Unravelling the Phytotoxic Effects of Glyphosate on Sensitive and Resistant Amaranthus palmeri Populations by GC–MS and LC–MS Metabolic Profiling
Abstract
:1. Introduction
2. Results and Discussion
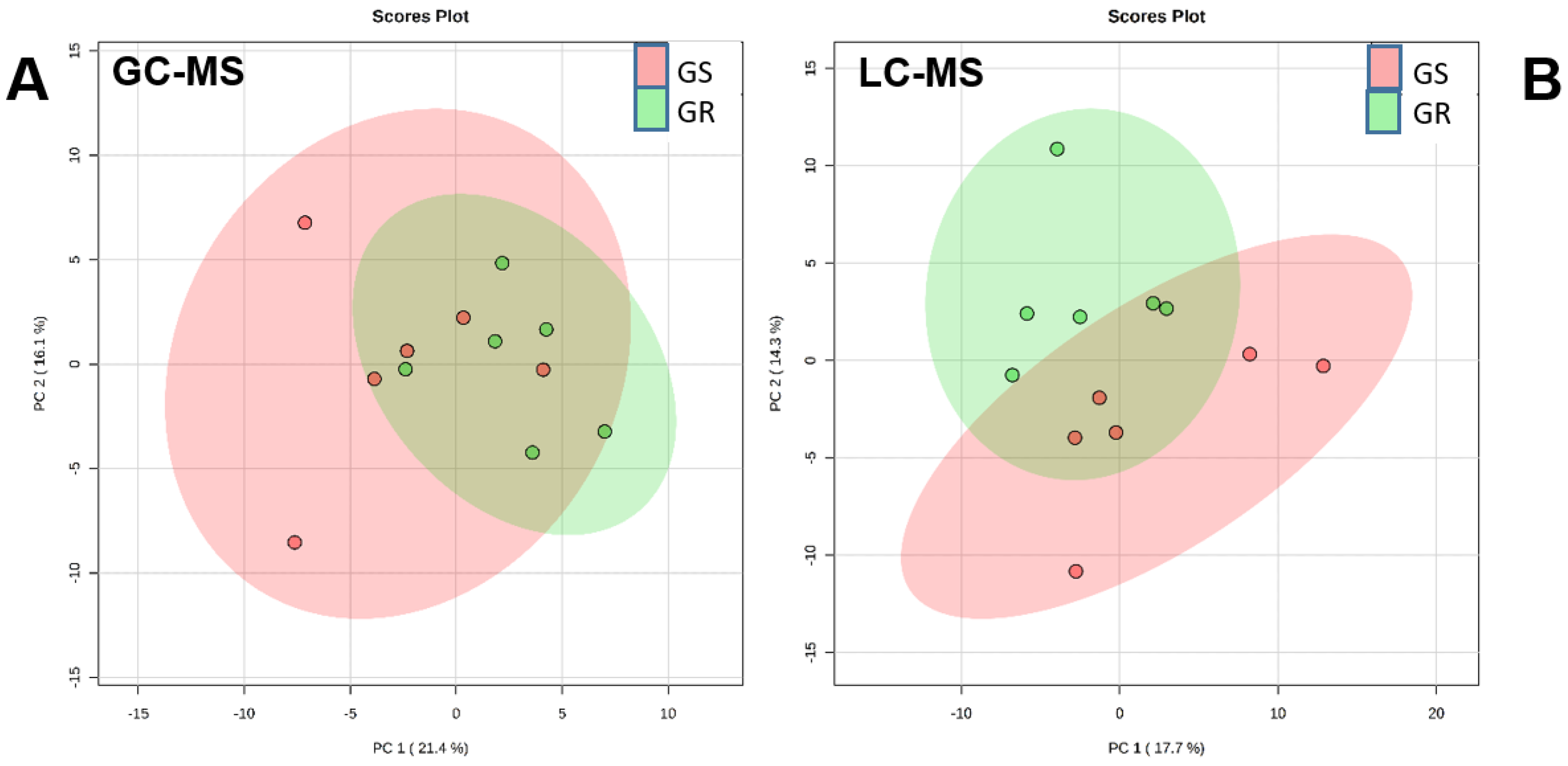
2.1. Metabolic Comparison between Nontreated Plants of Sensitive and Resistant Populations
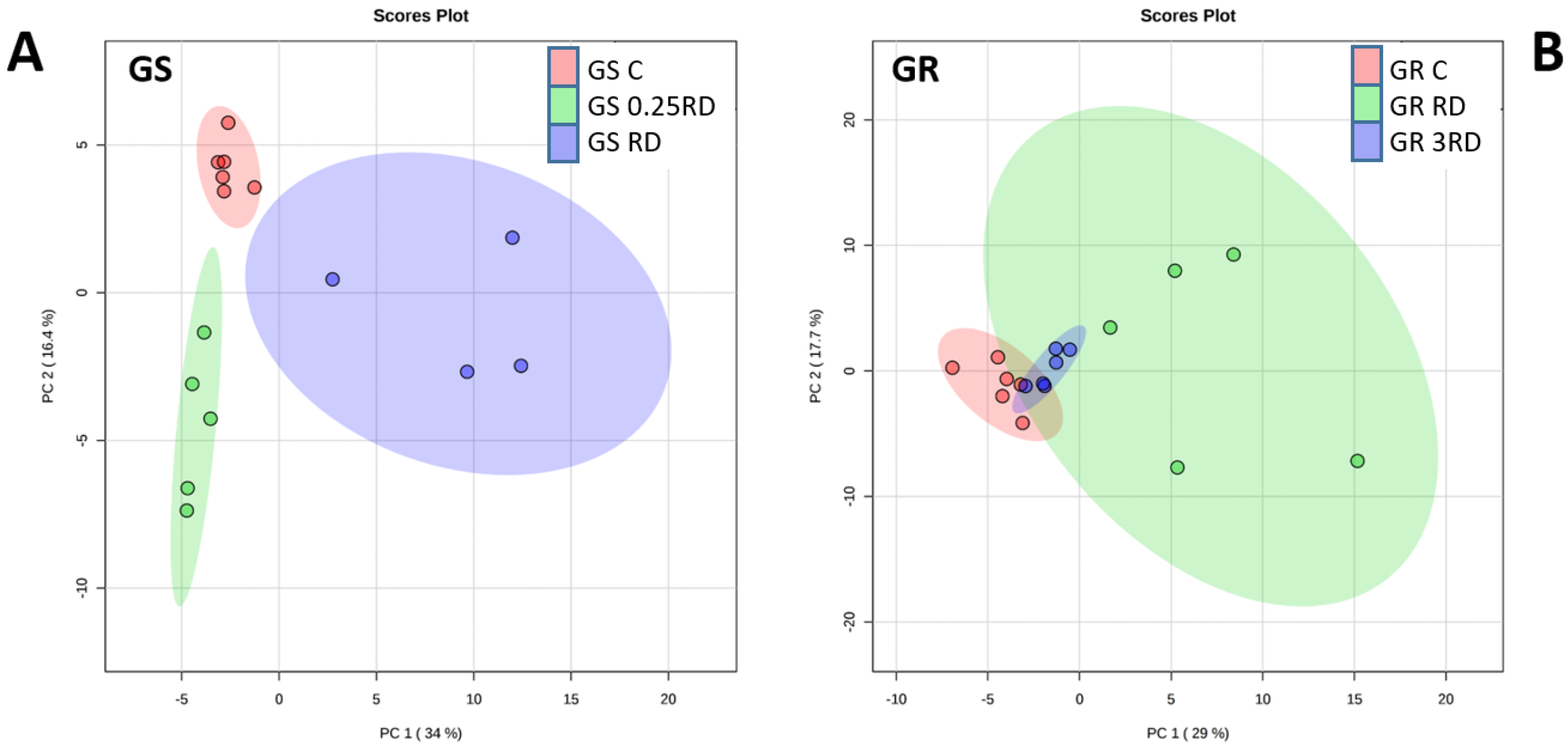
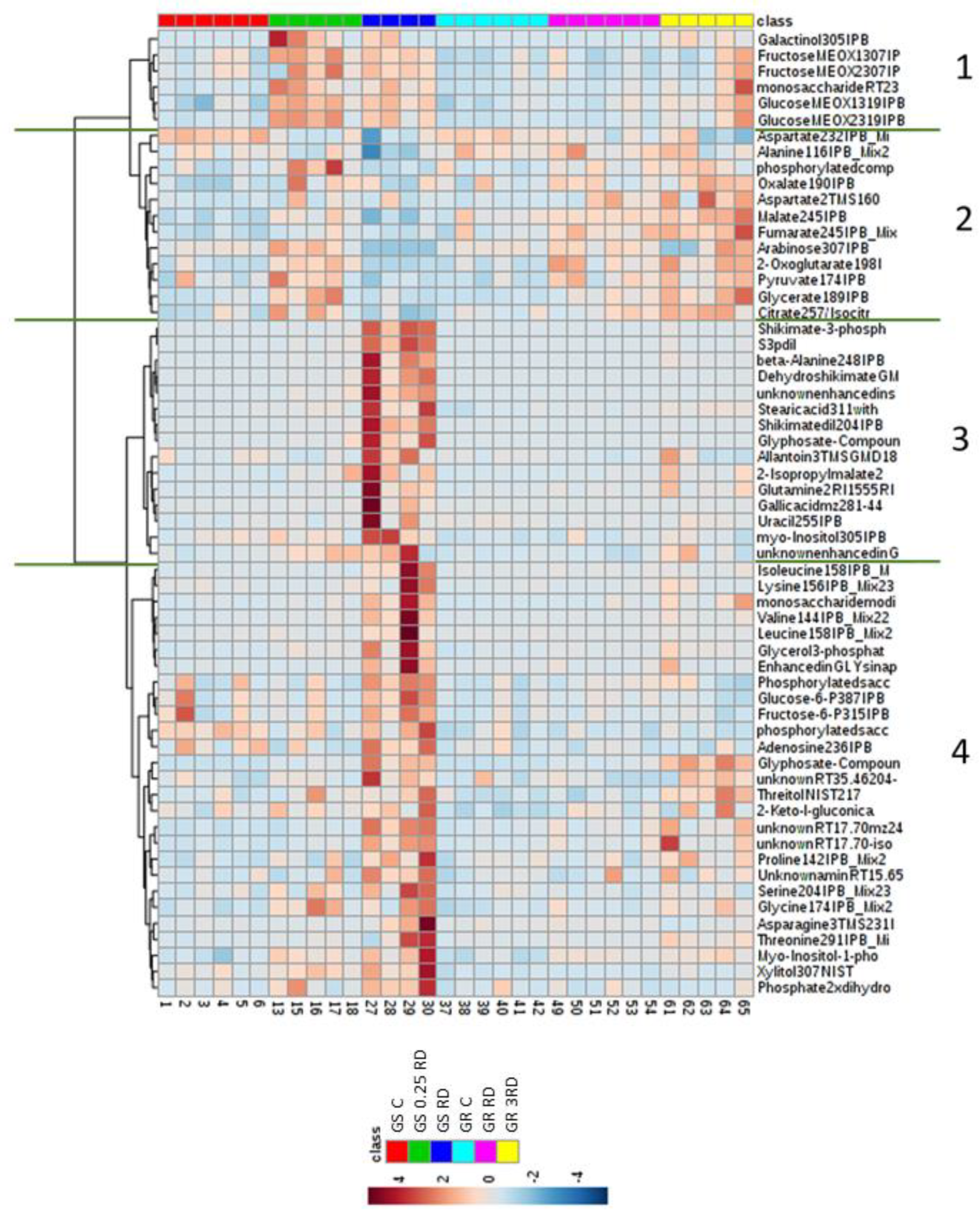
2.2. Metabolic Profiling of Glyphosate-Treated Plants by GC–MS
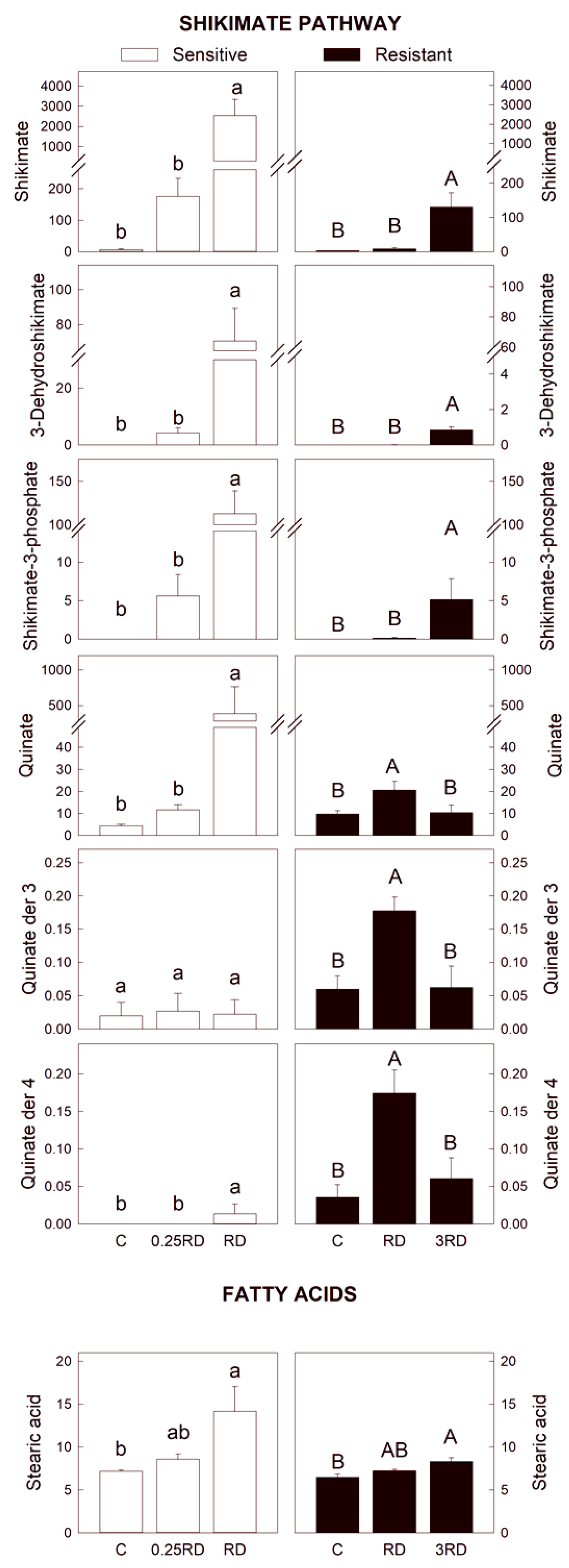
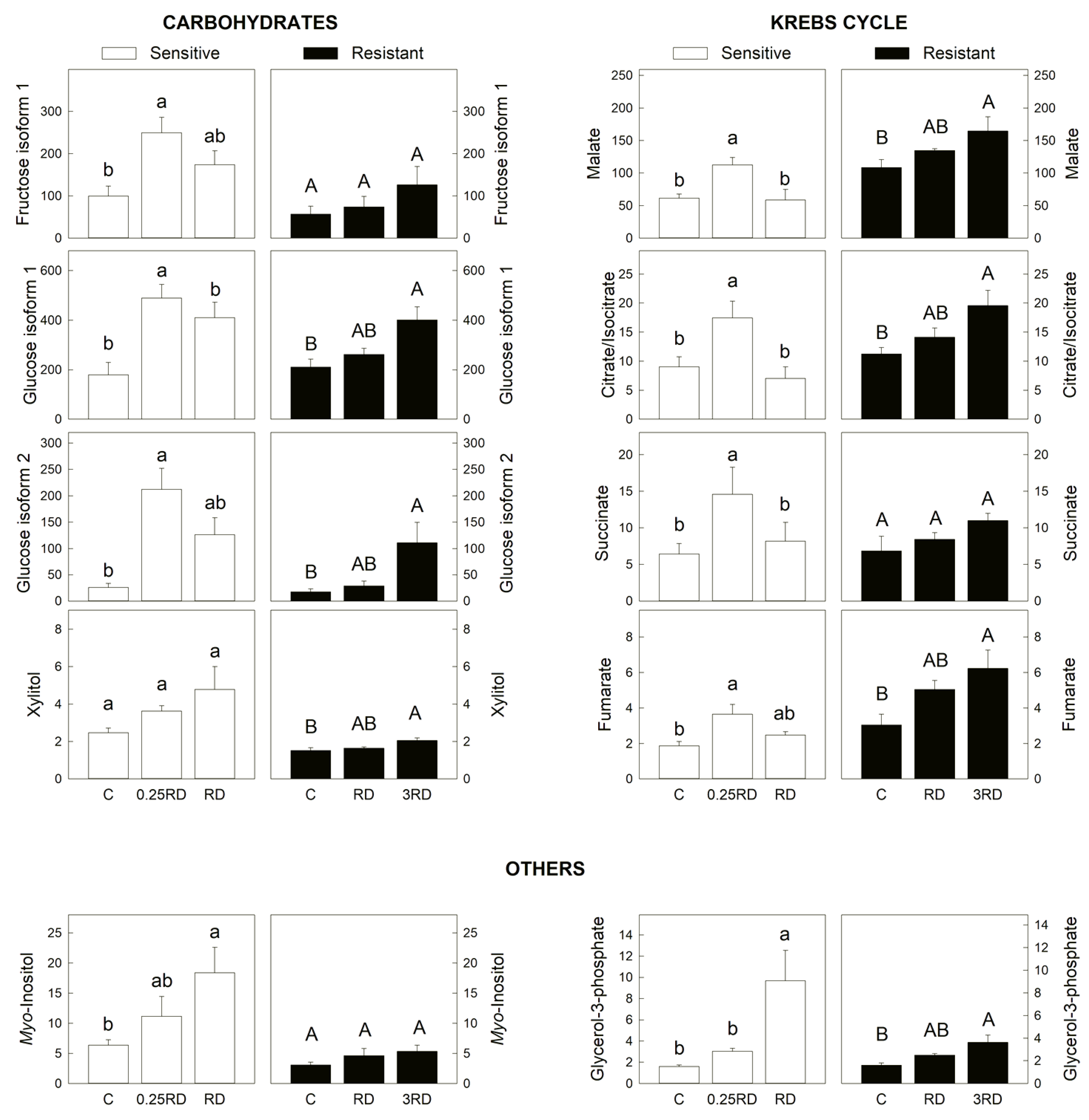
2.3. Selected Primary Metabolites Detected by GC–MS
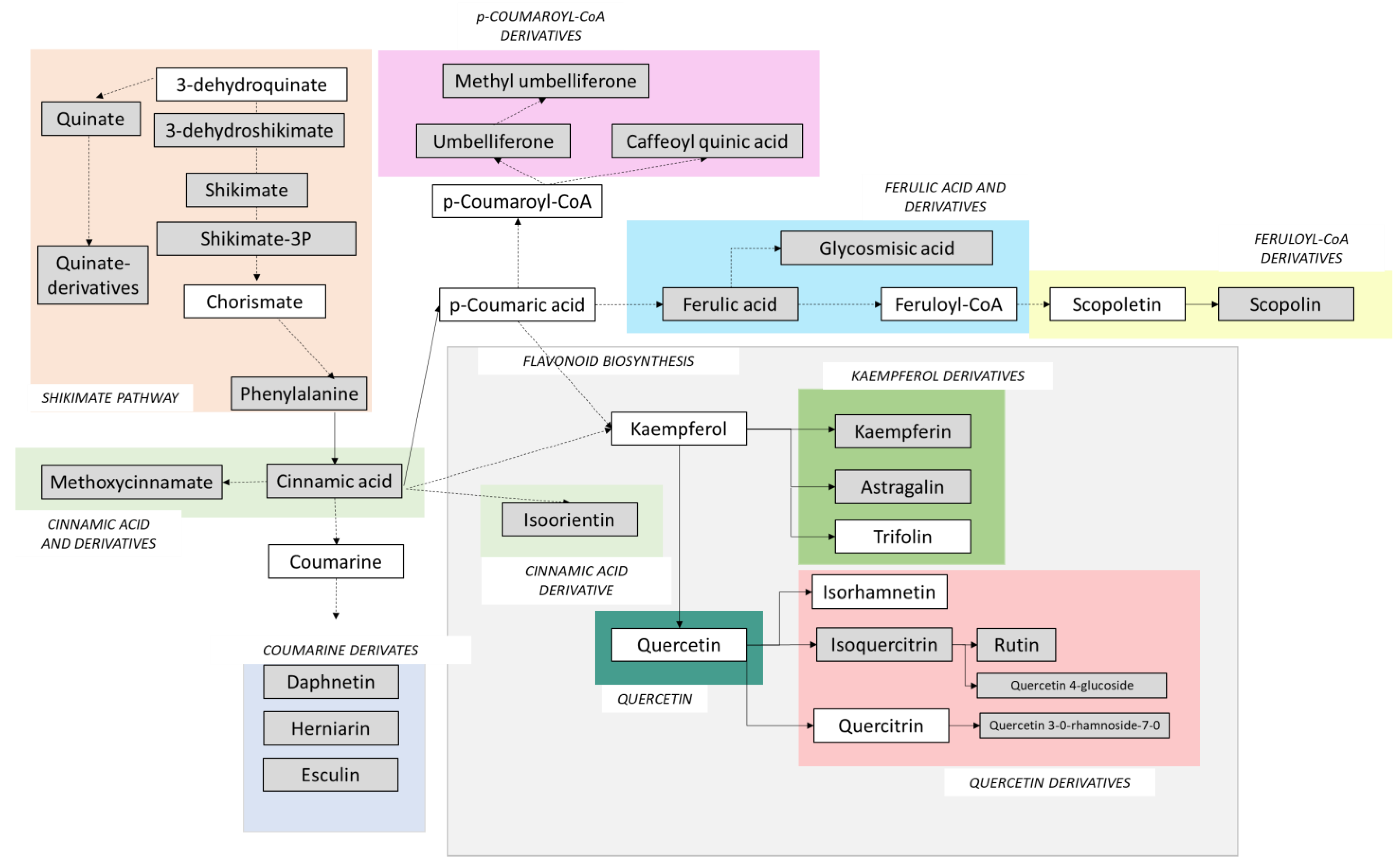
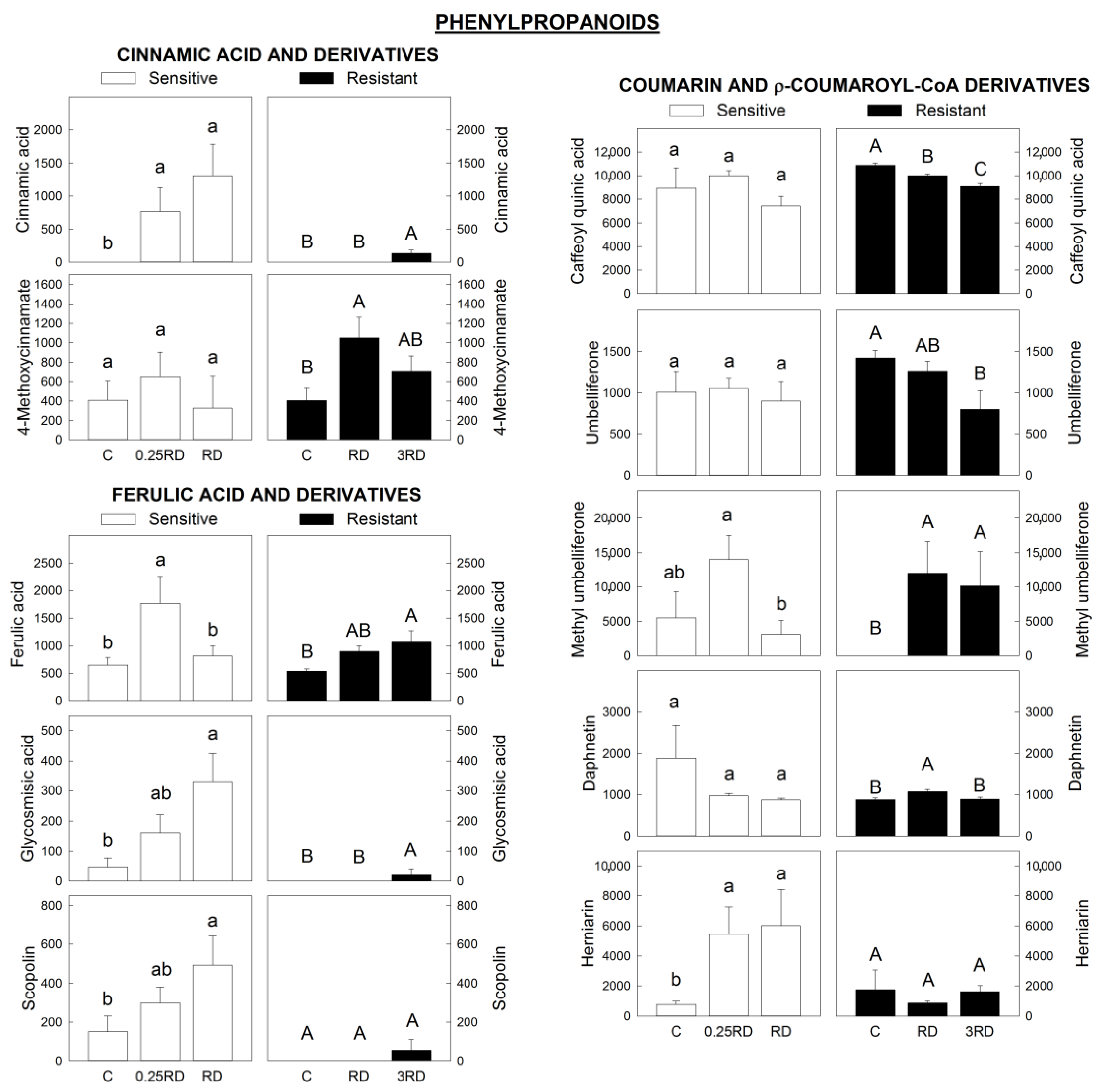
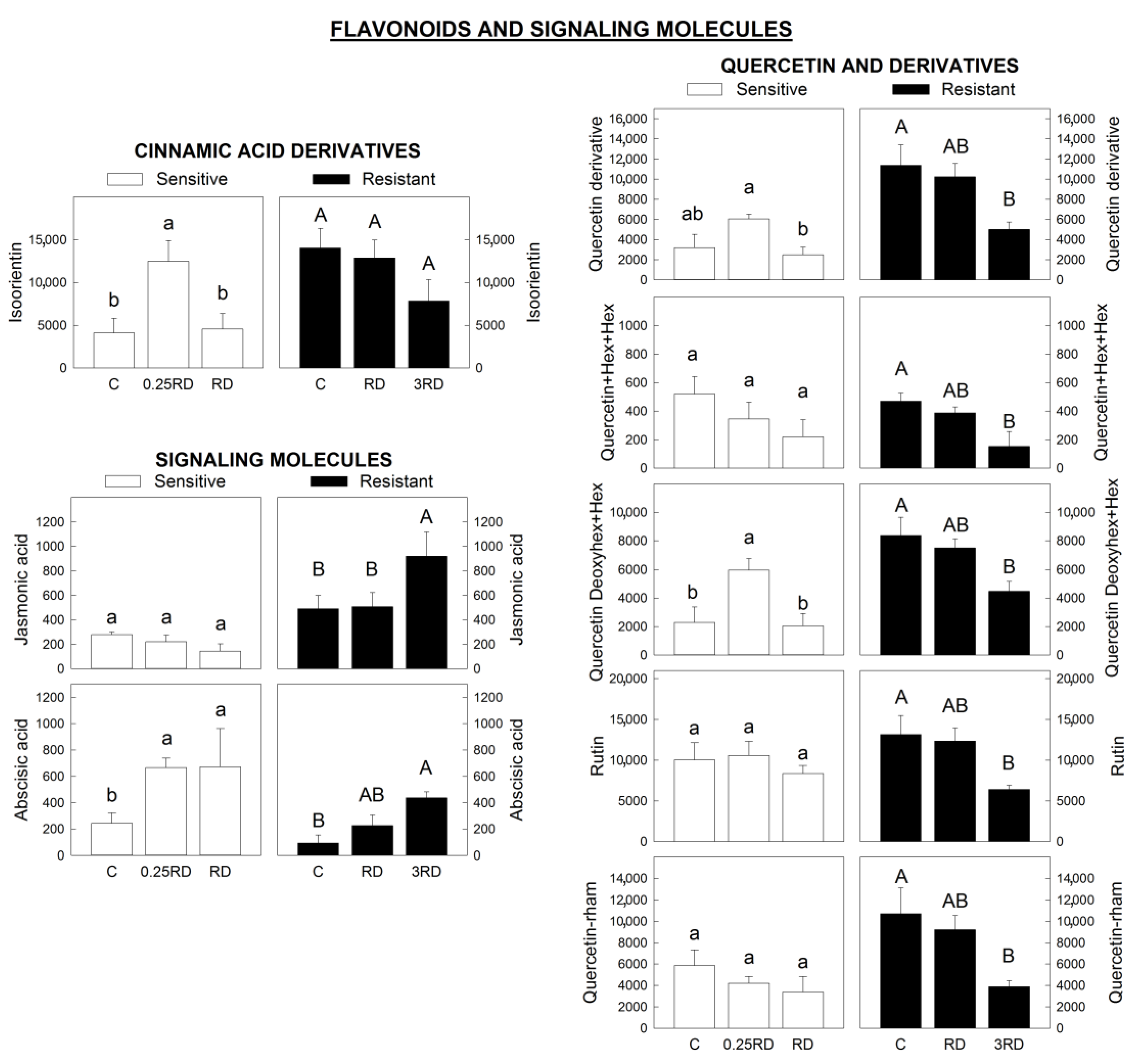
2.4. Phenylpropanoids, Flavonoids and Signalling Molecules Detected by LC–MS
3. Materials and Methods
3.1. General Experiment Procedures
3.2. Plant Material and Glyphosate Treatments
3.3. Metabolite Extraction for GS–MS and LC–MS
3.4. Non-Targeted Metabolic Profiling by GC–MS
3.5. LC–MS
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Steinrücken, H.C.; Amrhein, N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvylshikimic acid-3-phosphate synthase. Biochem. Biophys. Res. Commun. 1980, 94, 1207–1212. [Google Scholar] [CrossRef]
- Siehl, D.L. Inhibitors of EPSP Sinthase, Glutamine synthetase and histidine synthesis. In Hebicide Activity: Toxicology, Biochemistry and Molecular Biology; Roe, R.M., Burton, J.D., Kuhr, R.J., Eds.; IOS Press: Amsterdam, The Netherlands, 1997; pp. 37–67. [Google Scholar]
- Boudet, A.M. Evolution and current status of research in phenolic compounds. Phytochemistry 2007, 68, 2722–2735. [Google Scholar] [CrossRef]
- Tohge, T.; Watanabe, M.; Hoefgen, R.; Fernie, A.R. Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 2013, 4, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Duke, S.O.; Powles, S.B.; Sammons, R.D. Glyphosate—How it became a once in a hundred year herbicide and its future. Outlooks Pest Manag. 2018, 29, 247–251. [Google Scholar] [CrossRef]
- Westwood, J.H.; Charudattan, R.; Duke, S.O.; Fennimore, S.A.; Marrone, P.; Slaughter, D.C.; Swanton, C.; Zollinger, R. Weed management in 2050: Perspectives on the future of weed science. Weed Sci. 2018, 66, 275–285. [Google Scholar] [CrossRef] [Green Version]
- Gaines, T.A.; Patterson, E.L.; Neve, P. Molecular Mechanisms of Adaptive Evolution Revealed by Global Selection for Glyphosate Resistance. New Phytol. 2019, 223, 1770–1775. [Google Scholar] [CrossRef] [Green Version]
- Nakabayashi, R.; Saito, K. Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 2015, 24, 10–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres-García, J.R.; Tafoya-Razo, J.A.; Velázquez-Márquez, S.; Tiessen, A. Double herbicide-resistant biotypes of wild oat Avena fatua) display characteristic metabolic fingerprints before and after applying ACCase- and ALS-Inhibitors. Acta Physiol. Plant. 2018, 40, 119. [Google Scholar] [CrossRef]
- Trenkamp, S.; Eckes, P.; Busch, M.; Fernie, A.R. Temporally Resolved GC-MS-Based Metabolic profiling of herbicide treated plants treated reveals that changes in polar primary metabolites alone can distinguish herbicides of differing mode of action. Metabolomics 2009, 5, 277–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serra, A.A.; Couée, I.; Renault, D.; Gouesbet, G.; Sulmon, C. Metabolic profiling of Lolium perenne shows functional integration of metabolic responses to diverse subtoxic conditions of chemical stress. J. Exp. Bot. 2015, 66, 1801–1816. [Google Scholar] [CrossRef] [Green Version]
- Neuweger, H.; Albaum, S.P.; Dondrup, M.; Persicke, M.; Watt, T.; Niehaus, K.; Stoye, J.; Goesmann, A. MeltDB: A Software Platform for the analysis and integration of metabolomics experiment data. Bioinformatics 2008, 24, 2726–2732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maroli, A.S.; Nandula, V.K.; Duke, S.O.; Gerard, P.; Tharayil, N. Comparative metabolomic analyses of Ipomoea lacunosa biotypes with contrasting glyphosate tolerance captures herbicide-induced differential perturbations in cellular physiology. J. Agric. Food Chem. 2018, 66, 2027–2039. [Google Scholar] [CrossRef]
- Maroli, A.S.; Nandula, V.K.; Dayan, F.E.; Duke, S.O.; Gerard, P.; Tharayil, N. Metabolic profiling and enzyme analyses indicate a potential role of antioxidant systems in complementing glyphosate resistance in an Amaranthus palmeri biotype. J. Agric. Food Chem. 2015, 63, 9199–9209. [Google Scholar] [CrossRef] [PubMed]
- Maroli, A.S.; Nandula, V.K.; Duke, S.O.; Tharayil, N. stable isotope resolved metabolomics reveals the role of anabolic and catabolic processes in glyphosate-induced amino acid accumulation in Amaranthus palmeri biotypes. J. Agric. Food Chem. 2016, 64, 7040–7048. [Google Scholar] [CrossRef] [Green Version]
- Harrigan, G.G.; Venkatesh, T.V.; Leibman, M.; Blankenship, J.; Perez, T.; Halls, S.; Chassy, A.W.; Fiehn, O.; Xu, Y.; Goodacre, R. Evaluation of metabolomics profiles of grain from maize hybrids derived from near-isogenic GM positive and negative segregant inbreds demonstrates that observed differences cannot be attributed unequivocally to the GM trait. Metabolomics 2016, 12, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mesnage, R.; Agapito-Tenfen, S.Z.; Vilperte, V.; Renney, G.; Ward, M.; Séralini, G.-E.; Nodari, R.O.; Antoniou, M.N. An integrated multi-omics analysis of the NK603 Roundup-tolerant GM maize reveals metabolism disturbances caused by the transformation process. Sci. Rep. 2016, 6, 37855. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Escalada, M.; Zulet-González, A.; Gil-Monreal, M.; Zabalza, A.; Ravet, K.; Gaines, T.; Royuela, M. Effects of EPSPS Copy Number Variation (CNV) and glyphosate application on the aromatic and branched chain amino acid synthesis pathways in Amaranthus palmeri. Front. Plant Sci. 2017, 8, 1970. [Google Scholar] [CrossRef] [Green Version]
- Giacomini, D.; Westra, P.; Ward, S.M. Impact of Genetic Background in fitness cost studies: An example from glyphosate-resistant Palmer amaranth. Weed Sci. 2014, 62, 29–37. [Google Scholar] [CrossRef]
- Vila-Aiub, M.M.; Goh, S.S.; Gaines, T.A.; Han, H.; Busi, R.; Yu, Q.; Powles, S.B. No Fitness cost of glyphosate resistance endowed by massive epsps gene amplification in Amaranthus palmeri. Planta 2014, 239, 793–801. [Google Scholar] [CrossRef]
- Böttcher, C.; Centeno, D.; Freitag, J.; Höfgen, R.; Köhl, K.; Kopka, J.; Kroymann, J.; Matros, A.; Mock, H.P.; Neumann, S.; et al. Teaching (and Learning from) Metabolomics: The 2006 PlantMetaNet ETNA Metabolomics Research School. Physiol. Plant. 2008, 132, 136–149. [Google Scholar] [CrossRef]
- Böttcher, C.; Von Roepenack-Lahaye, E.; Schmidt, J.; Schmotz, C.; Neumann, S.; Scheel, D.; Clemens, S. Metabolome analysis of biosynthetic mutants reveals a diversity of metabolic changes and allows identification of a large number of new compounds in Arabidopsis. Plant Physiol. 2008, 147, 2107–2120. [Google Scholar] [CrossRef] [Green Version]
- Gomes, M.P.; Smedbol, E.; Chalifour, A.; Hénault-ethier, L.; Labrecque, M.; Lepage, L.; Lucotte, M.; Juneau, P. Alteration of plant physiology by glyphosate and its by-product aminomethylphosphonic acid: An Overview. J. Exp. Bot. 2014, 65, 4691–4703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zulet, A.; Zabalza, A.; Royuela, M. phytotoxic and metabolic effects of exogenous quinate on Pisum sativum L. J. Plant Growth Regul. 2013, 32, 779–788. [Google Scholar] [CrossRef] [Green Version]
- Zulet, A.; Gil-Monreal, M.; Zabalza, A.; van Dongen, J.T.; Royuela, M. Fermentation and alternative oxidase contribute to the action of amino acid biosynthesis-inhibiting herbicides. J. Plant Physiol. 2015, 175, 102–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Escalada, M.; Gil-Monreal, M.; Zabalza, A.; Royuela, M. Characterization of the Amaranthus palmeri physiological response to glyphosate in susceptible and resistant populations. J. Agric. Food Chem. 2016, 64, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Zabalza, A.; Orcaray, L.; Fernández-Escalada, M.; Zulet-González, A.; Royuela, M. the pattern of shikimate pathway and phenylpropanoids after inhibition by glyphosate or quinate feeding in pea roots. Pestic. Biochem. Physiol. 2017, 141, 96–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zulet, A.; Gil-Monreal, M.; Villamor, J.G.; Zabalza, A.; van der Hoorn, R.A.L.; Royuela, M. Proteolytic pathways induced by herbicides that inhibit amino acid biosynthesis. PLoS ONE 2013, 8, e73847. [Google Scholar] [CrossRef] [Green Version]
- Zabalza, A.; Zulet, A.; Gil-Monreal, M.; Igal, M.; Royuela, M. Branched-chain amino acid biosynthesis inhibitors: Herbicide efficacy is associated with an induced carbon-nitrogen imbalance. J. Plant Physiol. 2013, 170, 814–821. [Google Scholar] [CrossRef] [Green Version]
- Lydon, J.; Duke, S.O. Glyphosate induction of elevated levels of hydroxybenzoic acids in higher-plants. J. Agric. Food Chem. 1988, 36, 813–818. [Google Scholar] [CrossRef]
- Becerril, J.M.; Duke, S.O.; Lydon, J. Glyphosate effects on shikimate pathway products in leaves and flowers of velvetleaf. Phytochemistry 1989, 28, 695–699. [Google Scholar] [CrossRef]
- Hernandez, A.; Garcia-Plazaola, J.I.; Becerril, J.M. Glyphosate effects on phenolic metabolism of nodulated soybean (Glycine max L. Merr.). J. Agric. Food Chem. 1999, 47, 2920–2925. [Google Scholar] [CrossRef] [PubMed]
- de María, N.; Becerril, J.M.; García-Plazaola, J.I.; Hernandez, A.; De Felipe, M.R.; Fernandez-Pascual, M. New insights on glyphosate mode of action in nodular metabolism: Role of shikimate accumulation. J. Agric. Food Chem. 2006, 54, 2621–2628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orcaray, L.; Igal, M.; Marino, D.; Zabalza, A.; Royuela, M. The possible role of quinate in the mode of action of glyphosate and acetolactate synthase inhibitors. Pest Manag. Sci. 2010, 66, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Zulet-González, A.; Barco-Antoñanzas, M.; Gil-Monreal, M.; Royuela, M.; Zabalza, A. Increased glyphosate-induced gene expression in the shikimate pathway is abolished in the presence of aromatic amino acids and mimicked by shikimate. Front. Plant Sci. 2020, 11, 459. [Google Scholar] [CrossRef]
- Liu, X.; Ma, D.; Zhang, Z.; Wang, S.; Du, S.; Deng, X.; Yin, L. Plant lipid remodeling in response to abiotic stresses. Environ. Exp. Bot. 2019, 165, 174–184. [Google Scholar] [CrossRef]
- Orcaray, L.; Zulet, A.; Zabalza, A.; Royuela, M. Impairment of carbon metabolism induced by the herbicide glyphosate. J. Plant Physiol. 2012, 169, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Armendáriz, O.; Gil-Monreal, M.; Zulet, A.; Zabalza, A.; Royuela, M. Both foliar and residual applications of herbicides that inhibit amino acid biosynthesis induce alternative respiration and aerobic fermentation in pea roots. Plant Biol. 2016, 18, 382–390. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Escalada, M.; Zulet-González, A.; Gil-Monreal, M.; Royuela, M.; Zabalza, A. Physiological performance of glyphosate and imazamox mixtures on Amaranthus palmeri sensitive and resistant to glyphosate. Sci. Rep. 2019, 9, 18225. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Zhou, K.; Li, Y.; Chen, X.; Liu, B.; Li, C.; Gong, X.; Ma, F. Exogenous myo-inositol alleviates salinity-induced stress in Malus hupehensis Rehd. Plant Physiol. Biochem. 2018, 133, 116–126. [Google Scholar] [CrossRef]
- Deng, Y.; Lu, S. Biosynthesis and regulation of phenylpropanoids in plants. CRC. Crit. Rev. Plant Sci. 2017, 36, 257–290. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [Green Version]
- Hoagland, R.E. Effects of glyphosate on metabolism of phenolic compounds: VI. effects of glyphosine and glyphosate metabolites on phenylalanine ammonia-lyase activity, growth, and protein, chlorophyll, and anthocyanin levels in soybean (Glycine max) seedlings. Weed Sci. 1980, 28, 393–400. [Google Scholar] [CrossRef]
- Marchiosi, R.; Lucio Ferrarese, M.D.L.; Bonini, E.A.; Fernandes, N.G.; Ferro, A.P.; Ferrarese-Filho, O. Glyphosate-induced metabolic changes in susceptible and glyphosate-resistant soybean (Glycine max L.) Roots. Pestic. Biochem. Physiol. 2009, 93, 28–33. [Google Scholar] [CrossRef]
- Sharon, A.; Amsellem, Z.; Gressel, J. Glyphosate suppression of an elicited defense response: Increased susceptibility of Cassia obtusifolia to a mycoherbicide. Plant Physiol. 1992, 98, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mobin, M.; Wu, C.-H.; Tewari, R.K.; Paek, K.-Y. Studies on the glyphosate-induced amino acid starvation and addition of precursors on caffeic acid accumulation and profiles in adventitious roots of Echinacea purpurea (L.) Moench. Plant Cell Tissue Organ Cult. 2015, 120, 291–301. [Google Scholar] [CrossRef]
- Hura, T.; Hura, K.; Grzesiak, S. Leaf dehydration induces different content of phenolics and ferulic acid in drought-resistant and -sensitive genotypes of spring triticale. Z. Naturforsch.—Sect. C J. Biosci. 2009, 64, 85–95. [Google Scholar] [CrossRef]
- Ghimire, B.K.; Son, N.Y.; Kim, S.H.; Yu, C.Y.; Chung, I.M. Evaluating water deficit and glyphosate treatment on the accumulation of phenolic compounds and photosynthesis rate in transgenic Codonopsis lanceolata (Siebold & Zucc.) Trautv. over-expressing γ-tocopherol methyltransferase (γ-TMT) gene. 3 Biotech 2017, 7, 167. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, C.; Sebastiani, F.; Tattini, M. Review: ABA, flavonols, and the evolvability of land plants. Plant Sci. 2019, 280, 448–454. [Google Scholar] [CrossRef]
- Avanci, N.C.; Luche, D.D.; Goldman, G.H.; Goldman, M.H. Jasmonates are phytohormones with multiple functions, including plant defense and reproduction. Genet. Mol. Res. 2010, 9, 484–505. [Google Scholar] [CrossRef]
- Jiang, L.X.; Jin, L.G.; Guo, Y.; Tao, B.; Qiu, L.J. Glyphosate effects on the gene expression of the apical bud in soybean (Glycine max). Biochem. Biophys. Res. Commun. 2013, 437, 544–549. [Google Scholar] [CrossRef]
- Fuchs, B.; Saikkonen, K.; Helander, M. Glyphosate-modulated biosynthesis driving plant defense and species interactions. Trends Plant Sci. 2021, 26, 312–323. [Google Scholar] [CrossRef]
- Chandi, A.; Milla-Lewis, S.R.; Giacomini, D.; Westra, P.; Preston, C.; Jordan, D.L.; York, A.C.; Burton, J.D.; Whitaker, J.R. Inheritance of evolved glyphosate resistance in a north carolina Palmer amaranth (Amaranthus palmeri) biotype. Int. J. Agron. 2012, 2012, 176108. [Google Scholar] [CrossRef] [Green Version]
- Zulet-González, A.; Fernández-Escalada, M.; Zabalza, A.; Royuela, M. Enhancement of glyphosate efficacy on Amaranthus palmeri by exogenous quinate application. Pestic. Biochem. Physiol. 2019, 158, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Culpepper, A.S.; Grey, T.L.; Vencill, W.K.; Kichler, J.M.; Webster, T.M.; Brown, S.M.; York, A.C.; Davis, J.W.; Hanna, W.W. Glyphosate-resistant Palmer amaranth (Amaranthus palmeri) confirmed in Georgia. Weed Sci. 2006, 54, 620–626. [Google Scholar] [CrossRef]
- Gorzolka, K.; Lissel, M.; Kessler, N.; Loch-Ahring, S.; Niehaus, K. Metabolite fingerprinting of barley whole seeds, endosperms, and embryos during industrial malting. J. Biotechnol. 2012, 159, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Matern, A.; Böttcher, C.; Eschen-Lippold, L.; Westermann, B.; Smolka, U.; Döll, S.; Trempel, F.; Aryal, B.; Scheel, D.; Geisler, M.; Rosahl, S. A substrate of the ABC transporter PEN3 stimulates bacterial flagellin (flg22)-induced callose deposition in Arabidopsis thaliana. J. Biol. Chem. 2019, 294, 6857–6870. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zulet-Gonzalez, A.; Gorzolka, K.; Döll, S.; Gil-Monreal, M.; Royuela, M.; Zabalza, A. Unravelling the Phytotoxic Effects of Glyphosate on Sensitive and Resistant Amaranthus palmeri Populations by GC–MS and LC–MS Metabolic Profiling. Plants 2023, 12, 1345. https://doi.org/10.3390/plants12061345
Zulet-Gonzalez A, Gorzolka K, Döll S, Gil-Monreal M, Royuela M, Zabalza A. Unravelling the Phytotoxic Effects of Glyphosate on Sensitive and Resistant Amaranthus palmeri Populations by GC–MS and LC–MS Metabolic Profiling. Plants. 2023; 12(6):1345. https://doi.org/10.3390/plants12061345
Chicago/Turabian StyleZulet-Gonzalez, Ainhoa, Karin Gorzolka, Stefanie Döll, Miriam Gil-Monreal, Mercedes Royuela, and Ana Zabalza. 2023. "Unravelling the Phytotoxic Effects of Glyphosate on Sensitive and Resistant Amaranthus palmeri Populations by GC–MS and LC–MS Metabolic Profiling" Plants 12, no. 6: 1345. https://doi.org/10.3390/plants12061345
APA StyleZulet-Gonzalez, A., Gorzolka, K., Döll, S., Gil-Monreal, M., Royuela, M., & Zabalza, A. (2023). Unravelling the Phytotoxic Effects of Glyphosate on Sensitive and Resistant Amaranthus palmeri Populations by GC–MS and LC–MS Metabolic Profiling. Plants, 12(6), 1345. https://doi.org/10.3390/plants12061345





