Analysis of Potato Physiological and Molecular Adaptation in Response to Different Water and Nitrogen Combined Regimes
Abstract
1. Introduction
2. Results and Discussions
2.1. Sequencing Data Analysis and Validation
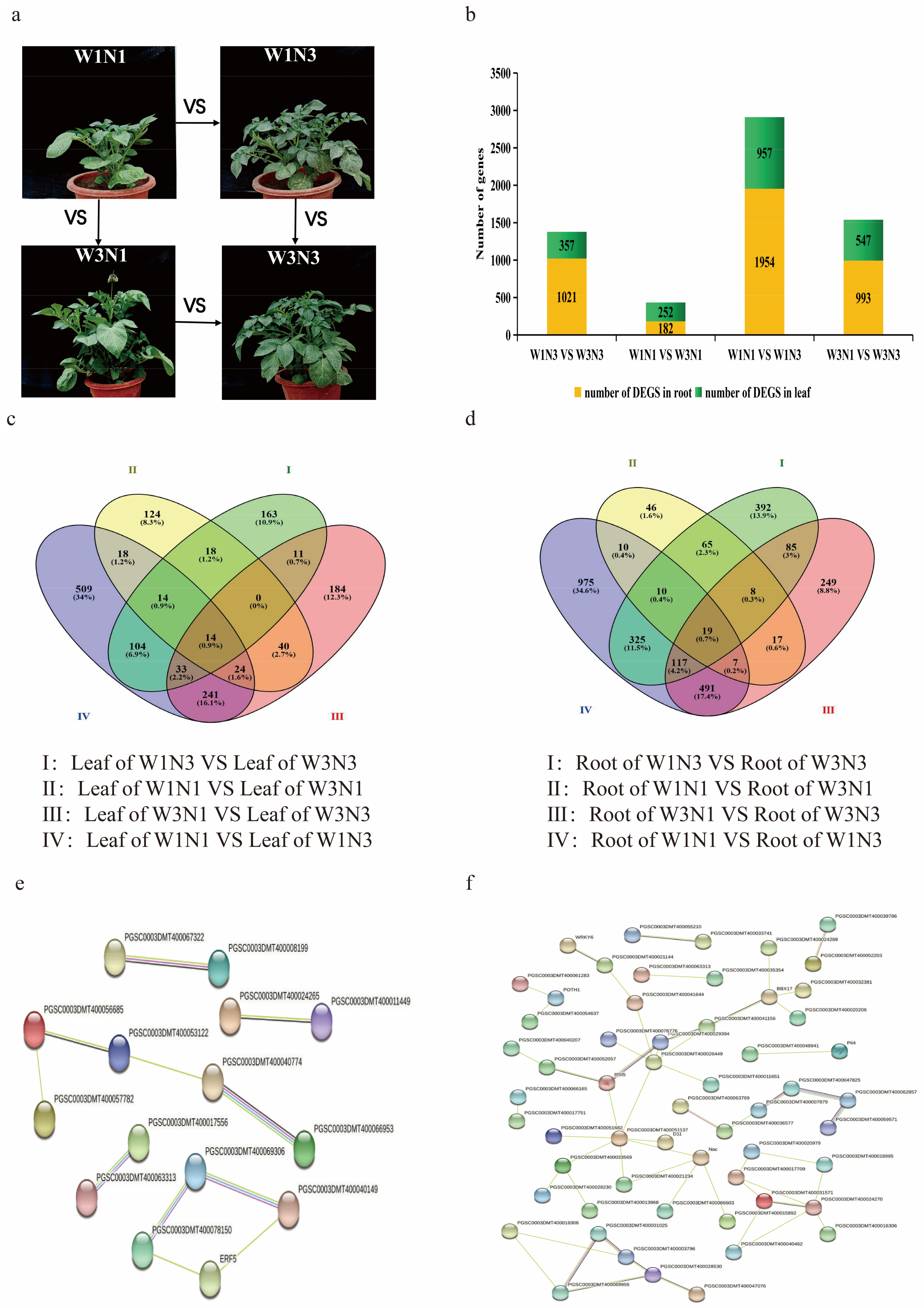
2.2. Tissue-Specific Variation in Water- and Nitrogen-Responsive Genes
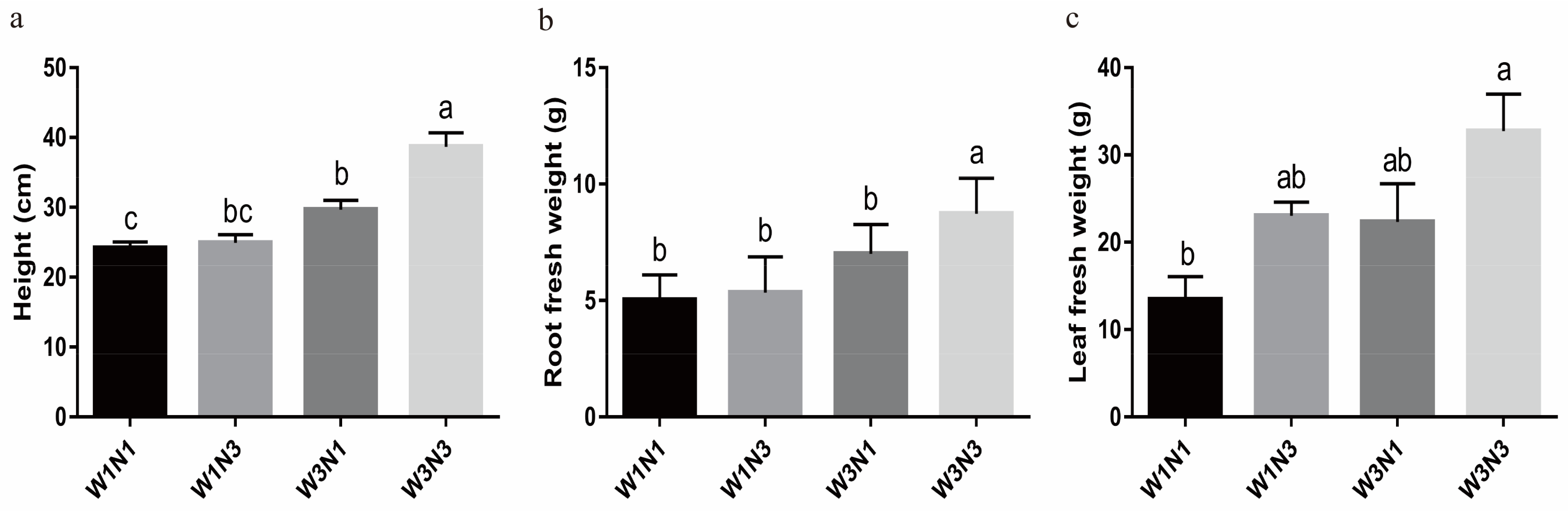
2.3. Potato Stress Response Was Increased under Drought and Nitrogen Deficit
2.4. Changes in Soil Nitrogen or Moisture Levels Led to Induction of Distinct Metabolic Pathways
2.4.1. Increased Nitrogen Application under Drought Conditions Can Improve the Efficiency of Light Energy Utilization in Leaves
2.4.2. Nitrogen Levels Could Affect Expression of Genes Related to Stress Resistance
2.4.3. Nitrogen Transporters Exhibited Tissue-Specific Expression in Potato
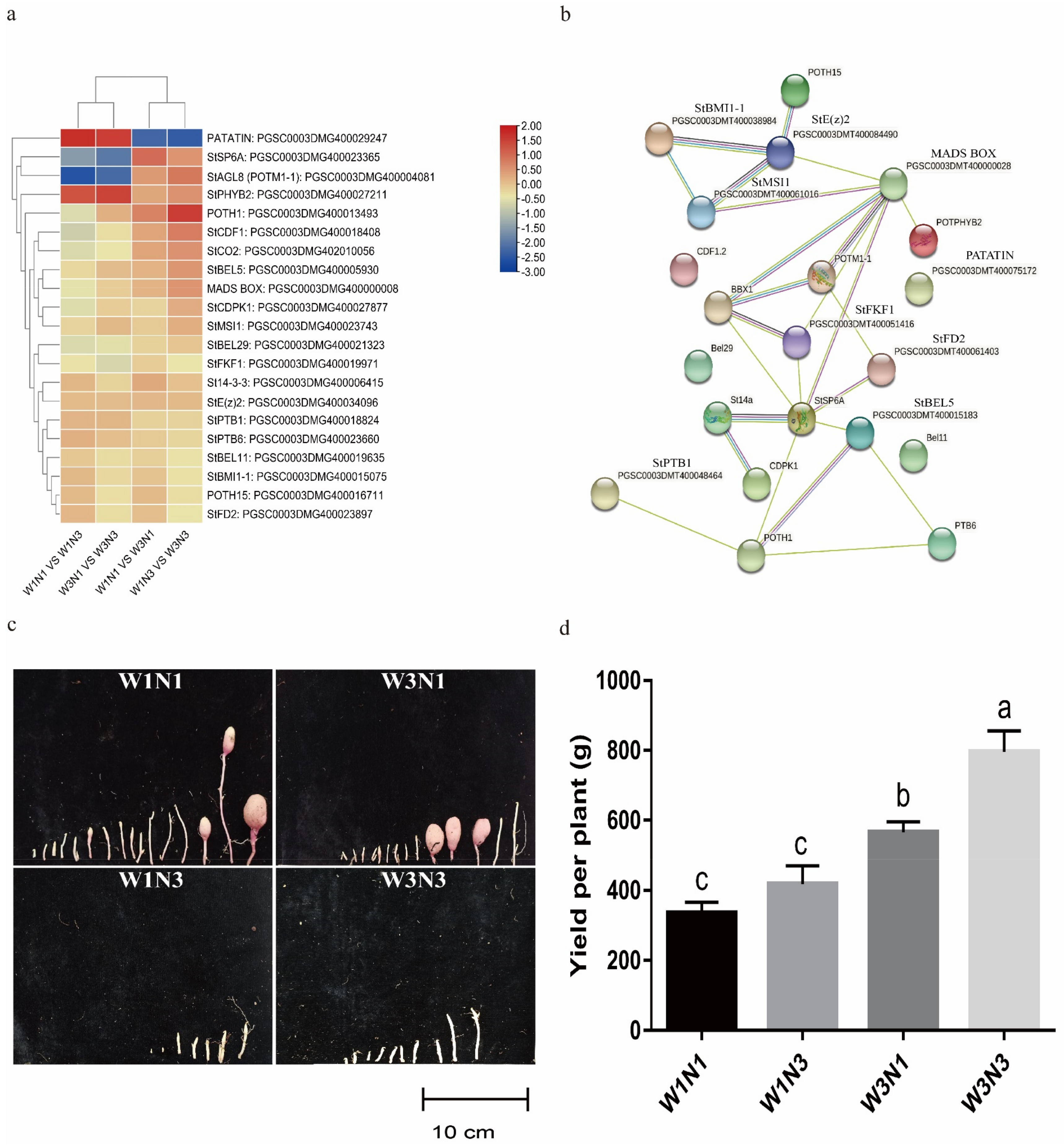
2.4.4. Increased Nitrogen Fertilization Can Delay Potato Tuber Expansion
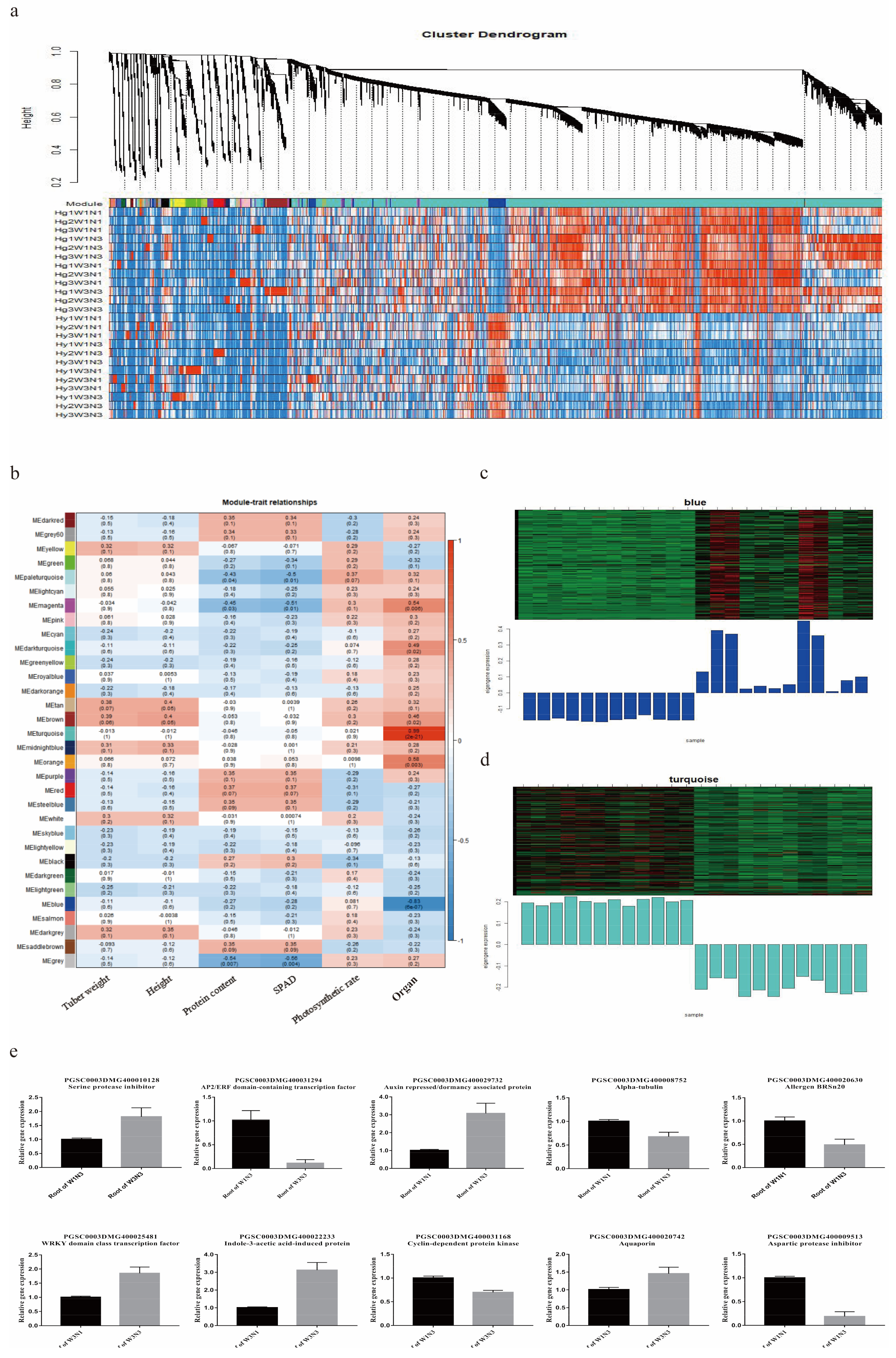
2.5. Screening of Key Genes Responding to Changes in Water and Nitrogen Levels
3. Materials and Methods
3.1. Experimental Location
3.2. Experimental Treatments
3.3. Measurement and Analysis of Physiological Indicators
3.4. RNA-Seq Experiments
3.5. Transcriptomic Data Analysis
3.6. Validation of Candidate Genes Via Reverse Transcription Quantitative PCR (RT-qPCR)
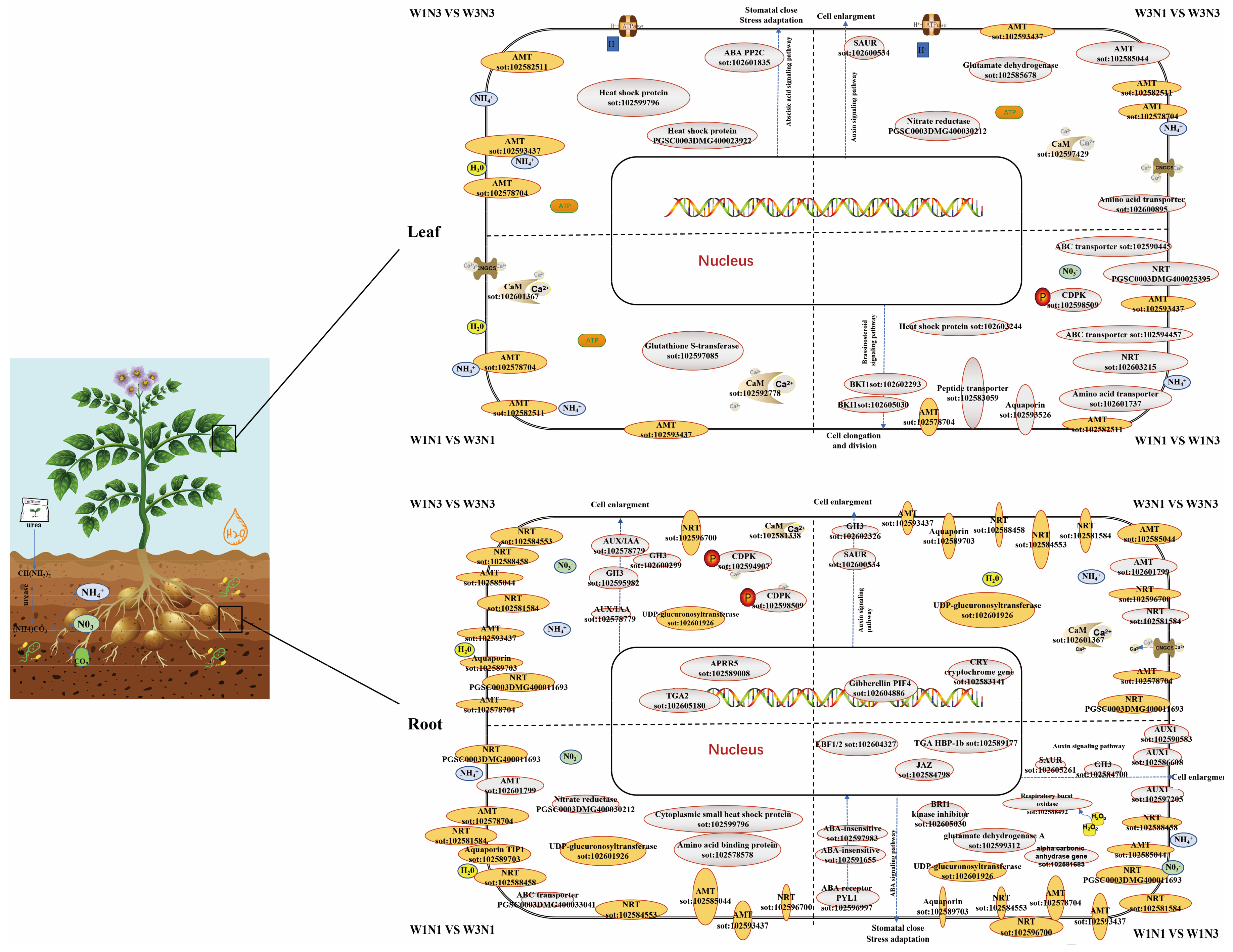
 are differentially expressed genes (DEGs) in response to changes in both nitrogen and water;
are differentially expressed genes (DEGs) in response to changes in both nitrogen and water;  are DEGs in response to changes in either water or nitrogen but not both. Dashed lines separate changes induced by different treatments.
are DEGs in response to changes in either water or nitrogen but not both. Dashed lines separate changes induced by different treatments.4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Coskun, D.; Britto, D.T.; Shi, W.; Kronzucker, H.J. Nitrogen transformations in modern agriculture and the role of biological nitrification inhibition. Nat. Plants 2017, 3, 17074. [Google Scholar] [CrossRef]
- Wang, M.; Shen, Q.; Xu, G.; Guo, S. New insight into the strategy for nitrogen metabolism in plant cells. Int. Rev. Cell Mol. Biol. 2014, 310, 1–37. [Google Scholar]
- Ishiyama, K.; Inoue, E.; Watanabe-Takahashi, A.; Obara, M.; Yamaya, T.; Takahashi, H. Kinetic properties and ammonium-dependent regulation of cytosolic isoenzymes of glutamine synthetase in Arabidopsis. J. Biol. Chem. 2014, 279, 16598–16605. [Google Scholar] [CrossRef] [PubMed]
- Besnard, J.; Pratelli, R.; Zhao, C.; Sonawala, U.; Collakova, E.; Pilot, G.; Okumoto, S. UMAMIT14 is an amino acid exporter involved in phloem unloading in Arabidopsis roots. J. Exp. Bot. 2016, 67, 6385–6397. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, W.; Chen, H.; Zhan, J.; He, C.; Wang, Q. Ammonium nitrogen tolerant chlorella strain screening and its damaging effects on photosynthesis. Front. Microbiol. 2019, 9, 3250. [Google Scholar] [CrossRef]
- Chen, A.; Lei, B.; Hu, W.; Lu, Y.; Mao, Y.; Duan, Z.; Shi, Z. Characteristics of ammonia volatilization on rice grown under different nitrogen application rates and its quantitative predictions in Erhai Lake Watershed, China. Nutr. Cycl. Agroecosyst. 2015, 101, 139–152. [Google Scholar] [CrossRef]
- Xie, Y.; Mao, Y.; Xu, S.; Zhou, H.; Duan, X.; Cui, W.; Zhang, J.; Xu, G. Heme-heme oxygenase 1 system is involved in ammonium tolerance by regulating antioxidant defence in Oryza sativa. Plant Cell Environ. 2015, 38, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Crespo, E.; Scalschi, L.; Llorens, E.; García-Agustín, P.; Camañes, G. NH4+ protects tomato plants against Pseudomonas syringae by activation of systemic acquired acclimation. J. Exp. Bot. 2015, 66, 6777–6790. [Google Scholar] [CrossRef] [PubMed]
- Zou, N.; Li, B.; Chen, H.; Su, Y.; Kronzucker, H.J.; Xiong, L.; Baluška, F.; Shi, W. GSA-1/ARG1 protects root gravitropism in Arabidopsis under ammonium stress. New Phytol. 2013, 200, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.Y.; Tian, S.S.; Liu, S.S.; Wang, W.Q.; Sui, N. Energy dissipation and antioxidant enzyme system protect photosystem II of sweet sorghum under drought stress. Photosynthetica 2018, 56, 861–872. [Google Scholar] [CrossRef]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef]
- Li, Y.; Chen, J.; Tian, L.; Shen, Z.; Amby, D.B.; Liu, F.; Gao, Q.; Wang, Y. Seedling-stage deficit irrigation with nitrogen application in three-year field study provides guidance for improving maize yield, water and nitrogen use efficiencies. Plants 2022, 11, 3007. [Google Scholar] [CrossRef]
- Yan, W.; Qin, J.; Duan, S.; Xu, J.; Jian, Y.; Jin, L.; Li, G. The effect of water-nitrogen coupling on potato photosynthesis, tuber formation and quality. Acta Hortic. Sin. 2022, 49, 1491–1504. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Nagel, A.; Thimm, O.; Redestig, H.; Blaesing, O.E.; Palacios-Rojas, N.; Selbig, J.; Hannemann, J.; Piques, M.C.; Steinhauser, D.; et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 2005, 138, 1195–1204. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kondhare, K.R.; Kumar, A.; Patil, N.S.; Malankar, N.N.; Saha, K.; Banerjee, A.K. Development of aerial and belowground tubers in potato is governed by photoperiod and epigenetic mechanism. Plant Physiol. 2021, 187, 1071–1086. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, K.H.; Sheen, J. Dynamic nutrient signaling networks in plants. Annu. Rev. Cell Dev. Biol. 2021, 37, 341–367. [Google Scholar] [CrossRef]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 functions as a nitrate sensor in plants. Cell 2009, 138, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Chiba, Y.; Shimizu, T.; Miyakawa, S.; Kanno, Y.; Koshiba, T. Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J. Plant Res. 2015, 128, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.; Bio, A.F.; Domínguez-Valdivia, M.D.; Aparicio-Tejo, P.M.; Lamsfus, C.; Martins-Loução, M.A. How does glutamine synthetase activity determine plant tolerance to ammonium? Planta 2006, 223, 1068–1080. [Google Scholar] [CrossRef]
- Guan, M.; de Bang, T.C.; Pedersen, C.; Schjoerring, J.K. Cytosolic glutamine synthetase Gln1; 2 is the main isozyme contributing to GS1 activity and can be up-regulated to relieve ammonium toxicity. Plant Physiol. 2016, 171, 1921–1933. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Tsay, Y.F. Influence of differing nitrate and nitrogen availability on flowering control in Arabidopsis. J. Exp. Bot. 2017, 68, 2603–2609. [Google Scholar] [CrossRef] [PubMed]
- Lanquar, V.; Loqué, D.; Hörmann, F.; Yuan, L.; Bohner, A.; Engelsberger, W.R.; Lalonde, S.; Schulze, W.X.; von Wirén, N.; Frommer, W.B. Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell 2009, 21, 3610–3622. [Google Scholar] [CrossRef] [PubMed]
- Engelsberger, W.R.; Schulze, W.X. Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J. 2012, 69, 978–995. [Google Scholar] [CrossRef]
- O’Brien, J.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutiérrez, R.A. Nitrate transport, sensing, and responses in plants. Mol. Plant 2016, 9, 837–856. [Google Scholar] [CrossRef] [PubMed]
- Patterson, K.; Cakmak, T.; Cooper, A.; Lager, I.; Rasmusson, A.G.; Escobar, M.A. Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ. 2010, 33, 1486–1501. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y.; Hanada, A.; Chiba, Y.; Ichikawa, T.; Nakazawa, M.; Matsui, M.; Koshiba, T.; Kamiya, Y.; Seo, M. Identifification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. USA 2012, 109, 9653–9658. [Google Scholar] [CrossRef] [PubMed]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef]
- Li, B.; Li, Q.; Xiong, L.; Kronzucker, H.J.; Krämer, U.; Shi, W. Arabidopsis plastid AMOS1/EGY1 integrates abscisic acid signaling to regulate global gene expression response to ammonium stress. Plant Physiol. 2012, 160, 2040–2051. [Google Scholar] [CrossRef]
- Ding, L.; Li, Y.; Wang, Y.; Gao, L.; Wang, M.; Chaumont, F.; Shen, Q.; Guo, S. Root ABA accumulation enhances rice seedling drought tolerance under ammonium supply: Interaction with aquaporins. Front. Plant Sci. 2016, 7, 1206. [Google Scholar] [CrossRef]
- Chen, J.; Liu, X.; Liu, S.; Fan, X.; Zhao, L.; Song, M.; Fan, X.; Xu, G. Co-overexpression of OsNAR2.1 and OsNRT2.3a increased agronomic nitrogen use efficiency in transgenic rice plants. Front. Plant Sci. 2020, 11, 1245. [Google Scholar] [CrossRef]
- Zhao, W.; Langfelder, P.; Fuller, T.; Dong, J.; Li, A.; Hovarth, S. Weighted gene coexpression network analysis: State of the art. J. Biopharm. Stat. 2010, 20, 281–300. [Google Scholar] [CrossRef]
- Wang, D.; Xu, T.; Yin, Z.; Wu, W.; Geng, H.; Li, L.; Yang, M.; Cai, H.; Lian, X. Overexpression of OsMYB305 in rice enhances the nitrogen uptake under low-nitrogen condition. Front. Plant Sci. 2020, 11, 369. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, H.; Gupta, N.; Singh, V.K.; Kumar, A.; Yadav, D. In silico analysis of PCR amplified DOF (DNA binding with one finger) transcription factor domain and cloned genes from cereals and millets. Online J. Bioinf. 2008, 9, 130–143. [Google Scholar]
- Fang, Z.; Wu, B.; Ji, Y. The amino acid transporter OsAAP4 contributes to rice tillering and grain yield by regulating neutral amino acid allocation through two splicing variants. Rice 2021, 14, 2. [Google Scholar] [CrossRef]
- Gao, L.; Lu, Z.; Ding, L.; Guo, J.; Wang, M.; Ling, N.; Guo, S.; Shen, Q. Role of aquaporins in determining carbon and nitrogen status in higher plants. Int. J. Mol. Sci. 2018, 19, 35. [Google Scholar] [CrossRef] [PubMed]

| Treatment | Level | Seedling Stage | Tuber Formation Stage | Tuber Expansion Stage | Starch Accumulation Stage |
|---|---|---|---|---|---|
| Urea/(g) | Limited (N1) | 2.24 | 0 | 0 | 0 |
| Sufficient (N3) | 7.78 | 2.22 | 1.11 | 0 | |
| Moisture content/(%) | Deficit (W1) | 35~45 | 45~55 | 50~60 | 30~40 |
| Adequate (W3) | 75~85 | 85~95 | 90~100 | 70~80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, W.; Qin, J.; Jian, Y.; Liu, J.; Bian, C.; Jin, L.; Li, G. Analysis of Potato Physiological and Molecular Adaptation in Response to Different Water and Nitrogen Combined Regimes. Plants 2023, 12, 1671. https://doi.org/10.3390/plants12081671
Yan W, Qin J, Jian Y, Liu J, Bian C, Jin L, Li G. Analysis of Potato Physiological and Molecular Adaptation in Response to Different Water and Nitrogen Combined Regimes. Plants. 2023; 12(8):1671. https://doi.org/10.3390/plants12081671
Chicago/Turabian StyleYan, Wenyuan, Junhong Qin, Yinqiao Jian, Jiangang Liu, Chunsong Bian, Liping Jin, and Guangcun Li. 2023. "Analysis of Potato Physiological and Molecular Adaptation in Response to Different Water and Nitrogen Combined Regimes" Plants 12, no. 8: 1671. https://doi.org/10.3390/plants12081671
APA StyleYan, W., Qin, J., Jian, Y., Liu, J., Bian, C., Jin, L., & Li, G. (2023). Analysis of Potato Physiological and Molecular Adaptation in Response to Different Water and Nitrogen Combined Regimes. Plants, 12(8), 1671. https://doi.org/10.3390/plants12081671






