In Depth Topological Analysis of Arabidopsis Mid-SUN Proteins and Their Interaction with the Membrane-Bound Transcription Factor MaMYB
Abstract
1. Introduction
2. Results
2.1. Mid-SUN Proteins Are Enriched in the ONM and ER
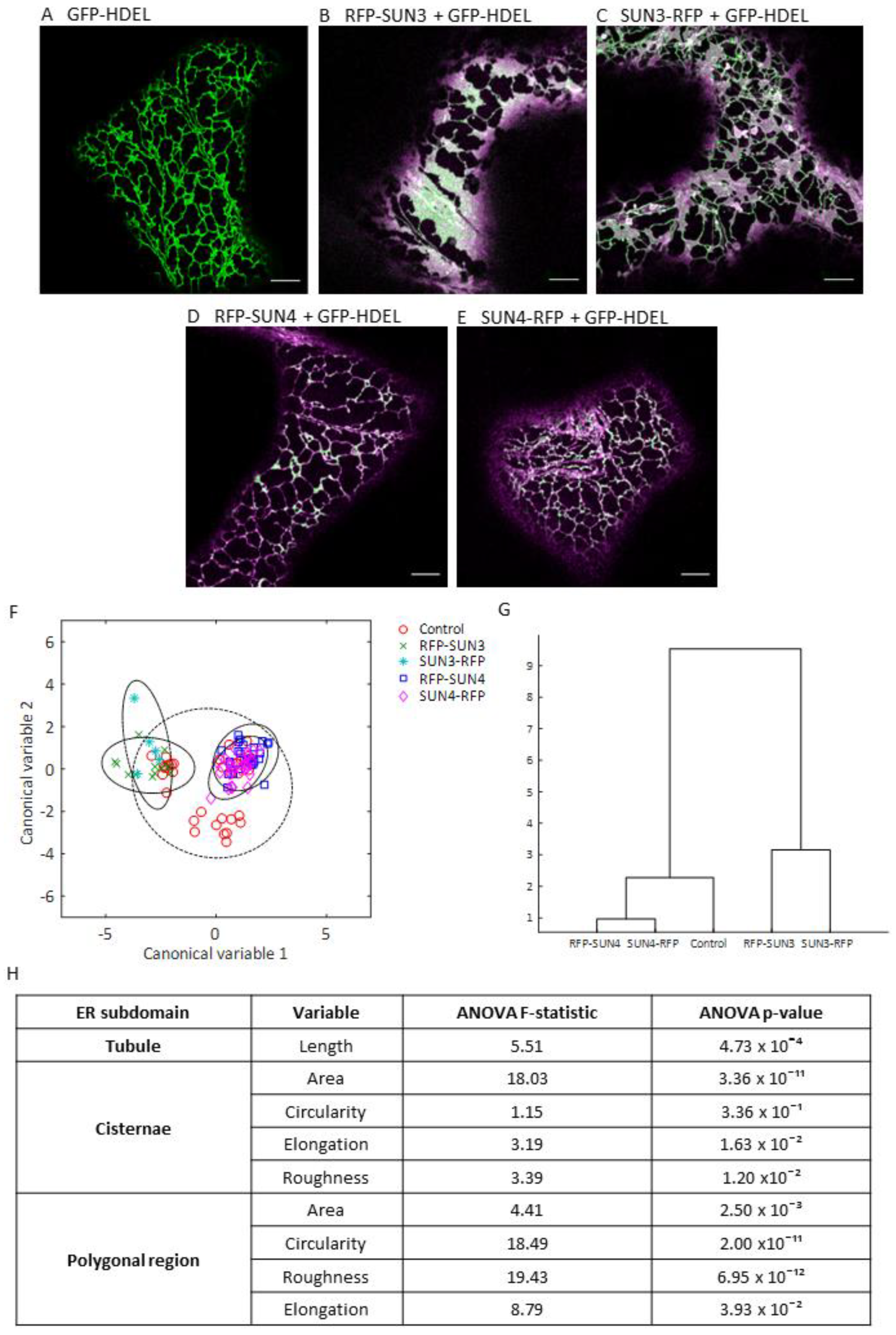
2.2. Quantifying the Effect of SUN3 and 4 OverExpression on ER Architecture
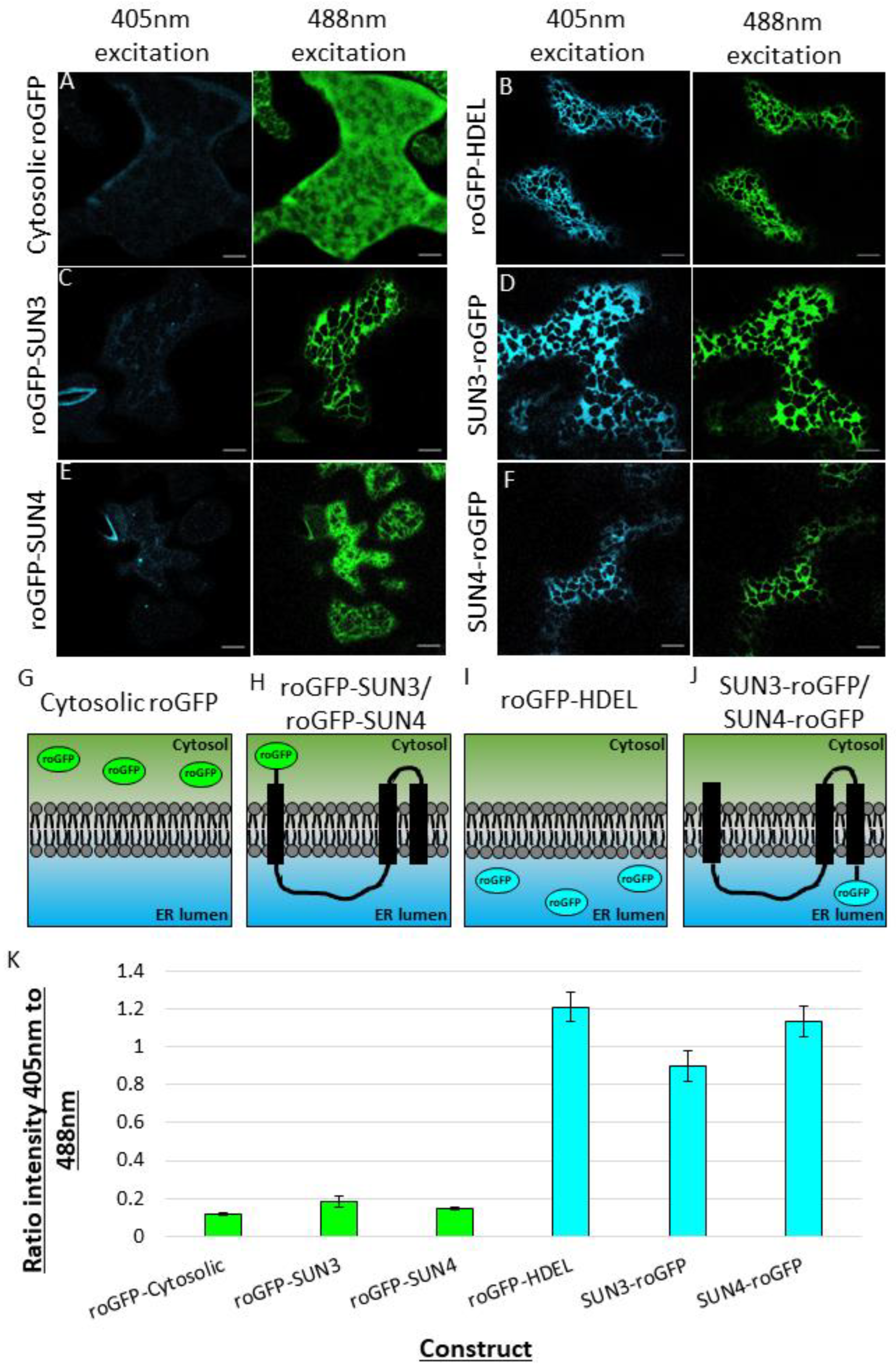
2.3. Using roGFP to Determine the Membrane Topology of SUN3 and 4
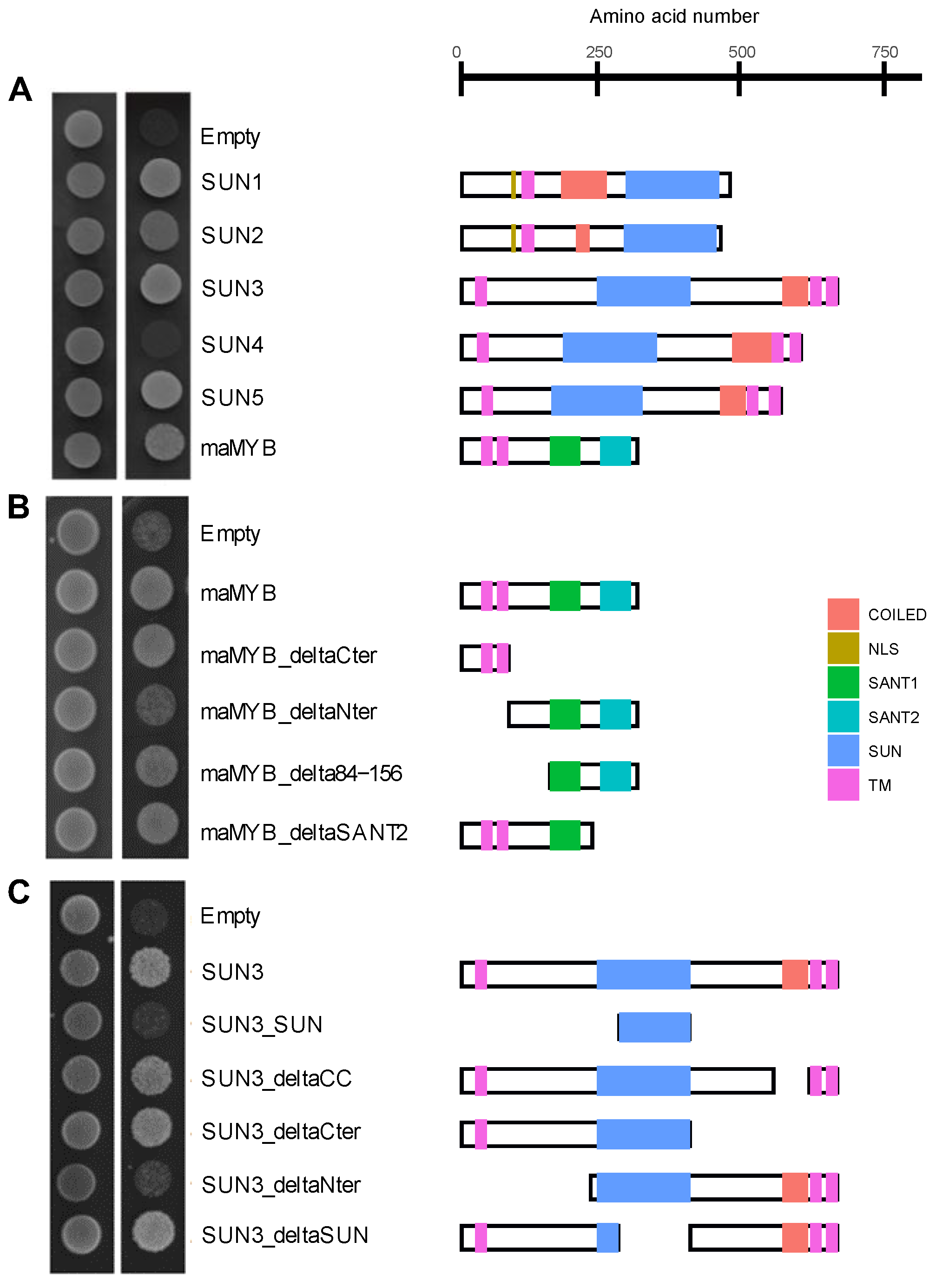
2.4. Arabidopsis Mid-SUN3 Interacts with the ER-Localised Membrane Transcription Factor maMYB
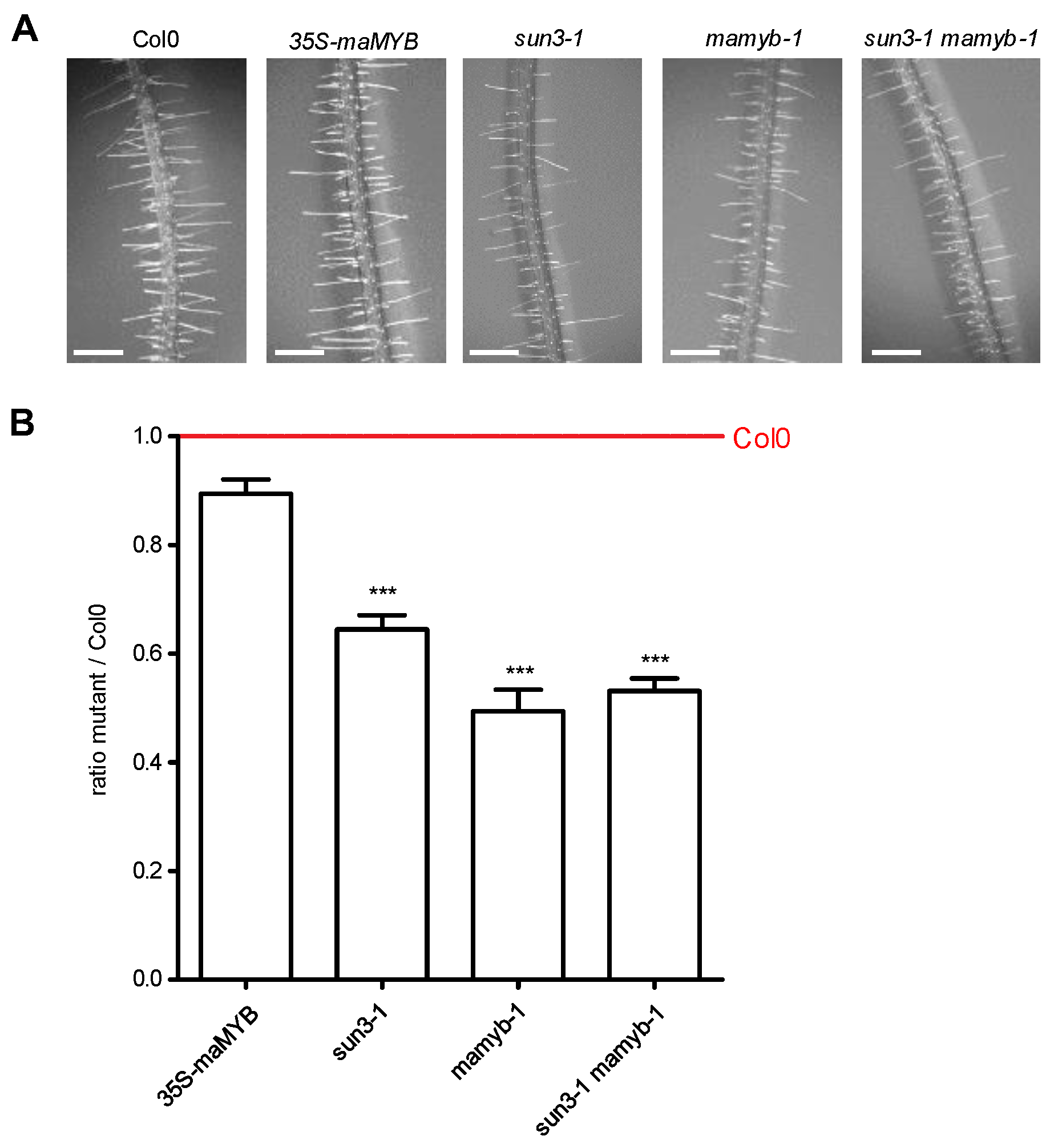
2.5. Disruption of SUN3 and maMYB Affects Root Hair Elongation
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Accessions Numbers, T-DNA Mutants and 35S::MaMYB-vYFP Line
4.3. Molecular Cloning and CRISPR/Cas9-Mediated Genome Editing
4.4. Subcellular Localisation of SUN3 and 4 Analysed via Proximity Distance
4.5. Quantifying Changes in ER Structure
4.6. Tubule Morphology Quantification
4.7. Determination of SUN3 and SUN4 Topology Using roGFP
4.8. Membrane Yeast Two-Hybrid (MYTH)
4.9. Conservation of MaMYB and mid-SUN3 and Phylogenetic Reconstruction
4.10. Root Hair Measurements
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Graumann, K.; Runions, J.; Evans, D.E. Characterization of SUN-domain Proteins at the Higher Plant Nuclear Envelope. Plant J. 2010, 61, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.P.; Simmons, C.R.; Bass, H.W. Structure and Expression of the Maize (Zea mays L.) SUN-Domain Protein Gene Family: Evidence for the Existence of Two Divergent Classes of SUN Proteins in Plants. BMC Plant Biol. 2010, 10, 269. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Fukuda, H. Dynamics of Arabidopsis SUN Proteins during Mitosis and Their Involvement in Nuclear Shaping. Plant J. 2011, 66, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Du, X.; Cai, Z.; Song, X.; Zhang, H.; Mizuno, T.; Suzuki, E.; Yee, M.R.; Berezov, A.; Murali, R.; et al. Structure of Sad1-UNC84 Homology (SUN) Domain Defines Features of Molecular Bridge in Nuclear Envelope. J. Biol. Chem. 2012, 287, 5317–5326. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Iwabuchi, K.; Fukao, Y.; Kondo, M.; Okamoto, K.; Ueda, H.; Nishimura, M.; Hara-Nishimura, I. Myosin XI-i Links the Nuclear Membrane to the Cytoskeleton to Control Nuclear Movement and Shape in Arabidopsis. Curr. Biol. 2013, 23, 1776–1781. [Google Scholar] [CrossRef]
- Zhou, X.; Groves, N.R.; Meier, I. Plant Nuclear Shape Is Independently Determined by the SUN-WIP-WIT2-Myosin XI-i Complex and CRWN1. Nucleus 2015, 6, 144–153. [Google Scholar] [CrossRef]
- Murphy, S.P.; Gumber, H.K.; Mao, Y.; Bass, H.W. A Dynamic Meiotic SUN Belt Includes the Zygotene-Stage Telomere Bouquet and Is Disrupted in Chromosome Segregation Mutants of Maize (Zea mays L.). Front. Plant Sci. 2014, 5, 314. [Google Scholar] [CrossRef]
- Varas, J.; Graumann, K.; Osman, K.; Pradillo, M.; Evans, D.E.; Santos, J.L.; Armstrong, S.J. Absence of SUN1 and SUN2 Proteins in Arabidopsis Thaliana Leads to a Delay in Meiotic Progression and Defects in Synapsis and Recombination. Plant J. 2015, 81, 329–346. [Google Scholar] [CrossRef]
- Pradillo, M.; Evans, D.; Graumann, K. The Nuclear Envelope in Higher Plant Mitosis and Meiosis. Nucleus 2019, 10, 55–66. [Google Scholar] [CrossRef]
- Sohaskey, M.L.; Jiang, Y.; Zhao, J.J.; Mohr, A.; Roemer, F.; Harland, R.M. Osteopotentia Regulates Osteoblast Maturation, Bone Formation, and Skeletal Integrity in Mice. J. Cell Biol. 2010, 189, 511–525. [Google Scholar] [CrossRef]
- Shimada, N.; Inouye, K.; Sawai, S.; Kawata, T. SunB, a Novel Sad1 and UNC-84 Domain-Containing Protein Required for Development of Dictyostelium Discoideum. Dev. Growth Differ. 2010, 52, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Friederichs, J.M.; Gardner, J.M.; Smoyer, C.J.; Whetstine, C.R.; Gogol, M.; Slaughter, B.D.; Jaspersen, S.L. Genetic Analysis of Mps3 SUN Domain Mutants in Saccharomyces Cerevisiae Reveals an Interaction with the SUN-Like Protein Slp1. G3 2012, 2, 1703–1718. [Google Scholar] [CrossRef] [PubMed]
- Graumann, K.; Vanrobays, E.; Tutois, S.; Probst, A.V.; Evans, D.E.; Tatout, C. Characterization of Two Distinct Subfamilies of SUN-Domain Proteins in Arabidopsis and Their Interactions with the Novel KASH-Domain Protein AtTIK. J. Exp. Bot. 2014, 65, 6499–6512. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, D.K.; Mishra, P.; Subba, P.; Rathi, D.; Chakraborty, S.; Chakraborty, N. Membrane-Associated Proteomics of Chickpea Identifies Sad1/UNC-84 Protein (CaSUN1), a Novel Component of Dehydration Signaling. Sci. Rep. 2014, 4, 4177. [Google Scholar] [CrossRef]
- Poulet, A.; Probst, A.V.; Graumann, K.; Tatout, C.; Evans, D. Exploring the Evolution of the Proteins of the Plant Nuclear Envelope. Nucleus 2017, 8, 46–59. [Google Scholar] [CrossRef]
- Vasnier, C.; de Muyt, A.; Zhang, L.; Tessé, S.; Kleckner, N.E.; Zickler, D.; Espagne, E. Absence of SUN-Domain Protein Slp1 Blocks Karyogamy and Switches Meiotic Recombination and Synapsis from Homologs to Sister Chromatids. Proc. Natl. Acad. Sci. USA 2014, 111, E4015–E4023. [Google Scholar] [CrossRef]
- Xue, Y.; Meng, J.-G.; Jia, P.-F.; Zhang, Z.-R.; Li, H.-J.; Yang, W.-C. POD1-SUN-CRT3 Chaperone Complex Guards the ER Sorting of LRR Receptor Kinases in Arabidopsis. Nat. Commun. 2022, 13, 2703. [Google Scholar] [CrossRef]
- Xue, Y.; Jia, P.-F.; Li, H.-J. SUN3/4/5 Proteins Regulate Endoplasmic Reticulum Tubule Formation and Luminal Spacing in Arabidopsis. J. Genet. Genom. 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, C.; Larrimore, K.E.; Ng, D.T.W. Slp1-Emp65: A Guardian Factor That Protects Folding Polypeptides from Promiscuous Degradation. Cell 2017, 171, 346–357.e12. [Google Scholar] [CrossRef]
- Yuan, L.; Pan, J.; Zhu, S.; Li, Y.; Yao, J.; Li, Q.; Fang, S.; Liu, C.; Wang, X.; Li, B.; et al. Evolution and Functional Divergence of SUN Genes in Plants. Front. Plant Sci. 2021, 12, 646622. [Google Scholar] [CrossRef]
- Liu, D.Y.T.; Smith, P.M.C.; Barton, D.A.; Day, D.A.; Overall, R.L. Characterisation of Arabidopsis Calnexin 1 and Calnexin 2 in the Endoplasmic Reticulum and at Plasmodesmata. Protoplasma 2017, 254, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Pain, C.; Kriechbaumer, V.; Kittelmann, M.; Hawes, C.; Fricker, M. Quantitative Analysis of Plant ER Architecture and Dynamics. Nat. Commun. 2019, 10, 984. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, I.; Tolley, N.; Aller, I.; Svozil, J.; Osterrieder, A.; Botchway, S.; Mueller, C.; Frigerio, L.; Hawes, C. Five Arabidopsis Reticulon Isoforms Share Endoplasmic Reticulum Location, Topology, and Membrane-Shaping Properties. Plant Cell 2010, 22, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Dooley, C.T.; Dore, T.M.; Hanson, G.T.; Jackson, W.C.; Remington, S.J.; Tsien, R.Y. Imaging Dynamic Redox Changes in Mammalian Cells with Green Fluorescent Protein Indicators. J. Biol. Chem. 2004, 279, 22284–22293. [Google Scholar] [CrossRef]
- Hanson, G.T.; Aggeler, R.; Oglesbee, D.; Cannon, M.; Capaldi, R.A.; Tsien, R.Y.; Remington, S.J. Investigating Mitochondrial Redox Potential with Redox-Sensitive Green Fluorescent Protein Indicators. J. Biol. Chem. 2004, 279, 13044–13053. [Google Scholar] [CrossRef]
- Meyer, A.J.; Brach, T.; Marty, L.; Kreye, S.; Rouhier, N.; Jacquot, J.-P.; Hell, R. Redox-Sensitive GFP in Arabidopsis Thaliana Is a Quantitative Biosensor for the Redox Potential of the Cellular Glutathione Redox Buffer. Plant J. 2007, 52, 973–986. [Google Scholar] [CrossRef]
- Schwarzländer, M.; Fricker, M.D.; Müller, C.; Marty, L.; Brach, T.; Novak, J.; Sweetlove, L.J.; Hell, R.; Meyer, A.J. Confocal Imaging of Glutathione Redox Potential in Living Plant Cells. J. Microsc. 2008, 231, 299–316. [Google Scholar] [CrossRef]
- Slabaugh, E.; Held, M.; Brandizzi, F. Control of Root Hair Development in Arabidopsis Thaliana by an Endoplasmic Reticulum Anchored Member of the R2R3-MYB Transcription Factor Family. Plant J. 2011, 67, 395–405. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Altenhoff, A.M.; Train, C.-M.; Gilbert, K.J.; Mediratta, I.; Mendes de Farias, T.; Moi, D.; Nevers, Y.; Radoykova, H.-S.; Rossier, V.; Warwick Vesztrocy, A.; et al. OMA Orthology in 2021: Website Overhaul, Conserved Isoforms, Ancestral Gene Order and More. Nucleic Acids Res. 2021, 49, D373–D379. [Google Scholar] [CrossRef] [PubMed]
- Möller, S.; Croning, M.D.R.; Apweiler, R. Evaluation of Methods for the Prediction of Membrane Spanning Regions. Bioinformatics 2001, 17, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-Based Nucleases and Nickases Can Be Used Efficiently for Genome Engineering in Arabidopsis Thaliana. Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB Transcription Factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Kim, S.-G.; Park, J.-E.; Park, H.-Y.; Lim, M.-H.; Chua, N.-H.; Park, C.-M. A Membrane-Bound NAC Transcription Factor Regulates Cell Division in Arabidopsis. Plant Cell 2006, 18, 3132–3144. [Google Scholar] [CrossRef]
- Park, J.; Kim, Y.-S.; Kim, S.-G.; Jung, J.-H.; Woo, J.-C.; Park, C.-M. Integration of Auxin and Salt Signals by the NAC Transcription Factor NTM2 during Seed Germination in Arabidopsis. Plant Physiol. 2011, 156, 537–549. [Google Scholar] [CrossRef]
- Shibata, Y.; Shemesh, T.; Prinz, W.A.; Palazzo, A.F.; Kozlov, M.M.; Rapoport, T.A. Mechanisms Determining the Morphology of the Peripheral ER. Cell 2010, 143, 774–788. [Google Scholar] [CrossRef]
- Kriechbaumer, V.; Breeze, E.; Pain, C.; Tolmie, F.; Frigerio, L.; Hawes, C. Arabidopsis Lunapark Proteins Are Involved in ER Cisternae Formation. New Phytol. 2018, 219, 990–1004. [Google Scholar] [CrossRef]
- Stefano, G.; Brandizzi, F. Unique and Conserved Features of the Plant ER-Shaping GTPase RHD3. Cell. Logist. 2014, 4, e28217. [Google Scholar] [CrossRef]
- Karimi, M.; Depicker, A.; Hilson, P. Recombinational Cloning with Plant Gateway Vectors. Plant Physiol. 2007, 145, 1144–1154. [Google Scholar] [CrossRef]
- Minkenberg, B.; Zhang, J.; Xie, K.; Yang, Y. CRISPR-PLANT v2: An Online Resource for Highly Specific Guide RNA Spacers Based on Improved off-Target Analysis. Plant Biotechnol. J. 2019, 17, 5–8. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral Dip: A Simplified Method for Agrobacterium-Mediated Transformation of Arabidopsis Thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, Transient Expression of Fluorescent Fusion Proteins in Tobacco Plants and Generation of Stably Transformed Plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Frigerio, L.; Tolley, N.; Hawes, C. The Plant Endoplasmic Reticulum: A Cell-Wide Web. Biochem. J. 2009, 423, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Kovesi, P. Phase Congruency: A Low-Level Image Invariant. Psychol. Res. 2000, 64, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Voisin, M.; Vanrobays, E.; Tatout, C. Investigation of Nuclear Periphery Protein Interactions in Plants Using the Membrane Yeast Two-Hybrid (MbY2H) System. Methods Mol. Biol. 2018, 1840, 221–235. [Google Scholar] [CrossRef]
- Oldenburg, K.R.; Vo, K.T.; Michaelis, S.; Paddon, C. Recombination-Mediated PCR-Directed Plasmid Construction in Vivo in Yeast. Nucleic Acids Res. 1997, 25, 451–452. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Nielsen, H.; Tsirigos, K.D.; Brunak, S.; von Heijne, G. A Brief History of Protein Sorting Prediction. Protein J. 2019, 38, 200–216. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andov, B.; Boulaflous-Stevens, A.; Pain, C.; Mermet, S.; Voisin, M.; Charrondiere, C.; Vanrobays, E.; Tutois, S.; Evans, D.E.; Kriechbaumer, V.; et al. In Depth Topological Analysis of Arabidopsis Mid-SUN Proteins and Their Interaction with the Membrane-Bound Transcription Factor MaMYB. Plants 2023, 12, 1787. https://doi.org/10.3390/plants12091787
Andov B, Boulaflous-Stevens A, Pain C, Mermet S, Voisin M, Charrondiere C, Vanrobays E, Tutois S, Evans DE, Kriechbaumer V, et al. In Depth Topological Analysis of Arabidopsis Mid-SUN Proteins and Their Interaction with the Membrane-Bound Transcription Factor MaMYB. Plants. 2023; 12(9):1787. https://doi.org/10.3390/plants12091787
Chicago/Turabian StyleAndov, Bisa, Aurelia Boulaflous-Stevens, Charlotte Pain, Sarah Mermet, Maxime Voisin, Camille Charrondiere, Emmanuel Vanrobays, Sylvie Tutois, David E. Evans, Verena Kriechbaumer, and et al. 2023. "In Depth Topological Analysis of Arabidopsis Mid-SUN Proteins and Their Interaction with the Membrane-Bound Transcription Factor MaMYB" Plants 12, no. 9: 1787. https://doi.org/10.3390/plants12091787
APA StyleAndov, B., Boulaflous-Stevens, A., Pain, C., Mermet, S., Voisin, M., Charrondiere, C., Vanrobays, E., Tutois, S., Evans, D. E., Kriechbaumer, V., Tatout, C., & Graumann, K. (2023). In Depth Topological Analysis of Arabidopsis Mid-SUN Proteins and Their Interaction with the Membrane-Bound Transcription Factor MaMYB. Plants, 12(9), 1787. https://doi.org/10.3390/plants12091787






