | 225(100), 201(30), 197(30), 183(30), 149(35) | 207(10), 197(100), 183(30), 181(70), 169(40) |
| 49 | 8.20 | Apigenin 7-O-(6”-caffeoyl)hexoside | C27H29O15– | 593.13006 | 593.12848 | 2.66 | 323(95), 269(100), 221(15), 179(10) | 225(100), 201(30), 183(10), 151(20), 149(35) | 225(10), 208(10), 197(40), 181(100), 169(15) |
| 50 | 8.38 | Apigenin derivative | C28H25O13– | 569.13006 | 569.12927 | 1.39 | 270(10), 269(100) | 225(100), 201(20), 197(30), 183(30), 149(25) | 225(5), 210(10), 197(100), 181(50), 169(40) |
| 51 | 8.50 | Apigenin 7-O-acetylhexoside isomer 3 | C23H21O11– | 473.10894 | 473.10726 | 3.55 | 413(5), 311(10), 270(15), 269(100), 268(50) | 225(100), 201(30), 197(25), 183(25), 149(30) | 197(70), 183(50), 181(100), 169(30) |
| 52 | 8.55 | Apigenin 7-O-diacetylhexoside isomer 1 | C25H23O12– | 515.11950 | 515.11713 | 4.60 | 455(30), 431(5), 413(10), 311(15), 269(100) | 225(100), 201(25), 197(25), 183(25), 149(30) | 210(10), 197(100), 183(30), 181(50), 169(40) |
| 53 | 8.63 | 6-Methoxyapigenin 7-O-(6”acetyl)hexoside | C24H23O12– | 503.11950 | 503.11816 | 2.66 | 488(100), 299(20), 284(10) | 429(10), 327(10), 313(40), 283(100), 255(30) | 255(100) |
| 54 | 8.68 | 6-Methoxyapigenin | C16H11O6– | 299.05611 | 299.05569 | 1.40 | 285(15), 284(100) | 256(100) | 238(30), 228(70), 211(60), 188(100) |
| 55 | 8.67 | Luteolin a | C15H9O6− | 285.04046 | 285.03992 | 1.89 | 257(40), 241(100), 217(50), 199(70), 175(70) | 255(50), 227(100), 211(75), 197(35), 183(85) | − |
| 56 | 8.74 | Apigenin 7-O-diacetylhexoside isomer 2 | C25H23O12– | 515.11950 | 515.11804 | 2.83 | 455(20), 270(10), 269(100), 268(40) | 225(100), 201(30), 197(20), 183(25), 149(40) | 197(100), 183(50), 181(80), 169(30) |
| 57 | 8.80 | Quercetin a | C15H9O7− | 301.03538 | 301.03483 | 1.83 | 271(50), 255(20), 179(100), 151(80), 107(5) | 151(100) | 107(100), 83(10) |
| 58 | 8.84 | 6-Methoxyluteolin | C16H11O7– | 315.05103 | 315.05063 | 1.27 | 301(20), 300(100), 166(5) | 283(40), 272(70), 255(50), 243(40), 216(100) | 201(25), 188(100), 173(20) |
| 59 | 9.05 | Apigenin 7-O-diacetylhexoside isomer 3 | C25H23O12– | 515.11950 | 515.11823 | 2.47 | 455(20), 293(10), 270(10), 269(100), 268(20) | 225(100), 201(25), 197(35), 183(30), 149(40) | 210(20), 197(100), 183(30), 181(60), 169(30) |
| 60 | 9.53 | Apigenin 7-O-diacetylhexoside isomer 4 | C25H23O12– | 515.11950 | 515.11761 | 3.67 | 455(30), 413(10), 311(15), 270(15), 269(100) | 225(100), 201(30), 197(30), 183(30), 149(35) | 207(10), 197(100), 183(30), 181(70), 169(40) |
| 61 | 9.56 | Apigenin a | C15H9O5− | 269.04554 | 269.04449 | 3.90 | 225(5), 177(15), 151(100) | 65(100) | − |
| 62 | 9.68 | Apigenin 7-O-diacetylhexoside isomer 5 | C25H23O12– | 515.11950 | 515.11835 | 2.23 | 455(10), 270(10), 269(100), 268(30) | 225(100), 201(30), 197(25), 183(25), 149(30) | 197(70), 183(50), 181(100), 169(30) |
| 63 | 9.73 | Kaempferol a | C15H9O6− | 285.04046 | 285.03969 | 2.70 | 255(100), 227(10) | 211(100), 195(5), 167(15) | 211(40), 137(100) |
| 64 | 9.81 | Chrysoeriol a | C16H11O6– | 299.05611 | 299.05565 | 1.54 | 285(10), 284(100) | 256(100) | 239(10), 227(100), 212(20), 200(15) |
| 65 | 9.95 | Isorhamnetin | C16H11O7– | 315.05103 | 315.05017 | 2.73 | 301(20), 300(100) | 283(30), 271(100), 255(40), 227(50), 151(90) | 243(100), 227(50), 215(10), 199(20) |
| 66 | 10.32 | Chrysosplenol | C18H15O8– | 359.07724 | 359.07703 | 0.58 | 345(10), 344(100), 287(10), 240(5) | 329(100), 301(5) | 314(100), 301(20), 286(30), 270(5), 175(5) |
| 67 | 11.74 | Chrysosplenetin | C19H17O8– | 373.09289 | 373.09277 | 0.32 | 359(10), 358(100) | 343(100) | 328(100), 315(15), 300(30), 284(10), 272(10) |
| Compound Name | mg/L |
|---|---|
| Phenolic acids and their derivatives | |
| Gallic acid | 0.072 |
| Protocatechuic acid | 0.649 |
| p-Hydroxybenzoic acid | 1.619 |
| Gentisic acid | 0.510 |
| Caffeic acid | 0.494 |
| p-Coumaric acid | 0.762 |
| Flavonoid aglycones and glycosides | |
| Quercetin 3-O-galactoside | 0.283 |
| Isorhamnetin 3-O-glucoside | 0.569 |
| Apigenin 7-O-glucoside | 2.408 |
| Naringenin | 0.142 |
| Luteolin | 0.191 |
| Quercetin | 0.861 |
| Apigenin | 1.542 |
| Kaempferol | 0.358 |
| Chrysoeriol | 0.286 |
| Parameters | Results |
|---|---|
| Antioxidant assays | |
| DPPH (mg TE/g) | 60.24 ± 2.79 |
| ABTS (mg TE/g) | 126.92 ± 9.50 |
| CUPRAC (mg TE/g) | 123.12 ± 0.93 |
| FRAP (mg TE/g) | 95.37 ± 1.98 |
| MCA (mg EDTAE/g) | 21.34 ± 0.06 |
| PBD (mmol TE/g) | 1.42 ± 0.06 |
| Enzyme inhibitory assays | |
| AChE inhibition (mg GALAE/g) | 0.85 ± 0.11 |
| Amylase inhibition (mmol ACAE/g) | 0.18 ± 0.01 |
| Glucosidase inhibition (mmol ACAE/g) | 13.11 ± 0.72 |
| Inputs | Output | ||
|---|---|---|---|
| Microwave Power (kW) | Extraction Time (min) | Solid-to-Solvent Ratio | TPC |
| 400 | 40 | 1:60 | 46.04 |
| 400 | 30 | 1:80 | 46.16 |
| 400 | 20 | 1:60 | 42.85 |
| 400 | 40 | 1:40 | 44.18 |
| 400 | 30 | 1:40 | 37.99 |
| 400 | 20 | 1:40 | 35.54 |
| 400 | 30 | 1:60 | 58.435 |
| 400 | 20 | 1:80 | 56.75 |
| 400 | 40 | 1:80 | 54.14 |
| 600 | 20 | 1:80 | 50.68 |
| 600 | 20 | 1:40 | 35.95 |
| 600 | 20 | 1:60 | 40.65 |
| 600 | 40 | 1:40 | 38.47 |
| 600 | 40 | 1:60 | 48.5 |
| 600 | 30 | 1:40 | 38.07 |
| 600 | 40 | 1:80 | 47.46 |
| 600 | 30 | 1:60 | 54.51 |
| 600 | 30 | 1:80 | 51.86 |
| 800 | 30 | 1:60 | 46.9 |
| 800 | 30 | 1:80 | 41.6 |
| 800 | 40 | 1:40 | 46.29 |
| 800 | 20 | 1:80 | 57.32 |
| 800 | 30 | 1:40 | 44.34 |
| 800 | 40 | 1:80 | 59.56 |
| 800 | 20 | 1:40 | 37.17 |
| 800 | 20 | 1:60 | 47.88 |
| 800 | 40 | 1:60 | 49.36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cvetanović Kljakić, A.; Radosavljević, M.; Zengin, G.; Yan, L.; Gašić, U.; Kojić, P.; Torbica, A.; Belović, M.; Zeković, Z. New Biological and Chemical Insights into Optimization of Chamomile Extracts by Using Artificial Neural Network (ANN) Model. Plants 2023, 12, 1211. https://doi.org/10.3390/plants12061211
Cvetanović Kljakić A, Radosavljević M, Zengin G, Yan L, Gašić U, Kojić P, Torbica A, Belović M, Zeković Z. New Biological and Chemical Insights into Optimization of Chamomile Extracts by Using Artificial Neural Network (ANN) Model. Plants. 2023; 12(6):1211. https://doi.org/10.3390/plants12061211
Chicago/Turabian StyleCvetanović Kljakić, Aleksandra, Miloš Radosavljević, Gokhan Zengin, Linlin Yan, Uroš Gašić, Predrag Kojić, Aleksandra Torbica, Miona Belović, and Zoran Zeković. 2023. "New Biological and Chemical Insights into Optimization of Chamomile Extracts by Using Artificial Neural Network (ANN) Model" Plants 12, no. 6: 1211. https://doi.org/10.3390/plants12061211
APA StyleCvetanović Kljakić, A., Radosavljević, M., Zengin, G., Yan, L., Gašić, U., Kojić, P., Torbica, A., Belović, M., & Zeković, Z. (2023). New Biological and Chemical Insights into Optimization of Chamomile Extracts by Using Artificial Neural Network (ANN) Model. Plants, 12(6), 1211. https://doi.org/10.3390/plants12061211












