Identification and Characterization of Common Bean (Phaseolus vulgaris) Non-Nodulating Mutants Altered in Rhizobial Infection
Abstract
:1. Introduction
2. Results
2.1. Screening for Altered Nodulation Phenotype in the Mutagenized Common Bean Population
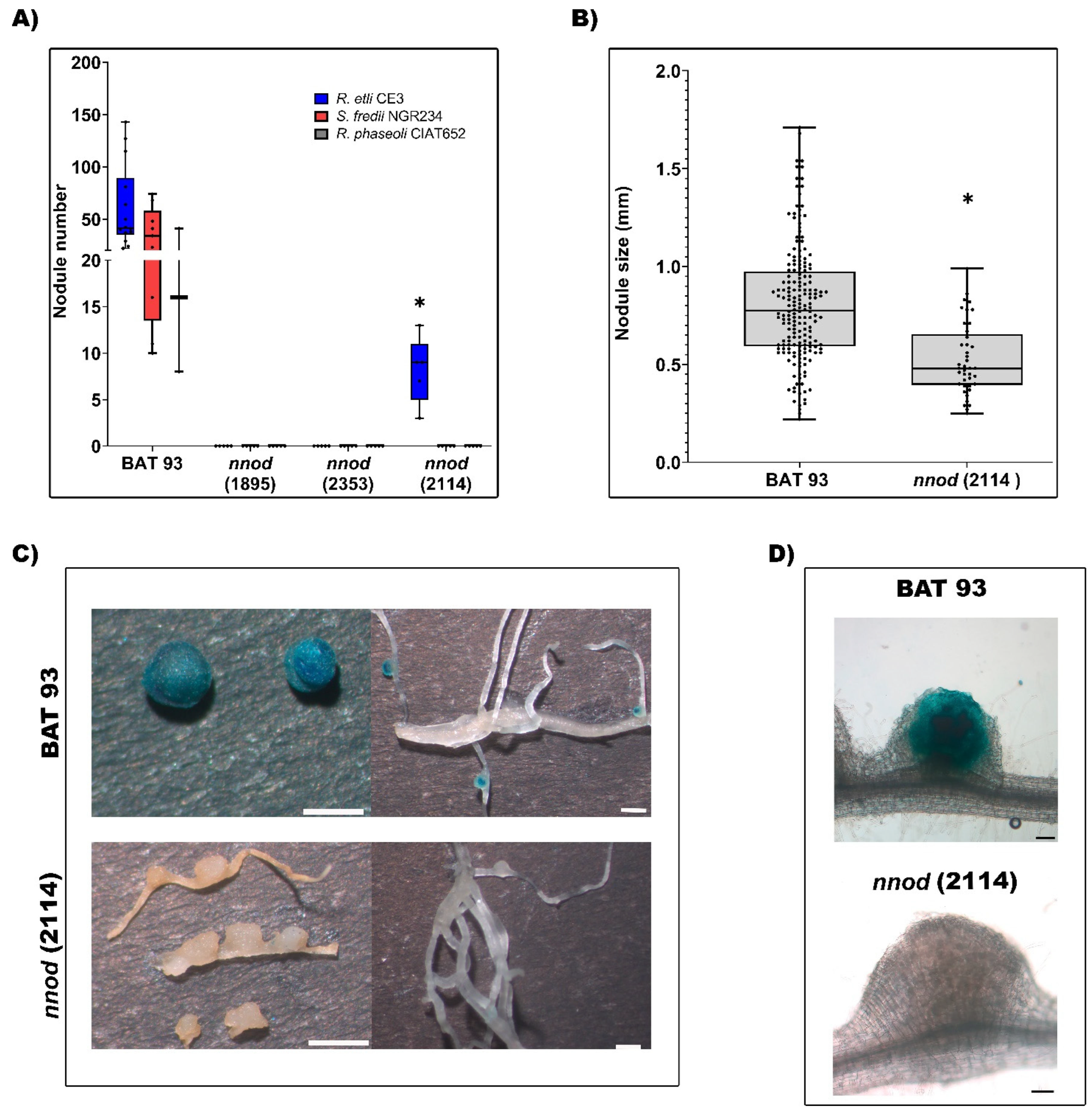
2.2. Growth, Root Phenotype and Nodulation Capacity of Nnod Mutants
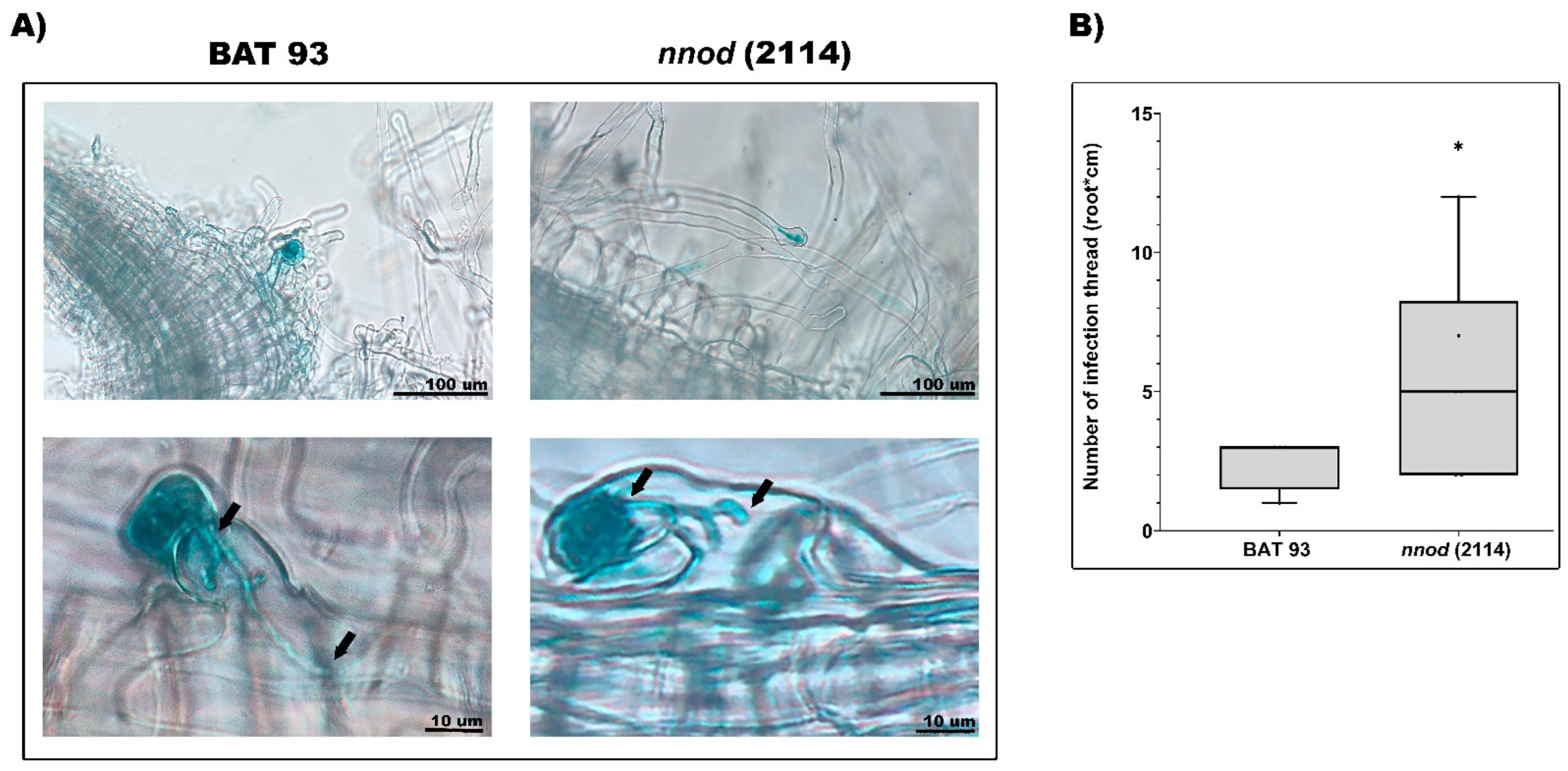
2.3. Comparative Microscopical Analysis of Inoculated BAT 93 and Nnod Mutants’ Inoculated Roots at the Early Stages of the Symbiotic Process
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Screening for Symbiotic Mutants
4.3. Rhizobial Strains and Culture Conditions
4.4. Observation (Analyses) of Infection and Nodulation Events
4.5. Root Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doyle, J.J.; Luckow, M.A. The Rest of the Iceberg. Legume Diversity and Evolution in a Phylogenetic Context. Plant Physiol. 2003, 131, 900–910. [Google Scholar] [CrossRef] [Green Version]
- Oldroyd, G.E.D. Speak, Friend, and Enter: Signalling Systems That Promote Beneficial Symbiotic Associations in Plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef]
- Venkateshwaran, M.; Volkening, J.D.; Sussman, M.R.; Ané, J.-M. Symbiosis and the Social Network of Higher Plants. Curr. Opin. Plant Biol. 2013, 16, 118–127. [Google Scholar] [CrossRef]
- Broughton, W.J.; Hernández, G.; Blair, M.; Beebe, S.; Gepts, P.; Vanderleyden, J. Beans (Phaseolus Spp.)–Model Food Legumes. Plant Soil. 2003, 252, 55–128. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, B.J.; Indrasumunar, A.; Hayashi, S.; Lin, M.-H.; Lin, Y.-H.; Reid, D.E.; Gresshoff, P.M. Molecular Analysis of Legume Nodule Development and Autoregulation. J. Integr. Plant. Biol. 2010, 52, 61–76. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 Years of Genetic Discoveries in Legume Nodulation and Symbiotic Nitrogen Fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouchi, H.; Imaizumi-Anraku, H.; Hayashi, M.; Hakoyama, T.; Nakagawa, T.; Umehara, Y.; Suganuma, N.; Kawaguchi, M. How Many Peas in a Pod? Legume Genes Responsible for Mutualistic Symbioses Underground. Plant Cell Physiol. 2010, 51, 1381–1397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzaki, T.; Yoro, E.; Kawaguchi, M. Leguminous Plants: Inventors of Root Nodules to Accommodate Symbiotic Bacteria. Int. Rev. Cell Mol. Biol. 2015, 316, 111–158. [Google Scholar] [CrossRef] [PubMed]
- Mergaert, P.; Kereszt, A.; Kondorosi, E. Gene Expression in Nitrogen-Fixing Symbiotic Nodule Cells in Medicago Truncatula and Other Nodulating Plants. Plant Cell 2020, 32, 42–68. [Google Scholar] [CrossRef]
- Borisov, A.; Morzhina, E.V.; Kulikov, O.A.; Tchetkova, S.A.; Lebsky, V.; Tikhonovich, I.A. New Symbiotic Mutants of Pea (Pisum Sativum L.) Affecting Either Nodule Initiation or Symbiosome Development. Symbiosis. 1993, 14, 297–313. [Google Scholar]
- Kneen, B.E.; LaRue, T.A. Induced Symbiosis Mutants of Pea (Pisum Sativum) and Sweetclover (Melilotus Alba Annua). Plant Sci. 1988, 58, 177–182. [Google Scholar] [CrossRef]
- Duc, G.; Messager, A. Mutagenesis of Pea (Pisum Sativum L.) and the Isolation of Mutants for Nodulation and Nitrogen Fixation. Plant Sci. 1989, 60, 207–213. [Google Scholar] [CrossRef]
- Bolon, Y.-T.; Haun, W.J.; Xu, W.W.; Grant, D.; Stacey, M.G.; Nelson, R.T.; Gerhardt, D.J.; Jeddeloh, J.A.; Stacey, G.; Muehlbauer, G.J.; et al. Phenotypic and Genomic Analyses of a Fast Neutron Mutant Population Resource in Soybean. Plant Physiol. 2011, 156, 240–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, B.J.; McNeil, D.L.; Gresshoff, P.M. Isolation and Properties of Soybean [Glycine Max (L.) Merr.] Mutants That Nodulate in the Presence of High Nitrate Concentrations. Proc. Natl. Acad. Sci. USA 1985, 82, 4162–4166. [Google Scholar] [CrossRef] [Green Version]
- Espina, M.J.; Ahmed, C.M.S.; Bernardini, A.; Adeleke, E.; Yadegari, Z.; Arelli, P.; Pantalone, V.; Taheri, A. Development and Phenotypic Screening of an Ethyl Methane Sulfonate Mutant Population in Soybean. Front. Plant Sci. 2018, 9, 394. [Google Scholar] [CrossRef] [Green Version]
- Dudley, M.E.; Long, S.R. A Non-Nodulating Alfalfa Mutant Displays Neither Root Hair Curling nor Early Cell Division in Response to Rhizobium Meliloti. Plant Cell 1989, 1, 65–72. [Google Scholar] [CrossRef] [Green Version]
- Davis, T.M. Two Genes That Confer Ineffective Nodulation in Chickpea (Cicer Arietinum L.). J. Hered. 1988, 79, 476–478. [Google Scholar] [CrossRef]
- O’Rourke, J.A.; Iniguez, L.P.; Bucciarelli, B.; Roessler, J.; Schmutz, J.; McClean, P.E.; Jackson, S.A.; Hernandez, G.; Graham, M.A.; Stupar, R.M.; et al. A Re-Sequencing Based Assessment of Genomic Heterogeneity and Fast Neutron-Induced Deletions in a Common Bean Cultivar. Front. Plant Sci. 2013, 4, 210. [Google Scholar] [CrossRef] [Green Version]
- Park, S.J.; Buttery, B.R. Nodulation mutants of white bean (Phaseolus Vulgaris L.) induced by ethyl-methane sulphonate. Can. J. Plant Sci. 1988, 68, 199–202. [Google Scholar] [CrossRef]
- Park, S.J.; Buttery, B.R. Ethyl-Methane Sulphonate (EMS) Induced Nodulation Mutants of Common Bean (Phaseolus Vulgaris L.) Lacking Effective Nodules. Plant Soil. 1992, 139, 295–298. [Google Scholar] [CrossRef]
- Porch, T.G.; Blair, M.W.; Lariguet, P.; Galeano, C.; Pankhurst, C.E.; Broughton, W.J. Generation of a Mutant Population for tilling Common Bean Genotype BAT 93. J. Am. Soc. Hortic. Sci. 2009, 134, 348–355. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, M.; Imaizumi-Anraku, H.; Koiwa, H.; Niwa, S.; Ikuta, A.; Syono, K.; Akao, S. Root, Root Hair, and Symbiotic Mutants of the Model Legume Lotus Japonicus. Mol. Plant-Microbe Interact. 2002, 15, 17–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, J.A.; Wang, T.L.; Welham, T.J.; Gardner, S.; Pike, J.M.; Yoshida, S.; Parniske, M. A TILLING Reverse Genetics Tool and a Web-Accessible Collection of Mutants of the Legume Lotus Japonicus. Plant Physiol. 2003, 131, 866–871. [Google Scholar] [CrossRef] [Green Version]
- Szczyglowski, K.; Shaw, R.S.; Wopereis, J.; Copeland, S.; Hamburger, D.; Kasiborski, B.; Dazzo, F.B.; de Bruijn, F.J. Nodule Organogenesis and Symbiotic Mutants of the Model Legume Lotus Japonicus. Mol. Plant-Microbe Interact. 1998, 11, 684–697. [Google Scholar] [CrossRef] [Green Version]
- Benaben, V.; Duc, G.; Lefebvre, V.; Huguet, T. TE7, An Inefficient Symbiotic Mutant of Medicago Truncatula Gaertn. Cv Jemalong. Plant Physiol. 1995, 107, 53–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penmetsa, R.V.; Cook, D.R. Production and Characterization of Diverse Developmental Mutants of Medicago Truncatula. Plant Physiol. 2000, 123, 1387–1398. [Google Scholar] [CrossRef] [Green Version]
- Tadege, M.; Wen, J.; He, J.; Tu, H.; Kwak, Y.; Eschstruth, A.; Cayrel, A.; Endre, G.; Zhao, P.X.; Chabaud, M.; et al. Large-Scale Insertional Mutagenesis Using the Tnt1 Retrotransposon in the Model Legume Medicago Truncatula. Plant J. 2008, 54, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Pislariu, C.I.D.; Murray, J.; Wen, J.; Cosson, V.; Muni, R.R.D.; Wang, M.; Benedito, A.V.; Andriankaja, A.; Cheng, X.; Jerez, I.T.; et al. A Medicago Truncatula Tobacco Retrotransposon Insertion Mutant Collection with Defects in Nodule Development and Symbiotic Nitrogen Fixation. Plant Physiol. 2012, 159, 1686–1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukai, E.; Stougaard, J.; Hayashi, M. Activation of an Endogenous Retrotransposon Associated with Epigenetic Changes in Lotus Japonicus: A Tool for Functional Genomics in Legumes. Plant Genome 2013, 6, plantgenome2013.04. [Google Scholar] [CrossRef] [Green Version]
- Małolepszy, A.; Mun, T.; Sandal, N.; Gupta, V.; Dubin, M.; Urbański, D.; Shah, N.; Bachmann, A.; Fukai, E.; Hirakawa, H.; et al. The LORE 1 Insertion Mutant Resource. Plant J. 2016, 88, 306–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buttery, B.R.; Park, S.J. Characterization of Some Non-Fixing Mutants of Common Bean (Phaseolus Vulgaris L.). Can. J. Plant Sci. 1993, 73, 977–983. [Google Scholar] [CrossRef]
- Park, S.J.; Buttery, B.R. Inheritance of Non-Nodulation and Ineffective Nodulation Mutants in Common Bean (Phaseolus Vulgaris L.). J. Hered. 1994, 85, 1–3. [Google Scholar] [CrossRef]
- Pedalino, M.; Giller, K.E.; Kipe-Nolt, J. Genetic and physiological characterization of the non-nodulating mutant of Phaseolus vulgaris L. -NOD125. J. Exp. Bot. 1992, 43, 843–849. [Google Scholar] [CrossRef]
- Ferguson, B.J.; Li, D.; Hastwell, A.H.; Reid, D.E.; Li, Y.; Jackson, S.A.; Gresshoff, P.M. The Soybean (Glycine Max) Nodulation-Suppressive CLE Peptide, GmRIC1, Functions Interspecifically in Common White Bean (Phaseolus Vulgari ), but Not in a Supernodulating Line Mutated in the Receptor PvNARK. Plant Biotechnol. J. 2014, 12, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.; Capella-Gutiérrez, S.; Rendón-Anaya, M.; Hernández-Oñate, M.; Minoche, A.E.; Erb, I.; Câmara, F.; Prieto-Barja, P.; Corvelo, A.; Sanseverino, W.; et al. Genome and Transcriptome Analysis of the Mesoamerican Common Bean and the Role of Gene Duplications in Establishing Tissue and Temporal Specialization of Genes. Genome Biol. 2016, 17, 32. [Google Scholar] [CrossRef] [Green Version]
- Cominelli, E.; Confalonieri, M.; Carlessi, M.; Cortinovis, G.; Daminati, M.G.; Porch, T.G.; Losa, A.; Sparvoli, F. Phytic Acid Transport in Phaseolus Vulgaris: A New Low Phytic Acid Mutant in the PvMRP1 Gene and Study of the PvMRPs Promoters in Two Different Plant Systems. Plant Sci. 2018, 270, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Panzeri, D.; Cassani, E.; Doria, E.; Tagliabue, G.; Forti, L.; Campion, B.; Bollini, R.; Brearley, C.A.; Pilu, R.; Nielsen, E.; et al. A Defective ABC Transporter of the MRP Family, Responsible for the Bean Lpa1 Mutation, Affects the Regulation of the Phytic Acid Pathway, Reduces Seed Myo -inositol and Alters ABA Sensitivity. New Phytol. 2011, 191, 70–83. [Google Scholar] [CrossRef] [Green Version]
- Segovia, L.; Young, J.P.W.; Martinez-Romero, E. Reclassification of American Rhizobium Leguminosarum Biovar Phaseoli Type I Strains as Rhizobium Etli Sp. Nov. Int. J. Syst. Bacteriol. 1993, 43, 374–377. [Google Scholar] [CrossRef] [Green Version]
- Pueppke, S.G.; Broughton, W.J. Rhizobium Sp. Strain NGR234 and R. Fredii USDA257 Share Exceptionally Broad, Nested Host Ranges. Mol. Plant-Microbe Interact. 1999, 12, 293–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, V.; Acosta, J.L.; Santamaría, R.I.; Bustos, P.; Fernández, J.L.; Hernández González, I.L.; Díaz, R.; Flores, M.; Palacios, R.; Mora, J.; et al. Conserved Symbiotic Plasmid DNA Sequences in the Multireplicon Pangenomic Structure of Rhizobium Etli. App. Environ. Microbiol. 2010, 76, 1604–1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyero-Saavedra, M.; Qiao, Z.; Sánchez-Correa, M.; Díaz-Pineda, M.; Reyes, J.; Covarrubias, A.; Libault, M.; Valdés-López, O. Gene Silencing of Argonaute5 Negatively Affects the Establishment of the Legume-Rhizobia Symbiosis. Genes 2017, 8, 352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fournier, J.; Teillet, A.; Chabaud, M.; Ivanov, S.; Genre, A.; Limpens, E.; de Carvalho-Niebel, F.; Barker, D.G. Remodeling of the Infection Chamber before Infection Thread Formation Reveals a Two-Step Mechanism for Rhizobial Entry into the Host Legume Root Hair. Plant Physiol. 2015, 167, 1233–1242. [Google Scholar] [CrossRef]
- Reid, D.E.; Ferguson, B.J.; Hayashi, S.; Lin, Y.-H.; Gresshoff, P.M. Molecular Mechanisms Controlling Legume Autoregulation of Nodulation. Ann. Bot. 2011, 108, 789–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buttery, B.R.; Park, S.J. Effects of nitrogen, inoculation and grafting on expression of supernodulation in a mutant of Phaseolus vulgaris L. Can. J. Plant Sci. 1990, 70, 375–381. [Google Scholar] [CrossRef]
- Cerri, M.R.; Frances, L.; Kelner, A.; Fournier, J.; Middleton, P.H.; Auriac, M.-C.; Mysore, K.S.; Wen, J.; Erard, M.; Barker, D.G.; et al. The Symbiosis-Related ERN Transcription Factors Act in Concert to Coordinate Rhizobial Host Root Infection. Plant Physiol. 2016, 171, 1037–1054. [Google Scholar] [CrossRef] [Green Version]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Grønlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; et al. Plant Recognition of Symbiotic Bacteria Requires Two LysM Receptor-like Kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef]
- Stracke, S.; Kistner, C.; Yoshida, S.; Mulder, L.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; Stougaard, J.; Szczyglowski, K.; et al. A Plant Receptor-like Kinase Required for Both Bacterial and Fungal Symbiosis. Nature 2002, 417, 959–962. [Google Scholar] [CrossRef]
- Kanamori, N.; Madsen, L.H.; Radutoiu, S.; Frantescu, M.; Quistgaard, E.M.H.; Miwa, H.; Downie, J.A.; James, E.K.; Felle, H.H.; Haaning, L.L.; et al. A Nucleoporin Is Required for Induction of Ca2+ Spiking in Legume Nodule Development and Essential for Rhizobial and Fungal Symbiosis. Proc. Natl. Acad. Sci. USA 2006, 103, 359–364. [Google Scholar] [CrossRef] [Green Version]
- Saito, K.; Yoshikawa, M.; Yano, K.; Miwa, H.; Uchida, H.; Asamizu, E.; Sato, S.; Tabata, S.; Imaizumi-Anraku, H.; Umehara, Y.; et al. Nucleoporin85 Is Required for Calcium Spiking, Fungal and Bacterial Symbioses, and Seed Production in Lotus Japonicus. Plant Cell 2007, 19, 610–624. [Google Scholar] [CrossRef] [Green Version]
- Kawaharada, Y.; Kelly, S.; Nielsen, M.W.; Hjuler, C.T.; Gysel, K.; Muszyński, A.; Carlson, R.W.; Thygesen, M.B.; Sandal, N.; Asmussen, M.H.; et al. Receptor-Mediated Exopolysaccharide Perception Controls Bacterial Infection. Nature 2015, 523, 308–312. [Google Scholar] [CrossRef] [Green Version]
- Yano, K.; Yoshida, S.; Müller, J.; Singh, S.; Banba, M.; Vickers, K.; Markmann, K.; White, C.; Schuller, B.; Sato, S.; et al. Cyclops, a Mediator of Symbiotic Intracellular Accommodation. Proc. Natl. Acad. Sci. USA 2008, 105, 20540–20545. [Google Scholar] [CrossRef] [Green Version]
- Sinharoy, S.; Liu, C.; Breakspear, A.; Guan, D.; Shailes, S.; Nakashima, J.; Zhang, S.; Wen, J.; Torres-Jerez, I.; Oldroyd, G.; et al. A Medicago Truncatula Cystathionine-β-Synthase-like Domain-Containing Protein Is Required for Rhizobial Infection and Symbiotic Nitrogen Fixation. Plant Physiol. 2016, 170, 2204–2217. [Google Scholar] [CrossRef] [Green Version]
- Liang, P.; Stratil, T.F.; Popp, C.; Marín, M.; Folgmann, J.; Mysore, K.S.; Wen, J.; Ott, T. Symbiotic Root Infections in Medicago Truncatula Require Remorin-Mediated Receptor Stabilization in Membrane Nanodomains. Proc. Natl. Acad. Sci. USA 2018, 115, 5289–5294. [Google Scholar] [CrossRef] [Green Version]
- Xie, F.; Murray, J.D.; Kim, J.; Heckmann, A.B.; Edwards, A.; Oldroyd, G.E.D.; Downie, J.A. Legume Pectate Lyase Required for Root Infection by Rhizobia. Proc. Natl. Acad. Sci. USA 2012, 109, 633–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.S.; Liao, J.; James, E.K.; Sato, S.; Tabata, S.; Jurkiewicz, A.; Madsen, L.H.; Stougaard, J.; Ross, L.; Szczyglowski, K. Lotus Japonicus ARPC1 Is Required for Rhizobial Infection. Plant Physiol. 2012, 160, 917–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, L.; Lin, J.; Xu, J.; Sato, S.; Parniske, M.; Wang, T.L.; Downie, J.A.; Xie, F. SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation. PLoS Genet. 2015, 11, e1005623. [Google Scholar] [CrossRef] [Green Version]
- Yokota, K.; Fukai, E.; Madsen, L.H.; Jurkiewicz, A.; Rueda, P.; Radutoiu, S.; Held, M.; Hossain, M.S.; Szczyglowski, K.; Morieri, G.; et al. Rearrangement of Actin Cytoskeleton Mediates Invasion of Lotus Japonicus Roots by Mesorhizobium Loti. Plant Cell 2009, 21, 267–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, J.D.; Muni, R.R.D.; Torres-Jerez, I.; Tang, Y.; Allen, S.; Andriankaja, M.; Li, G.; Laxmi, A.; Cheng, X.; Wen, J.; et al. Vapyrin, a Gene Essential for Intracellular Progression of Arbuscular Mycorrhizal Symbiosis, Is Also Essential for Infection by Rhizobia in the Nodule Symbiosis of Medicago Truncatula: VAPYRIN Is Required for Rhizobial and Mycorrhizal Symbioses. Plant J. 2011, 65, 244–252. [Google Scholar] [CrossRef]
- Yano, K.; Shibata, S.; Chen, W.-L.; Sato, S.; Kaneko, T.; Jurkiewicz, A.; Sandal, N.; Banba, M.; Imaizumi-Anraku, H.; Kojima, T.; et al. CERBERUS, a Novel U-Box Protein Containing WD-40 Repeats, Is Required for Formation of the Infection Thread and Nodule Development in the Legume- Rhizobium Symbiosis. Plant J. 2009, 60, 168–180. [Google Scholar] [CrossRef]
- Arrighi, J.-F.; Godfroy, O.; de Billy, F.; Saurat, O.; Jauneau, A.; Gough, C. The RPG Gene of Medicago Truncatula Controls Rhizobium -Directed Polar Growth during Infection. Proc. Natl. Acad. Sci. USA 2008, 105, 9817–9822. [Google Scholar] [CrossRef] [Green Version]
- Kovács, S.; Kiss, E.; Jenei, S.; Fehér-Juhász, E.; Kereszt, A.; Endre, G. The Medicago Truncatula IEF Gene Is Crucial for the Progression of Bacterial Infection During Symbiosis. MPMI 2022, 35, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Tuinstra, M.; Carraro, N.; Weil, C.; Dilkes, B.P. Whole-Genome Sequence Accuracy Is Improved by Replication in a Population of Mutagenized Sorghum. G3 Genes|Genomes|Genet. 2018, 8, 1079–1094. [Google Scholar] [CrossRef] [Green Version]
- Henry, I.M.; Nagalakshmi, U.; Lieberman, M.C.; Ngo, K.J.; Krasileva, K.V.; Vasquez-Gross, H.; Akhunova, A.; Akhunov, E.; Dubcovsky, J.; Tai, T.H.; et al. Efficient Genome-Wide Detection and Cataloging of EMS-Induced Mutations Using Exome Capture and Next-Generation Sequencing. Plant Cell 2014, 26, 1382–1397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isidra-Arellano, M.C.; Pozas-Rodríguez, E.A.; Rocío Reyero-Saavedra, M.; Arroyo-Canales, J.; Ferrer-Orgaz, S.; Socorro Sánchez-Correa, M.; Cardenas, L.; Covarrubias, A.A.; Valdés-López, O. Inhibition of Legume Nodulation by Pi Deficiency Is Dependent on the Autoregulation of Nodulation (AON) Pathway. Plant J. 2020, 103, 1125–1139. [Google Scholar] [CrossRef]
- Summerfield, R.J.; Huxley, P.A.; Minchin, F.R. Plant Husbandry and Management Techniques for Growing Grain Legumes Under Simulated Tropical Conditions in Controlled Environments*. Exp. Agric. 1977, 13, 81–92. [Google Scholar] [CrossRef]
- Isidra-Arellano, M.; Reyero-Saavedra, M.; Sánchez-Correa, M.; Pingault, L.; Sen, S.; Joshi, T.; Girard, L.; Castro-Guerrero, N.; Mendoza-Cozatl, D.; Libault, M.; et al. Phosphate Deficiency Negatively Affects Early Steps of the Symbiosis between Common Bean and Rhizobia. Genes 2018, 9, 498. [Google Scholar] [CrossRef] [Green Version]
- Figurski, D.H.; Helinski, D.R. Replication of an Origin-Containing Derivative of Plasmid RK2 Dependent on a Plasmid Function Provided in Trans. Proc. Natl. Acad. Sci. USA 1979, 76, 1648–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noel, K.D.; Sanchez, A.; Fernandez, L.; Leemans, J.; Cevallos, M.A. Rhizobium Phaseoli Symbiotic Mutants with Transposon Tn5 Insertions. J. Bacteriol. 1984, 158, 148–155. [Google Scholar] [CrossRef] [Green Version]
- Hynes, M.F.; McGregor, N.F. Two Plasmids Other than the Nodulation Plasmid Are Necessary for Formation of Nitrogen-Fixing Nodules by Rhizobium Leguminosarum. Mol. Microbiol. 1990, 4, 567–574. [Google Scholar] [CrossRef]
- Lobet, G.; Pagès, L.; Draye, X. A Novel Image-Analysis Toolbox Enabling Quantitative Analysis of Root System Architecture. Plant Physiol. 2011, 157, 29–39. [Google Scholar] [CrossRef] [Green Version]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyero-Saavedra, R.; Fuentes, S.I.; Leija, A.; Jiménez-Nopala, G.; Peláez, P.; Ramírez, M.; Girard, L.; Porch, T.G.; Hernández, G. Identification and Characterization of Common Bean (Phaseolus vulgaris) Non-Nodulating Mutants Altered in Rhizobial Infection. Plants 2023, 12, 1310. https://doi.org/10.3390/plants12061310
Reyero-Saavedra R, Fuentes SI, Leija A, Jiménez-Nopala G, Peláez P, Ramírez M, Girard L, Porch TG, Hernández G. Identification and Characterization of Common Bean (Phaseolus vulgaris) Non-Nodulating Mutants Altered in Rhizobial Infection. Plants. 2023; 12(6):1310. https://doi.org/10.3390/plants12061310
Chicago/Turabian StyleReyero-Saavedra, Rocío, Sara Isabel Fuentes, Alfonso Leija, Gladys Jiménez-Nopala, Pablo Peláez, Mario Ramírez, Lourdes Girard, Timothy G. Porch, and Georgina Hernández. 2023. "Identification and Characterization of Common Bean (Phaseolus vulgaris) Non-Nodulating Mutants Altered in Rhizobial Infection" Plants 12, no. 6: 1310. https://doi.org/10.3390/plants12061310




