Genetic Diversity and Population Structure of European Soybean Germplasm Revealed by Single Nucleotide Polymorphism
Abstract
1. Introduction
2. Results
2.1. Marker Quality and Filltering
2.2. Genetic Diversity
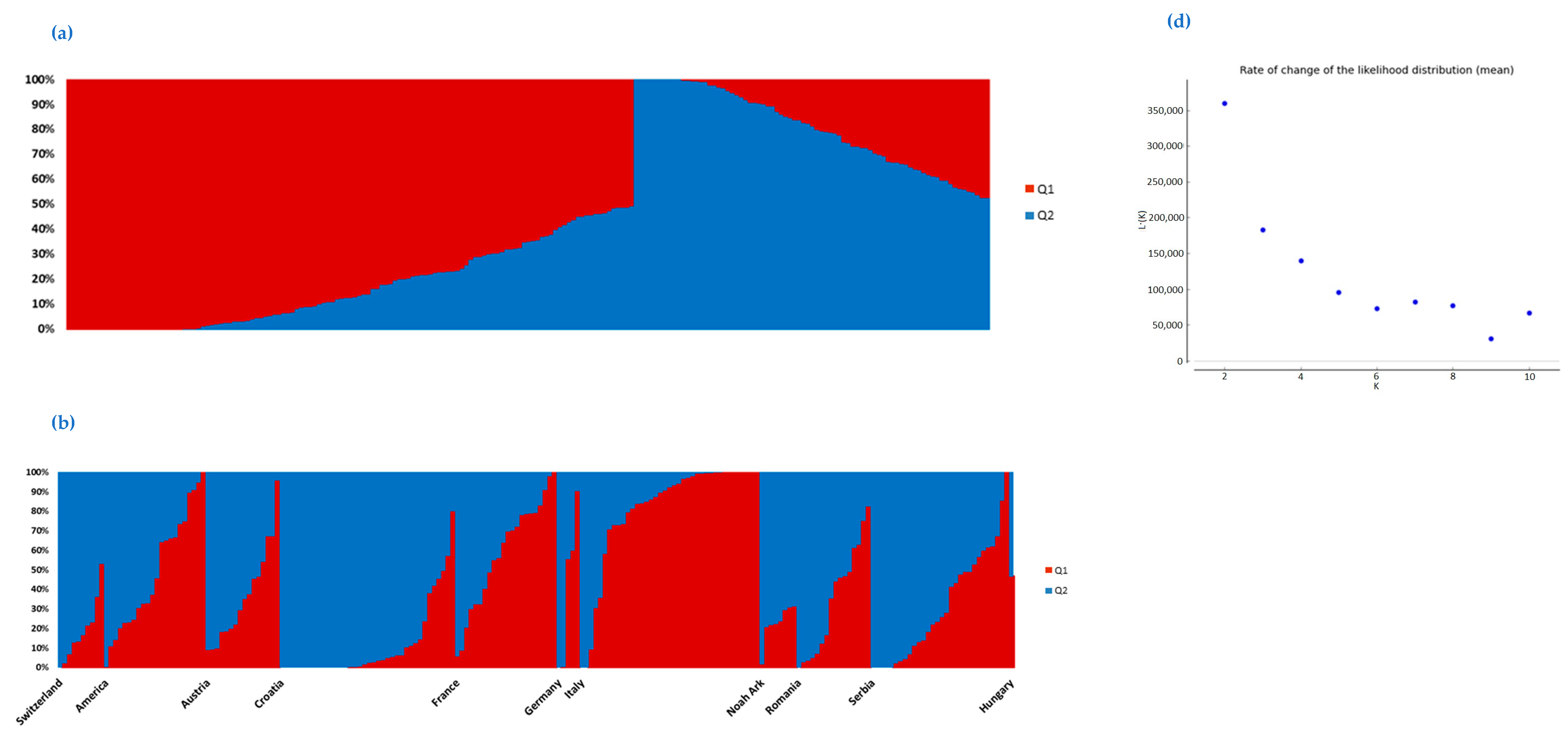
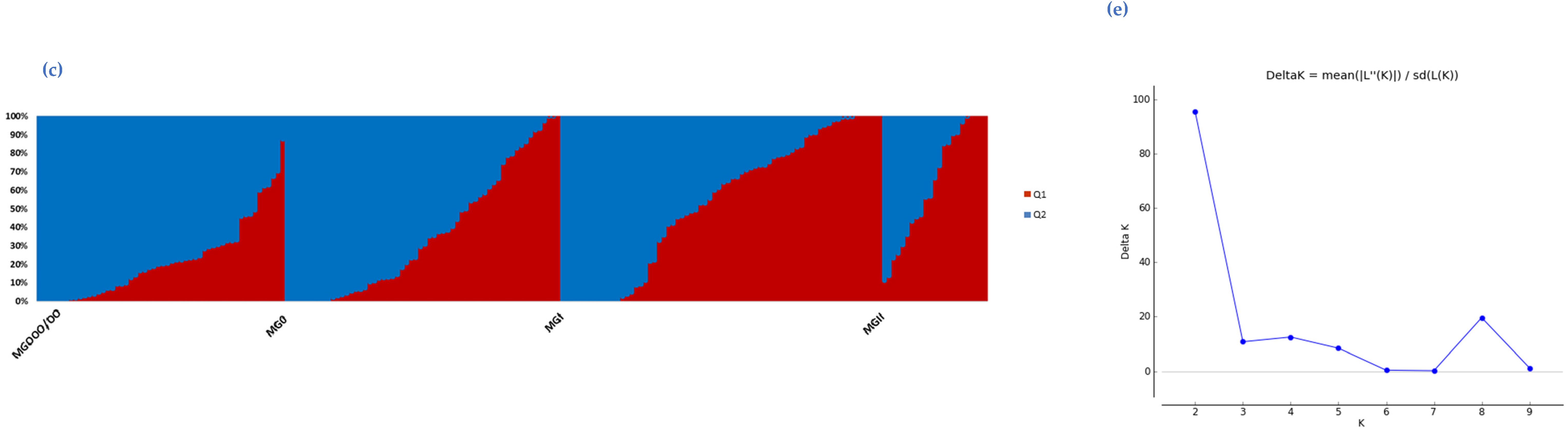
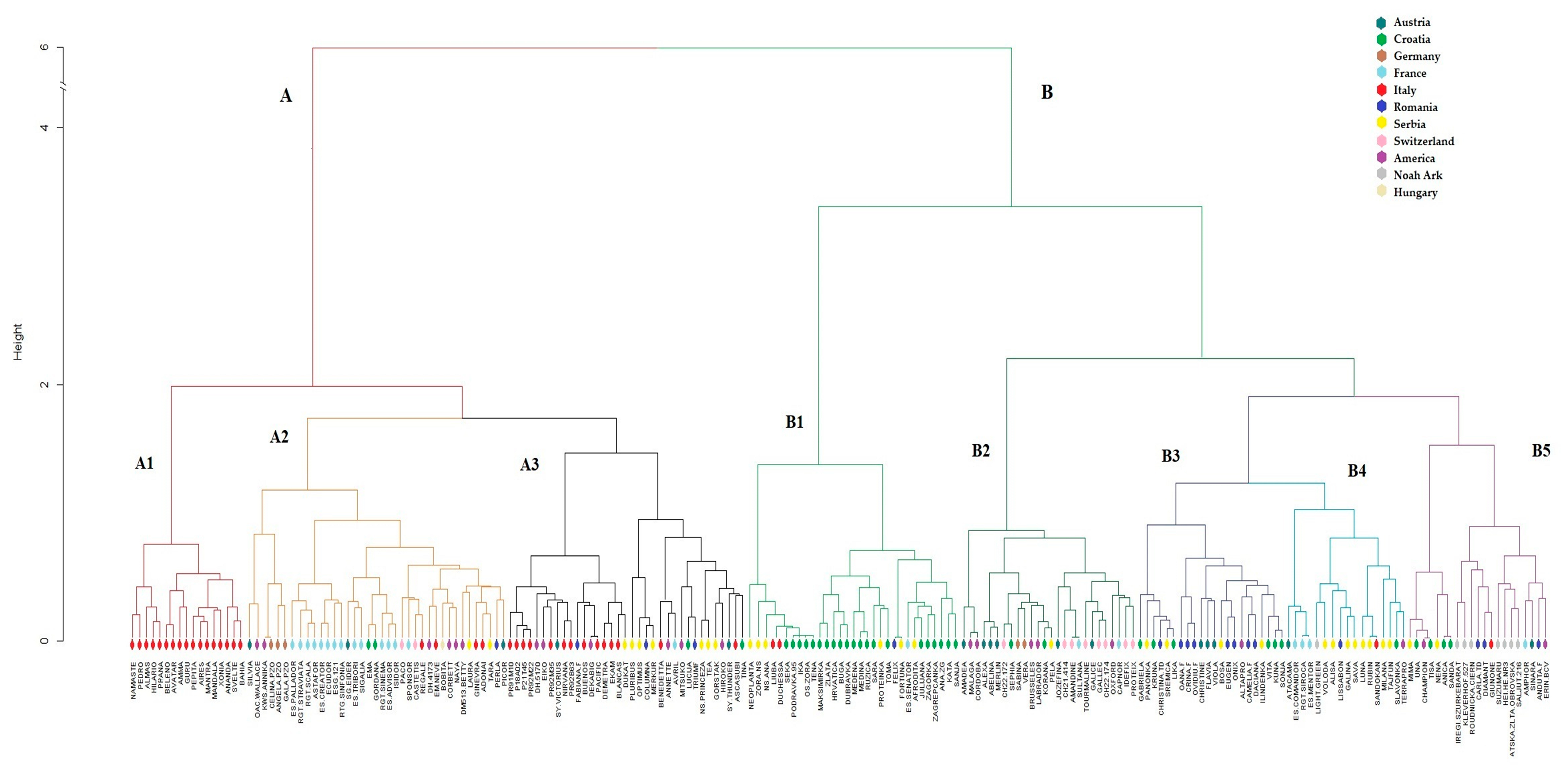
2.3. Population Structure of the Soybean Collection
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. SNP Genotyping and Data Filtering
4.3. Statistical Analysis and Genetic Differentiation of Soybean
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pagano, M.C.; Miransari, M. The importance of soybean production worldwide. Abiotic Biot. Stress. Soybean Prod. 2016, 1, 1–26. [Google Scholar] [CrossRef]
- Miladinović, J.; Hrustić, M.; Vidić, M. Soja, 1st ed.; Institut za Ratarstvo i Povrtarstvo, Sojaprotein, Bečej: Novi Sad, Serbia, 2008. [Google Scholar]
- Faostat; Food and Agriculture Organization of the United Nations: Rome, Italy, 2021; Volume 1.
- Hiel, R.; Geling, V.; de Vries, T.; Lan, C.-C.; Sleurink, N. European Soy Monitor; The Agency for a Healthy World: Geneva, Switzerland, 2020. [Google Scholar]
- Kurasch, A.K.; Hahn, V.; Leiser, W.L.; Vollmann, J.; Schori, A.; Bétrix, C.A.; Mayr, B.; Winkler, J.; Mechtler, K.; Aper, J.; et al. Identification of mega-environments in Europe and effect of allelic variation at maturity E loci on adaptation of European soybean. Plant Cell Environ. 2017, 40, 765–778. [Google Scholar] [CrossRef] [PubMed]
- Karges, K.; Bellingrath-Kimura, S.D.; Watson, C.A.; Stoddard, F.L.; Halwani, M.; Reckling, M. Agro-economic prospects for expanding soybean production beyond its current northerly limit in Europe. Eur. J. Agron. 2022, 133, 126415. [Google Scholar] [CrossRef]
- Bhanu, A.N. Assessment of Genetic Diversity in Crop Plants—An Overview. Adv. Plants Agric. Res. 2017, 7, 279–286. [Google Scholar] [CrossRef]
- Fang, J.; Zhu, X.; Wang, C.; Shangguan, L. Applications of DNA Technologies in Agriculture. Curr. Genomics 2016, 17, 379. [Google Scholar] [CrossRef] [PubMed]
- Tavaud-Pirra, M.; Sartre, P.; Nelson, R.; Santoni, S.; Texier, N.; Roumet, P. Genetic Diversity in a Soybean Collection. Crop Sci. 2009, 49, 895–902. [Google Scholar] [CrossRef]
- Rotzler, D.; Stamp, P.; Betrix, C.-A.; De Groote, J.-C.; Moullet, O.; Schori, A. Agronomic interest of lanceolate leaf in soybean. Agrarforschung 2009, 16, 472–477. [Google Scholar]
- Žulj Mihaljević, M.; Šarčević, H.; Lovrić, A.; Andrijanić, Z.; Sudarić, A.; Jukić, G.; Pejić, I. Genetic diversity of European commercial soybean [Glycine max (L.) Merr.] germplasm revealed by SSR markers. Genet. Resour. Crop Evol. 2020, 67, 1587–1600. [Google Scholar] [CrossRef]
- Vratarić, M.; Sudarić, A. Soja Glycine max. (L.) Merr., 2nd ed.; Poljoprivredni Institut Osijek: Osijek, Croatia, 2008. [Google Scholar]
- Ristova, D.; Šarčević, H.; Šimon, S.; Mihajlov, L.; Pejić, I. Genetic diversity in southeast european soybean germplasm revealed by SSR markers. Agric. Conspec. Sci. 2010, 75, 21–26. [Google Scholar]
- Hahn, V.; Würschum, T. Molecular genetic characterization of Central European soybean breeding germplasm. Plant Breed. 2014, 133, 748–755. [Google Scholar] [CrossRef]
- Miladinović, J.; Ćeran, M.; Đorđević, V.; Balešević-Tubić, S.; Petrović, K.; Đukić, V.; Miladinović, D. Allelic Variation and Distribution of the Major Maturity Genes in Different Soybean Collections. Front. Plant Sci. 2018, 9, 1286. [Google Scholar] [CrossRef] [PubMed]
- Saleem, A.; Muylle, H.; Aper, J.; Ruttink, T.; Wang, J.; Yu, D.; Roldán-Ruiz, I. A Genome-Wide Genetic Diversity Scan Reveals Multiple Signatures of Selection in a European Soybean Collection Compared to Chinese Collections of Wild and Cultivated Soybean Accessions. Front. Plant Sci. 2021, 12, 256. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Xu, J.Y.; Liu, Z.X.; Pachner, M.; Molin, E.M.; Rittler, L.; Hahn, V.; Leiser, W.; Gu, Y.Z.; Lu, Y.Q.; et al. Genetic diversity in early maturity Chinese and European elite soybeans: A comparative analysis. Euphytica 2023, 219, 17. [Google Scholar] [CrossRef]
- Bandillo, N.; Jarquin, D.; Song, Q.; Nelson, R.; Cregan, P.; Specht, J.; Lorenz, A. A Population Structure and Genome-Wide Association Analysis on the USDA Soybean Germplasm Collection. Plant Genome 2015, 8, plantgenome2015.04.0024. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, J.N.; Li, Z. Genomic signatures of North American soybean improvement inform diversity enrichment strategies and clarify the impact of hybridization. G3 Genes Genomes Genet. 2016, 6, 2693–2705. [Google Scholar] [CrossRef] [PubMed]
- Bruce, R.W.; Torkamaneh, D.; Grainger, C.; Belzile, F.; Eskandari, M.; Rajcan, I. Genome-wide genetic diversity is maintained through decades of soybean breeding in Canada. Theor. Appl. Genet. 2019, 132, 3089–3100. [Google Scholar] [CrossRef]
- Song, Q.; Hyten, D.L.; Jia, G.; Quigley, C.V.; Fickus, E.W.; Nelson, R.L.; Cregan, P.B. Development and Evaluation of SoySNP50K, a High-Density Genotyping Array for Soybean. PLoS ONE 2013, 8, e54985. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, H.; Wen, Z.; Fan, X.; Li, Y.; Guan, R.; Guo, Y.; Wang, S.; Wang, D.; Qiu, L. Comparison of genetic diversity between Chinese and american soybean (Glycine max (L.)) accessions revealed by high-density SNPs. Front. Plant Sci. 2017, 8, 2014. [Google Scholar] [CrossRef] [PubMed]
- Chander, S.; Garcia-Oliveira, A.L.; Gedil, M.; Shah, T.; Otusanya, G.O.; Asiedu, R.; Chigeza, G. Genetic diversity and population structure of soybean lines adapted to sub-saharan africa using single nucleotide polymorphism (Snp) markers. Agronomy 2021, 11, 604. [Google Scholar] [CrossRef]
- Abe, J.; Xu, D.H.; Suzuki, Y.; Kanazawa, A.; Shimamoto, Y. Soybean germplasm pools in Asia revealed by nuclear SSRs. Theor. Appl. Genet. 2003, 106, 445–453. [Google Scholar] [CrossRef]
- Li, A.Q.; Zhao, C.Z.; Wang, X.J.; Liu, Z.J.; Zhang, L.F.; Song, G.Q.; Yin, J.; Li, C.S.; Xia, H.; Bi, Y.P. Identification of SSR markers using soybean (Glycine max) ESTs from globular stage embryos. Electron. J. Biotechnol. 2010, 13, 6–7. [Google Scholar] [CrossRef]
- Shurtleff, W.; Aoyagi, A. Soy Oil and Soybean Meal—Part 1. A Chapter from the Unpublished Manuscript, History of Soybeans and Soyfoods, 1100 B.C. to the 1980s. In History of Soybean Crushing; Soyinfo Center: Lafayette, CA, USA, 2007. [Google Scholar]
- Signor, M. Experiences of soybean cropping and breeding in Northern Italy. In Proceedings of the Meeting Sustainable Soya—Sustainable Europe, Budapest, Hungary, 24–25 November 2016. [Google Scholar]
- Fu, Y.B.; Cober, E.R.; Morrison, M.J.; Marsolais, F.; Peterson, G.W.; Horbach, C. Patterns of genetic variation in a soybean germplasm collection as characterized with genotyping-by-sequencing. Plants 2021, 10, 1611. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Zhai, H.; Liu, B.; Kong, F.; Yuan, X.; Wu, H.; Cober, E.R.; Harada, K. Molecular identification of genes controlling flowering time, maturity, and photoperiod response in soybean. Plant Syst. Evol. 2012, 298, 1217–1227. [Google Scholar] [CrossRef]
- Miladinovié, J.; Nlihailovié, V.; Dordevic, V.; Vasiljevic, S.; Katanski, S.; Zivanov, D.; Randelovié, P. The importance of legume genetic resources for breeding. Ratar. Povrt. 2021, 58, 94–103. [Google Scholar] [CrossRef]
- Haupt, M.; Schmid, K. Combining focused identification of germplasm and core collection strategies to identify genebank accessions for central European soybean breeding. Plant Cell Environ. 2020, 43, 1421–1436. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 19, 395. [Google Scholar] [CrossRef]
- Adamack, A.T.; Gruber, B. PopGenReport: Simplifying basic population genetic analyses in R. Methods Ecol. Evol. 2014, 5, 384–387. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Goudet, J. HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef]

| No. of Cultivars | He | PIC | MAF | |
|---|---|---|---|---|
| Overall | 207 | 0.34 | 0.27 | 0.25 |
| Origin | ||||
| Austria | 16 | 0.31 | 0.25 | 0.23 |
| Croatia | 38 | 0.28 | 0.23 | 0.21 |
| France | 22 | 0.29 | 0.23 | 0.21 |
| Italy | 39 | 0.30 | 0.24 | 0.22 |
| Romania | 16 | 0.29 | 0.24 | 0.21 |
| Serbia | 30 | 0.31 | 0.25 | 0.22 |
| Switzerland | 10 | 0.24 | 0.19 | 0.18 |
| America | 22 | 0.33 | 0.26 | 0.24 |
| Maturity group | ||||
| MG000/00 | 54 | 0.31 | 0.25 | 0.23 |
| MG0 | 60 | 0.33 | 0.26 | 0.24 |
| MGI | 70 | 0.32 | 0.26 | 0.24 |
| MGII | 23 | 0.32 | 0.25 | 0.23 |
| Source of Variation | Country of Origin | Maturity Group | ||||
|---|---|---|---|---|---|---|
| df | Sigma | % | df | Sigma | % | |
| Between Populations | 7 | 172 | 9.8 | 3 | 80.1 | 4.5 |
| Within Populations | 185 | 1592 | 90.2 | 203 | 1684.7 | 95.5 |
| Total | 192 | 1764 | 100 | 206 | 1764.8 | 100 |
| Country of Origin | Austria | Croatia | France | Italy | Romania | Serbia | Switzerland | Average | Range |
|---|---|---|---|---|---|---|---|---|---|
| America | 0.05 | 0.11 | 0.07 | 0.06 | 0.06 | 0.06 | 0.19 | 0.08 | (0.05–0.19) |
| Austria | 0.12 | 0.10 | 0.09 | 0.08 | 0.08 | 0.20 | 0.10 | (0.05–0.20) | |
| Croatia | 0.15 | 0.15 | 0.15 | 0.12 | 0.23 | 0.15 | (0.11–0.23) | ||
| France | 0.12 | 0.13 | 0.10 | 0.24 | 0.13 | (0.07–0.24) | |||
| Italy | 0.12 | 0.09 | 0.23 | 0.12 | (0.06–0.23) | ||||
| Romania | 0.09 | 0.25 | 0.12 | (0.06–0.25) | |||||
| Serbia | 0.18 | 0.10 | (0.06–0.18) | ||||||
| Switzerland | 0.22 | (0.18–0.25) | |||||||
| FstAVG | 0.128 | ||||||||
| Maturity group | MG0 | MGI | MGII | Average | Range | ||||
| MG000/00 | 0.051 | 0.065 | 0.070 | 0.062 | (0.051–0.070) | ||||
| MG0 | 0.030 | 0.045 | 0.042 | (0.030–0.051) | |||||
| MGI | 0.058 | 0.051 | (0.030–0.065) | ||||||
| MGII | 0.058 | (0.045–0.070) | |||||||
| FstAVG | 0.053 |
| Country of Origin | No. | Maturity Group | No. |
|---|---|---|---|
| Austria | 16 | I | 70 |
| Croatia | 38 | II | 23 |
| France | 22 | 0 | 60 |
| Germany | 5 | 000/00 | 54 |
| Hungary | 1 | ||
| Italy | 39 | ||
| America | 22 | ||
| Noah Ark | 8 | ||
| Romania | 16 | ||
| Serbia | 30 | ||
| Switzerland | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrijanić, Z.; Nazzicari, N.; Šarčević, H.; Sudarić, A.; Annicchiarico, P.; Pejić, I. Genetic Diversity and Population Structure of European Soybean Germplasm Revealed by Single Nucleotide Polymorphism. Plants 2023, 12, 1837. https://doi.org/10.3390/plants12091837
Andrijanić Z, Nazzicari N, Šarčević H, Sudarić A, Annicchiarico P, Pejić I. Genetic Diversity and Population Structure of European Soybean Germplasm Revealed by Single Nucleotide Polymorphism. Plants. 2023; 12(9):1837. https://doi.org/10.3390/plants12091837
Chicago/Turabian StyleAndrijanić, Zoe, Nelson Nazzicari, Hrvoje Šarčević, Aleksandra Sudarić, Paolo Annicchiarico, and Ivan Pejić. 2023. "Genetic Diversity and Population Structure of European Soybean Germplasm Revealed by Single Nucleotide Polymorphism" Plants 12, no. 9: 1837. https://doi.org/10.3390/plants12091837
APA StyleAndrijanić, Z., Nazzicari, N., Šarčević, H., Sudarić, A., Annicchiarico, P., & Pejić, I. (2023). Genetic Diversity and Population Structure of European Soybean Germplasm Revealed by Single Nucleotide Polymorphism. Plants, 12(9), 1837. https://doi.org/10.3390/plants12091837







