Citrus Bright Spot Virus: A New Dichorhavirus, Transmitted by Brevipalpus azores, Causing Citrus Leprosis Disease in Brazil
Abstract
:1. Introduction
2. Results
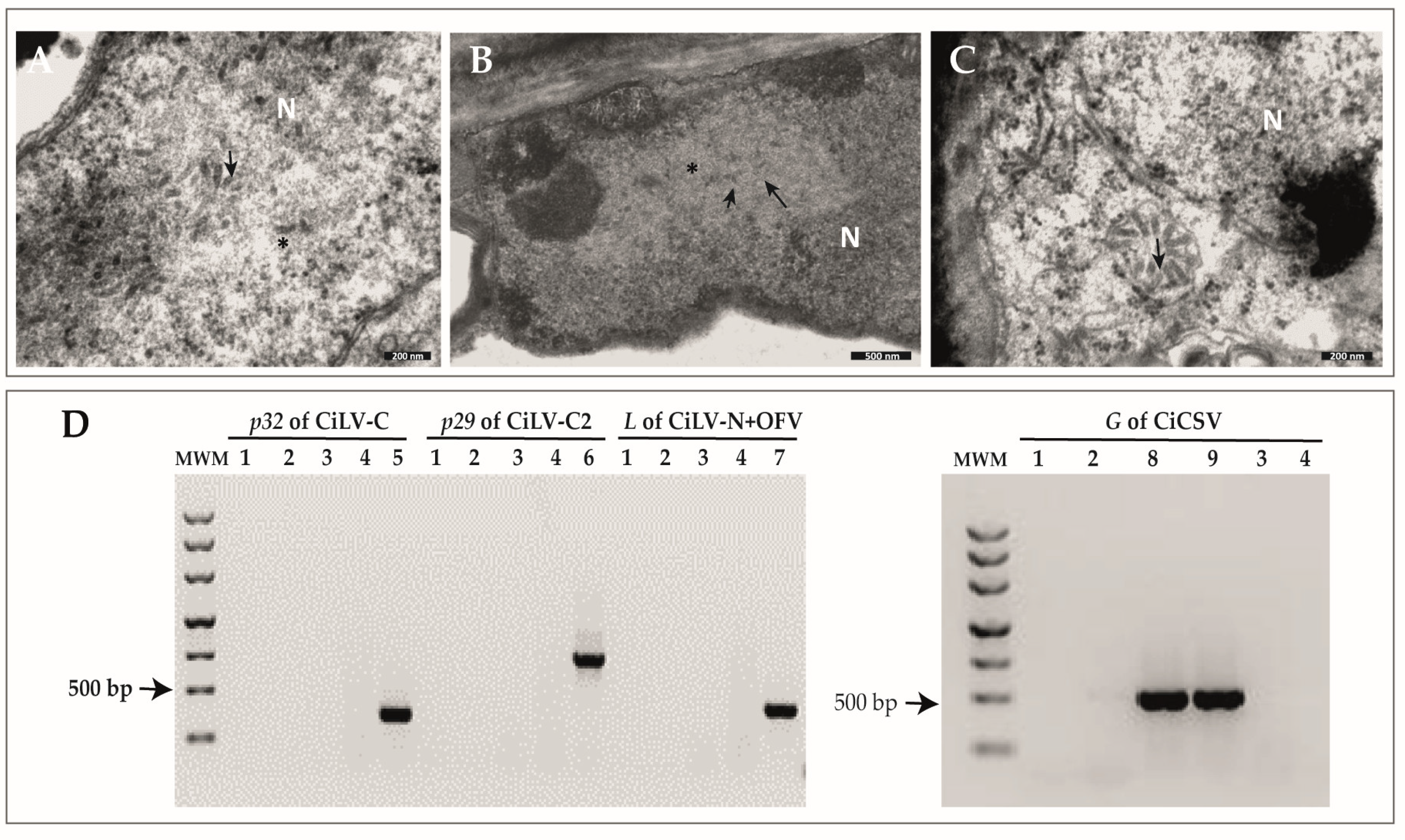
2.1. Identification of Rod-like Particles Associated with Citrus Leprosis Symptoms
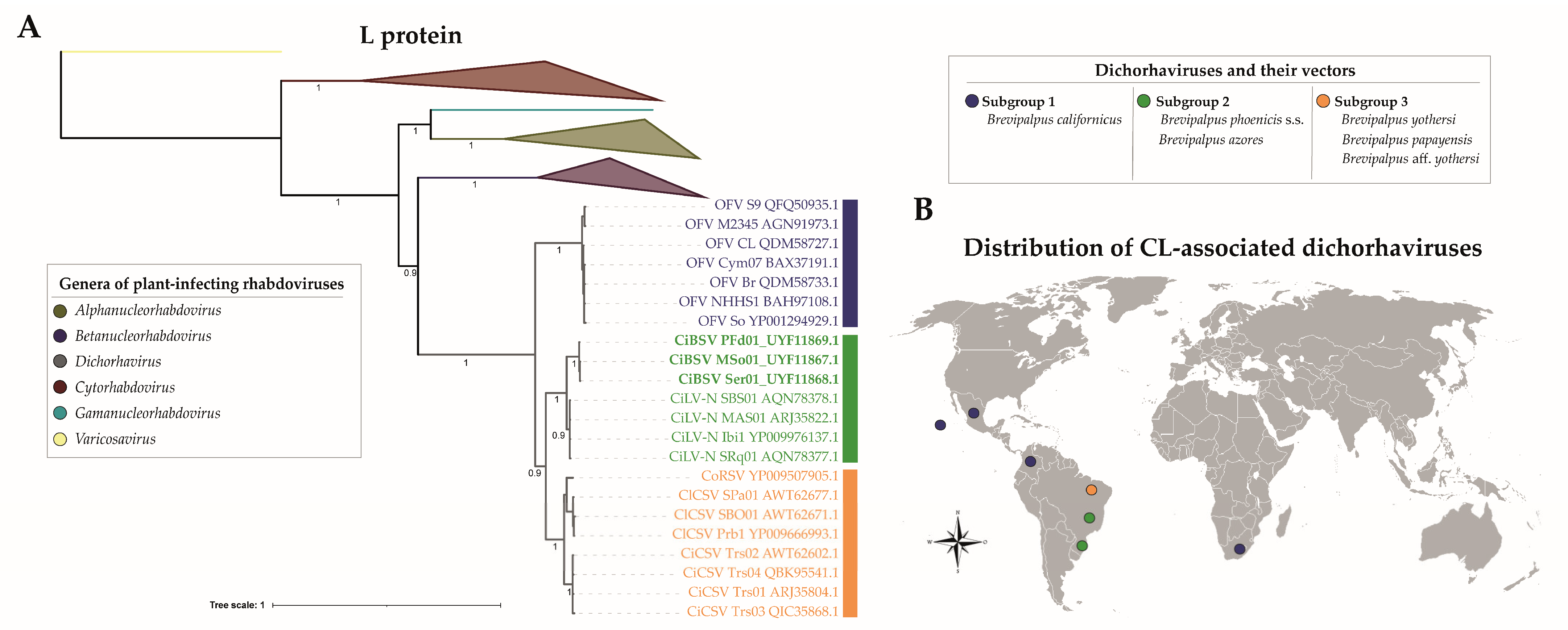
2.2. Characterization of Three Genome Sequences Confirms the Infection by Putative Novel Plant Rhabdoviruses
2.3. Brevipalpus Azores Mites Are Able to Transmit CiBSV Isolates
2.4. Primers for the Detection of CiBSV Are Specific
3. Discussion
4. Materials and Methods
4.1. Symptomatic Citrus Trees Samples
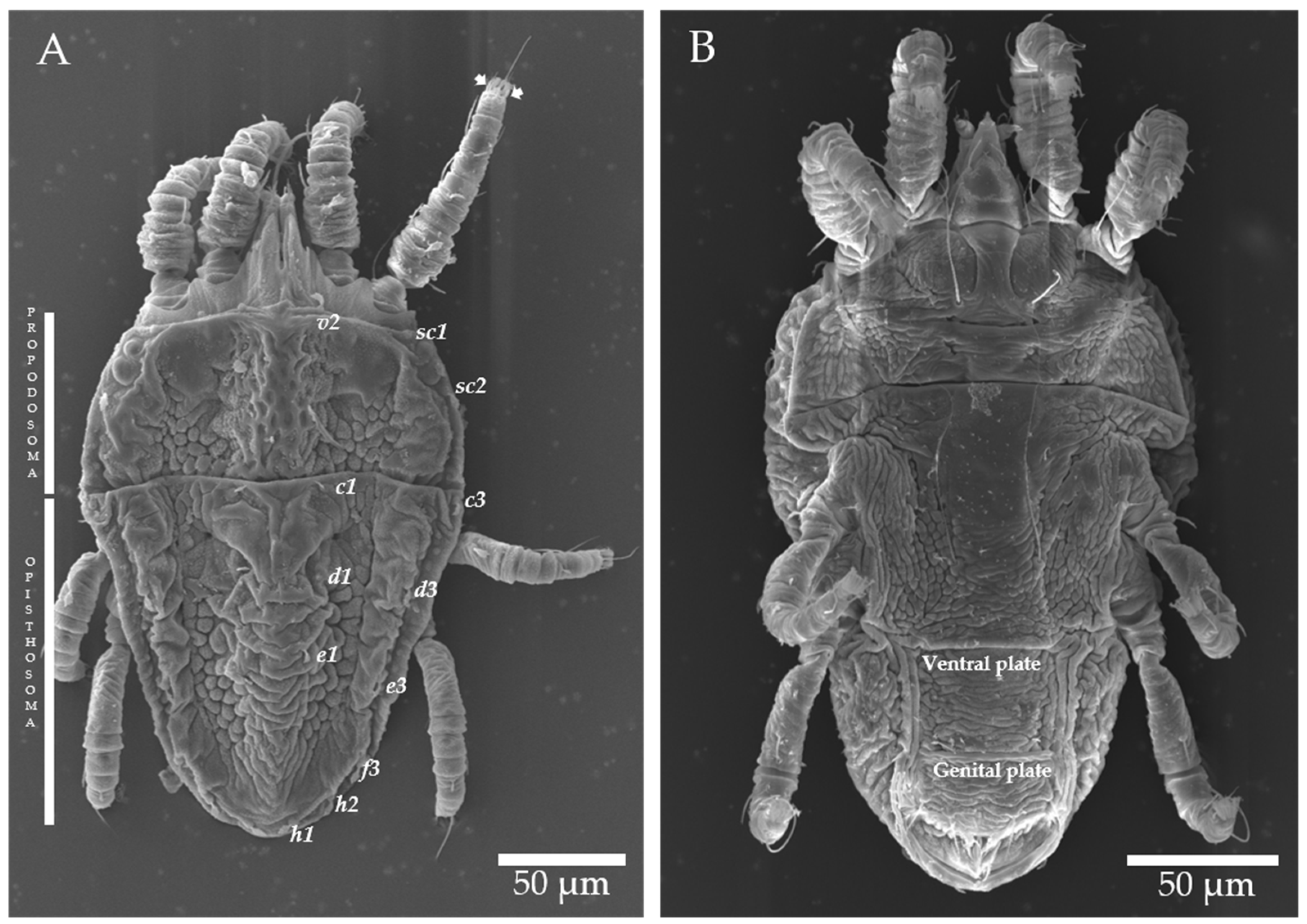
4.2. Morphological and Genetic Identification of Brevipalpus Mites
4.3. Virus Transmission Mediated by Brevipalpus Mites
4.4. TEM and Molecular Analyses
4.5. High Throughput Sequencing, Validation, and Annotation of Viral Genomes
4.6. Detection of Recombination and Reassortment Events
4.7. Phylogenetic Analysis Based on N and L Proteins of Plant Rhabdoviruses
4.8. Specific Detection of New Dichochaviruses by RT-PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dietzgen, R.G.; Kuhn, J.H.; Clawson, A.N.; Freitas-Astúa, J.; Goodin, M.M.; Kitajima, E.W.; Kondo, H.; Wetzel, T.; Whitfield, A.E. Dichorhavirus: A Proposed New Genus for Brevipalpus Mite-Transmitted, Nuclear, Bacilliform, Bipartite, Negative-Strand RNA Plant Viruses. Arch. Virol. 2014, 159, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Freitas-Astua, J.; Bejerman, N.; Blasdell, K.R.; Breyta, R.; Dietzgen, R.G.; Fooks, A.R.; Kondo, H.; Kurath, G.; Kuzmin, I.V.; et al. ICTV Virus Taxonomy Profile: Rhabdoviridae 2022. J. Gen. Virol. 2022, 103, 001689. [Google Scholar] [CrossRef] [PubMed]
- Dietzgen, R.G.; Freitas-Astúa, J.; Chabi-Jesus, C.; Ramos-González, P.L.; Goodin, M.M.; Kondo, H.; Tassi, A.D.; Kitajima, E.W. Dichorhaviruses in Their Host Plants and Mite Vectors. In Advances in Virus Research; Academic Press Inc.: Cambridge, MA, USA, 2018; Volume 102, pp. 119–148. ISBN 9780128151945. [Google Scholar]
- Kuhn, J.H.; Adkins, S.; Alkhovsky, S.V.; Avšič-Županc, T.; Ayllón, M.A.; Bahl, J.; Balkema-Buschmann, A.; Ballinger, M.J.; Bandte, M.; Beer, M.; et al. 2022 Taxonomic Update of Phylum Negarnaviricota (Riboviria: Orthornavirae), Including the Large Orders Bunyavirales and Mononegavirales. Arch. Virol. 2022, 167, 2857–2906. [Google Scholar] [CrossRef]
- Kitajima, E.W.; Rodrigues, J.C.V.; Freitas-Astua, J. An Annotated List of Ornamentals Naturally Found Infected by Brevipalpus Mite-Transmitted Viruses. Sci. Agric. 2010, 67, 348–371. [Google Scholar] [CrossRef] [Green Version]
- Kondo, H.; Maeda, T.; Tamada, T. Orchid Fleck Virus: Brevipalpus californicus Mite Transmission, Biological Properties and Genome Structure. Exp. Appl. Acarol. 2003, 30, 215–223. [Google Scholar] [CrossRef] [PubMed]
- de Lillo, E.; Freitas-Astúa, J.; Kitajima, E.W.; Ramos-González, P.L.; Simoni, S.; Tassi, A.D.; Valenzano, D. Phytophagous Mites Transmitting Plant Viruses: Update and Perspectives. Entomol. Gen. 2021, 41, 439–462. [Google Scholar] [CrossRef]
- Roy, A.; Hartung, J.S.; Schneider, W.L.; Shao, J.; Leon M, G.; Melzer, M.J.; Beard, J.J.; Otero-Colina, G.; Bauchan, G.R.; Ochoa, R.; et al. Role Bending: Complex Relationships between Viruses, Hosts, and Vectors Related to Citrus Leprosis, an Emerging Disease. Phytopathology 2015, 105, 872–884. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Leon, M.G.; Stone, A.L.; Schneider, W.L.; Hartung, J.; Brlansky, R.H. First Report of Citrus Leprosis Virus Nuclear Type in Sweet Orange in Colombia. Plant Dis. 2014, 98, 1162. [Google Scholar] [CrossRef]
- Cruz-Jaramillo, J.L.; Ruiz-Medrano, R.; Rojas-Morales, L.; López-Buenfil, J.A.; Morales-Galván, O.; Chavarín-Palacio, C.; Ramírez-Pool, J.A.; Xoconostle-Cázares, B. Characterization of a Proposed Dichorhavirus Associated with the Citrus Leprosis Disease and Analysis of the Host Response. Viruses 2014, 6, 2602–2622. [Google Scholar] [CrossRef] [Green Version]
- Cook, G.; Kirkman, W.; Clase, R.; Steyn, C.; Basson, E.; Fourie, P.H.; Moore, S.D.; Carstens, T.G.E.; Hattingh, V. Orchid Fleck Virus Associated with the First Case of Citrus Leprosis-N in South Africa. Plant Pathol. 2019, 155, 1373–1379. [Google Scholar] [CrossRef]
- Olmedo Velarde, A.; Roy, A.; Padmanabhan, C.; Nunziata, S.; Nakhla, M.K.K.M.K.; Melzer, M.J.; Olmedo-Velarde, A.; Roy, A.; Padmanabhan, C.; Nunziata, S.; et al. First Report of Orchid Fleck Virus Associated with Citrus Leprosis Symptoms in Rough Lemon (Citrus jambhiri) and Mandarin (C. reticulata) the United States. Plant Dis. Note 2021, 2. [Google Scholar] [CrossRef] [PubMed]
- Castro, E.B.; Mesa, N.C.; Feres, R.J.F.; de Moraes, G.J.; Ochoa, R.; Beard, J.J.; Demite, P.R. A Newly Available Database of an Important Family of Phytophagous Mites: Tenuipalpidae Database. Zootaxa 2020, 4868, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Chabi-Jesus, C.; Ramos-González, P.L.; Tassi, A.D.; Guerra-Peraza, O.; Kitajima, E.W.; Harakava, R.; Beserra, J.E.A.; Salaroli, R.B.; Freitas-Astúa, J. Identification and Characterization of Citrus Chlorotic Spot Virus, a New Dichorhavirus Associated with Citrus Leprosis-like Symptoms. Plant Dis. 2018, 102, 1588–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chabi-Jesus, C.; Ramos-González, P.L.; Tassi, A.D.; Barguil, B.M.; Beserra Junior, J.E.A.; Harakava, R.; Kitajima, E.W.; Freitas-Astúa, J. First Report of Citrus Chlorotic Spot Virus Infecting the Succulent Plant Agave desmettiana. Plant Dis. 2019, 103, 1438. [Google Scholar] [CrossRef]
- Ramos-González, P.L.; Chabi-Jesus, C.; Guerra-Peraza, O.; Tassi, A.D.; Kitajima, E.W.; Harakava, R.; Salaroli, R.B.; Freitas-Astúa, J. Citrus Leprosis Virus N: A New Dichorhavirus Causing Citrus Leprosis Disease. Phytopathology 2017, 107, 963–976. [Google Scholar] [CrossRef] [Green Version]
- Bassanezi, R.B.; Czermainski, A.B.C.; Laranjeira, F.F.; Moreira, A.S.; Ribeiro, P.J.; Krainski, E.T.; Amorim, L. Spatial Patterns of the Citrus Leprosis Virus and Its Associated Mite Vector in Systems without Intervention. Plant Pathol. 2019, 68, 85–93. [Google Scholar] [CrossRef] [Green Version]
- Moreira, R.R.; Machado, F.J.; Lanza, F.E.; Trombin, V.G.; Bassanezi, R.B.; de Miranda, M.P.; Barbosa, J.C.; da Silva Junior, G.J.; Behlau, F. Impact of Diseases and Pests on Premature Fruit Drop in Sweet Orange Orchards in São Paulo State Citrus Belt, Brazil. Pest Manag. Sci. 2022, 78, 2643–2656. [Google Scholar] [CrossRef]
- Ramos-González, P.L.; Chabi-Jesus, C.; Arena, G.D.; Tassi, A.D.; Kitajima, E.W.; Freitas-Astúa, J.; Freita-Astua, J. Citrus Leprosis: A Unique Multietiologic Disease. Cítricos en las Américas 2018, 1, 1–9. [Google Scholar]
- Chabi-Jesus, C.; Ramos-González, P.L.; Postclam-Barro, M.; Fontenele, R.S.; Harakava, R.; Bassanezi, R.B.; Moreira, A.S.; Kitajima, E.W.; Varsani, A.; Freitas-Astúa, J. Molecular Epidemiology of Citrus Leprosis Virus C: A New Viral Lineage and Phylodynamic of the Main Viral Subpopulations in the Americas. Front. Microbiol. 2021, 12, 641252. [Google Scholar] [CrossRef]
- Sauvêtre, P.; Veniant, E.; Croq, G.; Tassi, A.D.; Kitajima, E.W.; Chabi-Jesus, C.; Ramos-Gonzalez, P.; Freitas-Astua, J.; Navia Magalhães Ferreira, D. First Report of Orchid Fleck Virus in Orchid Collection of Jardin Du Luxembourg, Paris, France. Plant Dis. 2018, 102, 2670. [Google Scholar] [CrossRef]
- Carvalho, S.A.; Girardi, E.A.; Filho, F.d.A.A.M.; Ferrarezi, R.S.; Filho, H.D.C. Advances in Citrus Propagation in Brazil. Rev. Bras. Frutic. 2019, 41, e-422. [Google Scholar] [CrossRef] [Green Version]
- Bastianel, M.; Novelli, V.M.; Kitajima, E.W.; Kubo, K.S.; Bassanezi, R.B.; Machado, M.A.; Freitas-Astúa, J. Citrus Leprosis: Centennial of an Unusual Mite–Virus Pathosystem. Plant Dis. 2010, 94, 284–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos-González, P.L.; Chabi-Jesus, C.; Banguela-Castillo, A.; Tassi, A.D.; Rodrigues, M.d.C.; Kitajima, E.W.; Harakava, R.; Freitas-Astúa, J. Unveiling the Complete Genome Sequence of Clerodendrum Chlorotic Spot Virus, a Putative Dichorhavirus Infecting Ornamental Plants. Arch. Virol. 2018, 163, 2519–2524. [Google Scholar] [CrossRef] [PubMed]
- Beard, J.J.; Ochoa, R.; Braswell, W.E.; Bauchan, G.R. Brevipalpus phoenicis (Geijskes) Species Complex (Acari: Tenuipalpidae)-a Closer Look. Zootaxa 2015, 3944, 1–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, K.M.; Dietzgen, R.G.; Wang, R.; Goodin, M.M. Lettuce Necrotic Yellows Cytorhabdovirus Protein Localization and Interaction Map, and Comparison with Nucleorhabdoviruses. J. Gen. Virol. 2012, 93, 906–914. [Google Scholar] [CrossRef]
- Sun, K.; Zhou, X.; Lin, W.; Zhou, X.; Jackson, A.O.; Li, Z. Matrix-Glycoprotein Interactions Required for Budding of a Plant Nucleorhabdovirus and Induction of Inner Nuclear Membrane Invagination. Mol. Plant Pathol. 2018, 19, 2288–2301. [Google Scholar] [CrossRef] [Green Version]
- Siddell, S.G.; Walker, P.J.; Lefkowitz, E.J.; Mushegian, A.R.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; Knowles, N.J.; Kropinski, A.M.; et al. Binomial Nomenclature for Virus Species: A Consultation. Arch. Virol. 2020, 165, 519–525. [Google Scholar] [CrossRef] [Green Version]
- Ramos-González, P.L.; Chabi-Jesus, C.; Tassi, A.D.; Calegario, R.F.; Harakava, R.; Nome, C.F.; Kitajima, E.W.; Freitas-Astua, J. A Novel Lineage of Cile-Like Viruses Discloses the Phylogenetic Continuum Across the Family Kitaviridae. Front. Microbiol. 2022, 13, 836076. [Google Scholar] [CrossRef]
- Nunes, M.A.; de Carvalho Mineiro, J.L.; Rogerio, L.A.; Ferreira, L.M.; Tassi, A.; Novelli, V.M.; Kitajima, E.W.; Freitas-Astúa, J. First Report of Brevipalpus papayensis as Vector of Coffee Ringspot Virus and Citrus Leprosis Virus C. Plant Dis. 2018, 102, 1046. [Google Scholar] [CrossRef]
- Sánchez-Velázquez, E.J.; Santillán-Galicia, M.T.; Novelli, V.M.; Nunes, M.A.; Mora-Aguilera, G.; Valdez-Carrasco, J.M.; Otero-Colina, G.; Freitas-Astúa, J. Diversity and Genetic Variation among Brevipalpus Populations from Brazil and Mexico. PLoS ONE 2015, 10, e0133861. [Google Scholar] [CrossRef] [Green Version]
- Andrade, D.J.; Lorençon, J.R.; Siqueira, D.S.; Novelli, V.M.; Bassanezi, R.B. Space–Time Variability of Citrus Leprosis as Strategic Planning for Crop Management. Pest Manag. Sci. 2018, 74, 1798–1803. [Google Scholar] [CrossRef] [PubMed]
- Bobot, T.d.E.; Franklin, E.; Navia, D.; Gasnier, T.R.J.; Lofego, A.C.; Oliveira, B.M.d. Mites (Arachnida, Acari) on Citrus sinensis L. Osbeck Orange Trees in the State of Amazonas, Northern Brazil. Acta Amaz. 2011, 41, 557–566. [Google Scholar] [CrossRef] [Green Version]
- Locali-Fabris, E.C.; Freitas-Astúa, J.; Souza, A.A.; Takita, M.A.; Astúa-Monge, G.; Antonioli-Luizon, R.; Rodrigues, V.; Targon, M.L.P.N.; Machado, M.A. Complete Nucleotide Sequence, Genomic Organization and Phylogenetic Analysis of Citrus Leprosis Virus Cytoplasmic Type. J. Gen. Virol. 2006, 87, 2721–2729. [Google Scholar] [CrossRef] [PubMed]
- Ramos-González, P.L.; Chabi-Jesus, C.; Guerra-Peraza, O.; Breton, M.C.; Arena, G.D.G.D.G.D.; Nunes, M.A.M.A.; Kitajima, E.W.E.W.E.W.; Machado, M.A.M.A.M.A.; Freitas-Astúa, J. Phylogenetic and Molecular Variability Studies Reveal a New Genetic Clade of Citrus Leprosis Virus C. Viruses 2016, 8, 153. [Google Scholar] [CrossRef] [Green Version]
- Salinas-Vargas, D.; Santillán-Galicia, M.T.; Guzmán-Franco, A.W.; Hernández-López, A.; Ortega-Arenas, L.D.; Mora-Aguilera, G. Analysis of Genetic Variation in Brevipalpus yothersi (Acari: Tenuipalpidae) Populations from Four Species of Citrus Host Plants. PLoS ONE 2016, 11, e0164552. [Google Scholar] [CrossRef] [Green Version]
- Amaral, I.; de Moraes, G.J.; Melville, C.C.; Andrade, D.J. Factors Affecting Prevailing Population Levels of Brevipalpus yothersi (Acari: Tenuipalpidae) in Citrus Areas Affected by Citrus Leprosis in the State of Sao Paulo, Brazil. Exp. Appl. Acarol. 2018, 74, 395–402. [Google Scholar] [CrossRef] [Green Version]
- Navia, D.; Mendonça, R.S.; Ferragut, F.; Miranda, L.C.; Trincado, R.C.; Michaux, J.; Navajas, M. Cryptic Diversity in Brevipalpus Mites (Tenuipalpidae). Zool. Scr. 2013, 42, 406–426. [Google Scholar] [CrossRef]
- Omoto, C. Acaricide Resistance Management of Leprosis Mite (Brevipalpus phoenicis) in Brazilian Citrus. Pestic. Sci. 1998, 52, 189–191. [Google Scholar] [CrossRef]
- Hinomoto, N.; Takafuji, A. Genetic Diversity and Phylogeny of the Kanzawa Spider Mite, Tetranychus Kanzawai, in Japan. Exp. Appl. Acarol. 2001, 25, 355–370. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [Green Version]
- Arena, G.D.; Ramos-González, P.L.; Nunes, M.A.; Jesus, C.C.; Calegario, R.F.; Kitajima, E.W.; Novelli, V.M.; Freitas-Astúa, J.; Arena, G.D.; Ramos-González, P.L.; et al. Arabidopsis thaliana as a Model Host for Brevipalpus Mite-Transmitted Viruses. Sci. Agric. 2017, 74, 85–89. [Google Scholar] [CrossRef] [Green Version]
- Tassi, A.D.; Garita-Salazar, L.C.; Amorim, L.; Novelli, V.M.; Freitas-Astúa, J.; Childers, C.C.; Kitajima, E.W. Virus-Vector Relationship in the Citrus Leprosis Pathosystem. Exp. Appl. Acarol. 2017, 71, 227–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubo, K.S.; Freitas-Astúa, J.; Machado, M.A.; Kitajima, E.W. Orchid Fleck Symptoms May Be Caused Naturally by Two Different Viruses Transmitted by Brevipalpus. J. Gen. Plant Pathol. 2009, 75, 250–255. [Google Scholar] [CrossRef]
- Kitajima, E.W.; Chagas, C.M.; Braghini, M.T.; Fazuoli, L.C.; Locali-Fabris, E.C.; Salaroli, R.B. Natural Infection of Several Coffea Species and Hybrids and Psilanthus ebracteolatus by the Coffee Ringspot Virus (CoRSV). Sci. Agric. 2011, 68, 503–507. [Google Scholar] [CrossRef] [Green Version]
- Kubo, K.S.; Novelli, V.M.; Bastianel, M.; Locali-Fabris, E.C.; Antonioli-Luizon, R.; Machado, M.A.; Freitas-Astúa, J. Detection of Brevipalpus-Transmitted Viruses in Their Mite Vectors by RT–PCR. Exp. Appl. Acarol. 2011, 54, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Choudhary, N.; Guillermo, L.M.; Shao, J.; Govindarajulu, A.; Achor, D.; Wei, G.; Picton, D.D.; Levy, L.; Nakhla, M.K.; et al. A Novel Virus of the Genus Cilevirus Causing Symptoms Similar to Citrus Leprosis. Phytopathology 2013, 103, 488–500. [Google Scholar] [CrossRef] [Green Version]
- Ramos-González, P.L.; Santos, G.F.d.; Chabi-Jesus, C.; Harakava, R.; Kitajima, E.W.; Freitas-Astúa, J. Passion Fruit Green Spot Virus Genome Harbors a New Orphan ORF and Highlights the Flexibility of the 5′-End of the RNA2 Segment Across Cileviruses. Front. Microbiol. 2020, 11, 206. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-Length Human Immunodeficiency Virus Type 1 Genomes from Subtype C-Infected Seroconverters in India, with Evidence of Intersubtype Recombination.Pdf. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef] [Green Version]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating Signal Peptides from Transmembrane Regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of Protein Subcellular Localization Using Deep Learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosakovsky Pond, S.L.; Posada, D.; Gravenor, M.B.; Woelk, C.H.; Frost, S.D.W. GARD: A Genetic Algorithm for Recombination Detection. Bioinformatics 2006, 22, 3096–3098. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]



| Genus | Virus 1 | Target | Sequence 5′-3′ | Amplicon Size (bp) | Reference | |
|---|---|---|---|---|---|---|
| Dichorhavirus | OFV | N | F: | TGTCATAGCCGACATAAACACC | 326 | [44] |
| R: | TGTAGAGCTTGCGAGATACAGG | |||||
| CiLV-N | N | F: | CCGTACCCATTGTGAAAATA | 420 | [16] | |
| R: | GAACCCCTTTGAGGAATG | |||||
| OFV + CiLV-N | L | F: | CAASTGTCATGCCTGCATGG | 362 | ||
| R: | TTGATRCATGATGCRAGRCTGTATG | |||||
| CiCSV | G | F: | CTGTTTTGCCCATGCTAC | 500 | [14] | |
| R: | CCTCCTCTTCTAGCGTCAT | |||||
| CoRSV | L | F: | GGACCATGAGACAGGAGGTG | 394 | [45] | |
| R: | CTCTGCCAGTCCTCAATGTG | |||||
| ClCSV | L | F: | AGTGTACCGCCTCACAGAAG | 219 | [46] | |
| R: | CGGGGTCTTGTTGTTCATAG | |||||
| Tentative dichorhavirus | CiBSV | N | F: | CAGTCACTATAGATTACTCAGCAG | 296 | This study |
| R: | TGCTACTCCTACCATCAC | |||||
| L | F: | GCCTACGGAGAGGAAGAT | 938 | |||
| R: | GGGTCTACTGAGCTGTATATGA | |||||
| Cilevirus | CiLV-C | p24 | F: | CGCAGTTTCCTAATAACACC | 322 | [20] |
| R: | GGGAGTTCAGCATAAAGC | |||||
| CiLV-C2 | p29 | F: | ATGAGTAACATTGTGTCGTTTTCGTTGT | 795 | [47] | |
| R: | TCACTCTTCCTGTTCATCAACCTGTT | |||||
| PfGSV | p29 | F: | ACACCAAGAGTACTATCGATC | 452 | [48] | |
| R: | CATCAAGTGGAGCAAGTTC | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chabi-Jesus, C.; Ramos-González, P.L.; Tassi, A.D.; Rossetto Pereira, L.; Bastianel, M.; Lau, D.; Canale, M.C.; Harakava, R.; Novelli, V.M.; Kitajima, E.W.; et al. Citrus Bright Spot Virus: A New Dichorhavirus, Transmitted by Brevipalpus azores, Causing Citrus Leprosis Disease in Brazil. Plants 2023, 12, 1371. https://doi.org/10.3390/plants12061371
Chabi-Jesus C, Ramos-González PL, Tassi AD, Rossetto Pereira L, Bastianel M, Lau D, Canale MC, Harakava R, Novelli VM, Kitajima EW, et al. Citrus Bright Spot Virus: A New Dichorhavirus, Transmitted by Brevipalpus azores, Causing Citrus Leprosis Disease in Brazil. Plants. 2023; 12(6):1371. https://doi.org/10.3390/plants12061371
Chicago/Turabian StyleChabi-Jesus, Camila, Pedro Luis Ramos-González, Aline Daniele Tassi, Laura Rossetto Pereira, Marinês Bastianel, Douglas Lau, Maria Cristina Canale, Ricardo Harakava, Valdenice Moreira Novelli, Elliot Watanabe Kitajima, and et al. 2023. "Citrus Bright Spot Virus: A New Dichorhavirus, Transmitted by Brevipalpus azores, Causing Citrus Leprosis Disease in Brazil" Plants 12, no. 6: 1371. https://doi.org/10.3390/plants12061371
APA StyleChabi-Jesus, C., Ramos-González, P. L., Tassi, A. D., Rossetto Pereira, L., Bastianel, M., Lau, D., Canale, M. C., Harakava, R., Novelli, V. M., Kitajima, E. W., & Freitas-Astúa, J. (2023). Citrus Bright Spot Virus: A New Dichorhavirus, Transmitted by Brevipalpus azores, Causing Citrus Leprosis Disease in Brazil. Plants, 12(6), 1371. https://doi.org/10.3390/plants12061371











