Identification, Characterization, and Cytological Analysis of Several Unexpected Hybrids Derived from Reciprocal Crosses between Raphanobrassica and Its Diploid Parents
Abstract
1. Introduction
2. Results
2.1. Development of Hybrids from Reciprocal Crosses
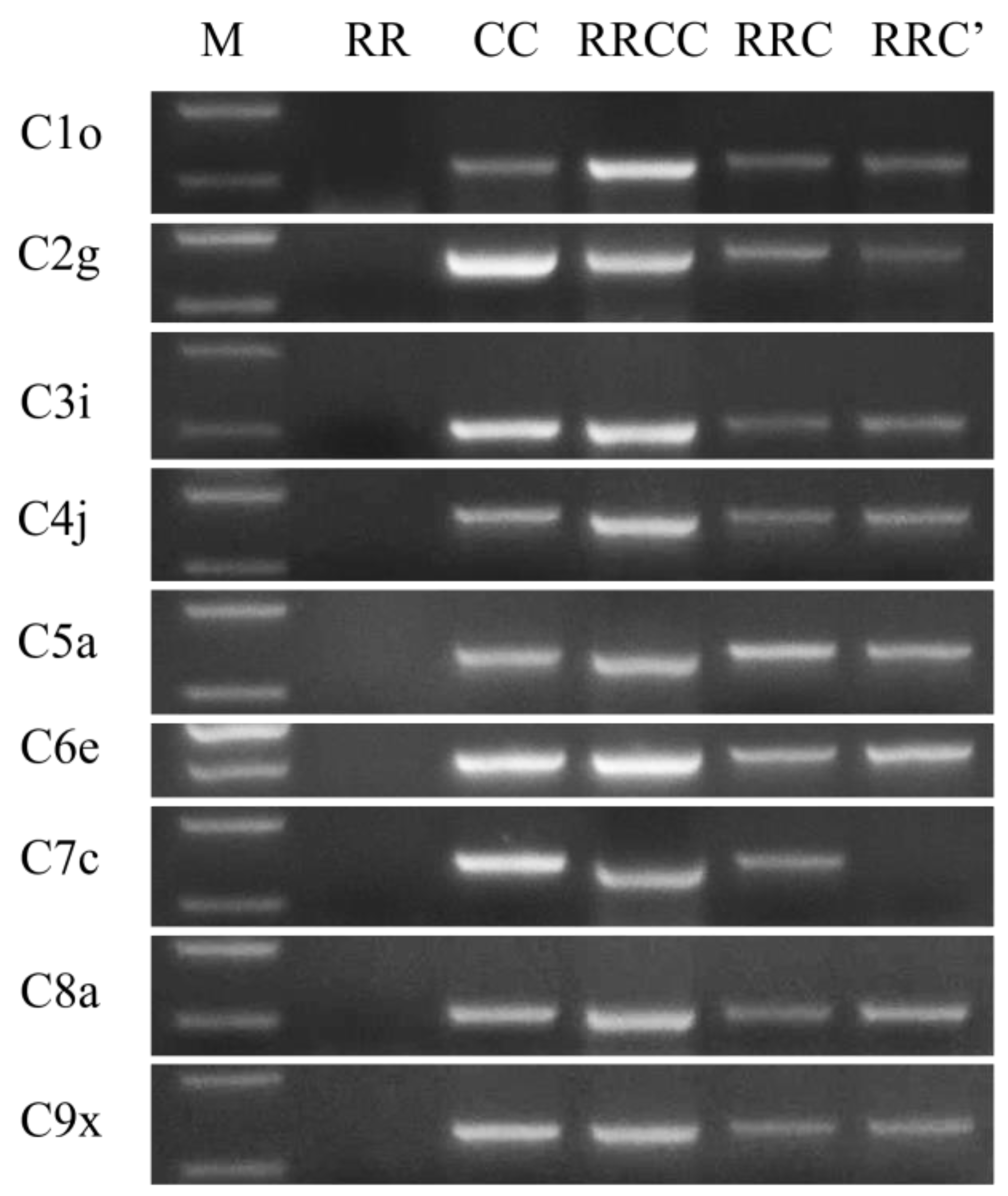
2.2. Determination of Chromosome Constitutions of the Unexpected Hybrids
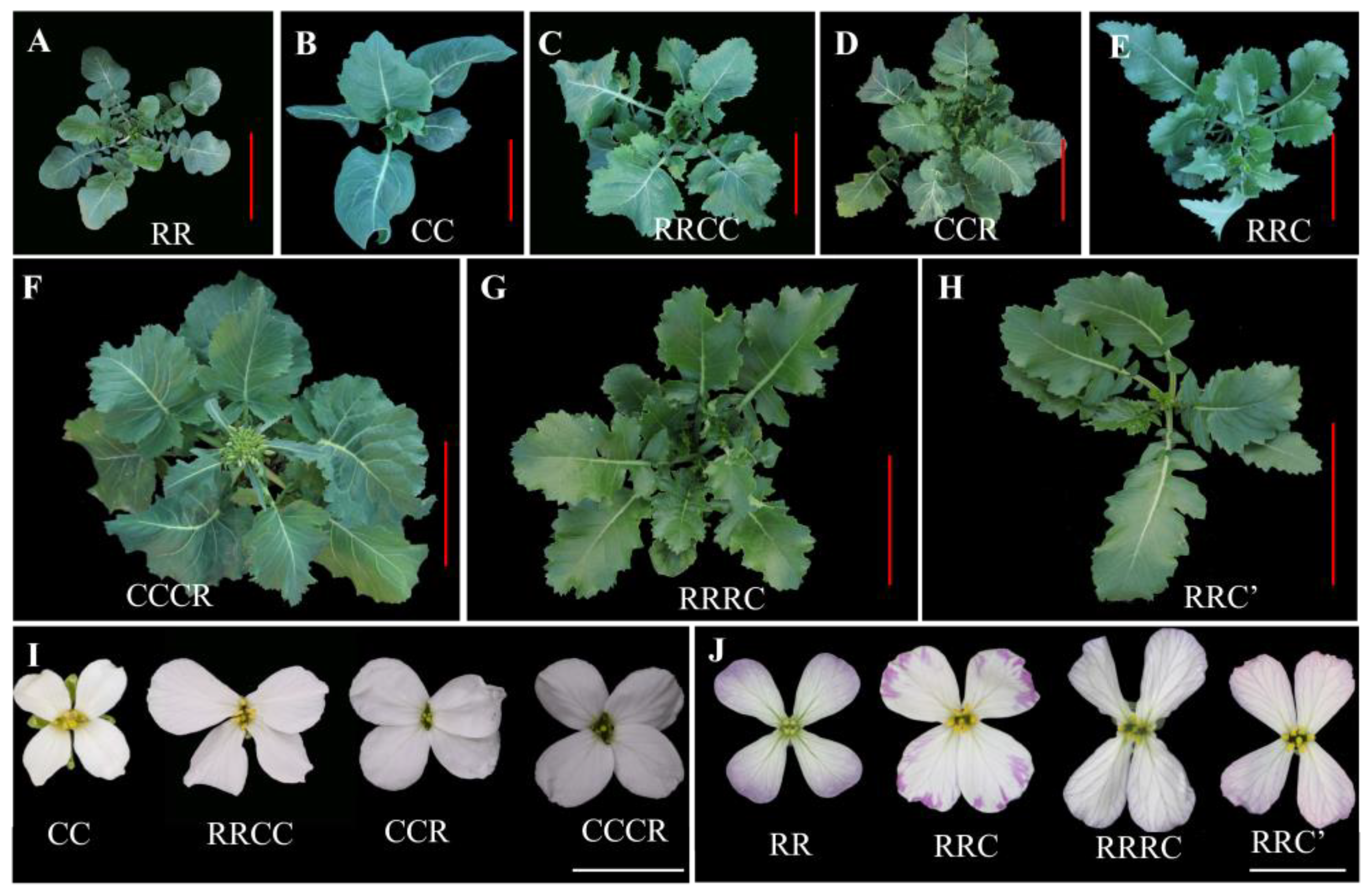
2.3. Morphology and Fertility of the Unexpected Hybrids
2.4. Chromosome Behaviours of the Triploid and Unexpected Hybrids
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Crosses
4.2. PCR Screening of C Chromosome-Specific Primers
4.3. Morphological and Cytological Analysis of These Hybrids
4.4. Probe Preparation and Fluorescence In Situ Hybridization (FISH) Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Soltis, P.S.; Soltis, D.E. Polyploidy and Genome Evolution; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y. Double the genome, double the fun: Genome duplications n angiosperms. Mol. Plant 2018, 11, 357–358. [Google Scholar] [CrossRef]
- Zhu, B.; Tu, Y.; Zeng, P.; Ge, X.; Li, Z. Extraction of the constituent subgenomes of the natural allopolyploid rapeseed (Brassica napus L.). Genetics 2016, 204, 1015–1027. [Google Scholar] [CrossRef]
- Katche, E.; Quezada-Martinez, D.; Katche, E.I.; Vasquez-Teuber, P.; Mason, A.S. Interspecific hybridization for Brassica crop improvement. Crop Breed. Genet. Genom. 2019, 1. [Google Scholar] [CrossRef]
- An, H.; Qi, X.; Gaynor, M.L.; Hao, Y.; Gebken, S.C.; Mabry, M.E.; McAlvay, A.C.; Teakle, G.R.; Conant, G.C.; Barker, M.S.; et al. Transcriptome and organellar sequencing highlights the complex origin and diversification of allotetraploid Brassica napus. Nat. Commun. 2019, 10, 2878. [Google Scholar] [CrossRef] [PubMed]
- Park, H.R.; Park, J.E.; Kim, J.H.; Shin, H.; Yu, S.H.; Son, S.; Yi, G.; Lee, S.S.; Kim, H.H.; Huh, J.H. Meiotic chromosome stability and suppression of crossover between non-homologous chromosomes in x Brassicoraphanus, an intergeneric allotetraploid derived from a cross between Brassica rapa and Raphanus sativus. Front. Plant Sci. 2020, 11, 851. [Google Scholar] [CrossRef]
- Feng, Q.; Yu, J.; Yang, X.; Lv, X.; Lu, Y.; Yuan, J.; Du, X.; Zhu, B.; Li, Z. Development and characterization of an allooctaploid (AABBCCRR) incorporating Brassica and radish genomes via two rounds of interspecific hybridizations. Sci. Hortic. 2022, 293, 110730. [Google Scholar] [CrossRef]
- Zhang, L.; Miao, L.; He, J.; Li, H.; Li, M. The Transcriptome and Metabolome Reveal the Potential Mechanism of Lodging Resistance in Intergeneric Hybrids between Brassica napus and Capsella bursa-pastoris. Int. J. Mol. Sci. 2022, 23, 4481. [Google Scholar] [CrossRef]
- Kaminishi, A.; Miyohashi, F.; Kita, N. Diversity of morphological traits and glucosinolate composition in backcross progenies of Brassica rapa L. × Eruca vesicaria (L.) Cav. hybrids. Euphytica 2022, 218, 114. [Google Scholar] [CrossRef]
- Zhou, J.; Tan, C.; Cui, C.; Ge, X.; Li, Z. Distinct subgenome stabilities in synthesized Brassica allohexaploids. Theor. Appl. Genet. 2016, 129, 1257–1271. [Google Scholar] [CrossRef]
- Li, Q.; Xu, B.; Du, Y.; Peng, A.; Ren, X.; Si, J.; Song, H. Development of Ogura CMS restorers in Brassica oleracea subspecies via direct Rfo B gene transformation. Theor. Appl. Genet. 2021, 134, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Zhang, D.; Zhao, H.; Su, T.; Zhao, X.; Wang, W.; Li, P.; Yu, Y.; Wang, J.; Yu, S. Identification of a Monosomic Alien Chromosome Addition Line Responsible for the Purple Color Trait in Heading Chinese Cabbage. Horticulturae 2023, 9, 146. [Google Scholar] [CrossRef]
- Mason, A.; Chèvre, A.-M. Optimization of Recombination in Interspecific Hybrids to Introduce New Genetic Diversity into Oilseed Rape (Brassica napus L.); CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Quezada-Martinez, D.; Addo Nyarko, C.P.; Schiessl, S.V.; Mason, A.S. Using wild relatives and related species to build climate resilience in Brassica crops. Theor. Appl. Genet. 2021, 134, 1711–1728. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Banga, S.S. Heteroploidy in Brassica juncea: Basics and Applications. In The Brassica juncea Genome; Springer: Berlin/Heidelberg, Germany, 2022; pp. 115–145. [Google Scholar]
- Roberts, M.A.; Reader, S.M.; Dalgliesh, C.; Miller, T.E.; Foote, T.N.; Fish, L.J.; Snape, J.; Moore, G. Induction and characterization of Ph1 wheat mutants. Genetics 1999, 153, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
- Serra, H.; Svačina, R.; Baumann, U.; Whitford, R.; Sutton, T.; Bartoš, J.; Sourdille, P. Ph2 encodes the mismatch repair protein MSH7-3D that inhibits wheat homoeologous recombination. Nat. Commun. 2021, 12, 803. [Google Scholar] [CrossRef] [PubMed]
- Jenczewski, E.; Eber, F.; Grimaud, A.; Huet, S.; Lucas, M.O.; Monod, H.; Chevre, A.M. PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 2003, 164, 645–653. [Google Scholar] [CrossRef]
- Cui, C.; Ge, X.; Gautam, M.; Kang, L.; Li, Z. Cytoplasmic and genomic effects on meiotic pairing in Brassica hybrids and allotetraploids from pair crosses of three cultivated diploids. Genetics 2012, 191, 725–738. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef]
- Warwick, S.I. Brassicaceae in agriculture. Genet. Genom. Brassicaceae 2011, 33–65. [Google Scholar] [CrossRef]
- Nagaharu, U.J.J.J.B.; Nagaharu, N. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Yang, Y.W.; Tai, P.Y.; Chen, Y.; Li, W.H. A study of the phylogeny of Brassica rapa, B. nigra, Raphanus sativus, and their related genera using noncoding regions of chloroplast DNA. Mol. Phylogenetics Evol. 2002, 23, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Zeng, T.; Feng, Q.; Hu, L.; Luo, X.; Weng, Q.; He, J.; Zhu, B. The complete chloroplast genome sequence of yellow mustard (Sinapis alba L.) and its phylogenetic relationship to other Brassicaceae species. Gene 2020, 731, 144340. [Google Scholar] [CrossRef] [PubMed]
- Kolte, S.J. The search for resistance to major diseases of rapeseed and mustard in India. GCIRC Rapeseed Congr. 1991, 1991, 219–225. [Google Scholar]
- Siemens, J. Interspecific hybridisation between wild relatives and Brassica napus to introduce new resistance traits into the oilseed rape gene pool. Czech J. Genet. Plant Breed. 2002, 38, 155–157. [Google Scholar] [CrossRef]
- Zhan, Z.; Nwafor, C.C.; Hou, Z.; Gong, J.; Zhu, B.; Jiang, Y.; Zhou, Y.; Wu, J.; Piao, Z.; Tong, Y.; et al. Cytological and morphological analysis of hybrids between Brassicoraphanus, and Brassica napus for introgression of clubroot resistant trait into Brassica napus L. PLoS ONE 2017, 12, e0177470. [Google Scholar] [CrossRef]
- Krämer, R.; Scholze, P.; Marthe, F.; Ryschka, U.; Klocke, E.; Schumann, G. Improvement of disease resistance in cabbage: 1. Turnip mosaic virus, TuMV. Gesunde Pflanz. 2003, 55, 193–198. [Google Scholar] [CrossRef]
- Peterka, H.; Budahn, H.; Schrader, O.; Ahne, R.; Schütze, W. Transfer of resistance against the beet cyst nematode from radish (Raphanus sativus) to rape (Brassica napus) by monosomic chromosome addition. Theor. Appl. Genet. 2004, 109, 30–41. [Google Scholar] [CrossRef]
- Prakash, S.; Bhat, S.R.; Quiros, C.F.; Kirti, P.B.; Chopra, V.L. Brassica and its close allies: Cytogenetics and evolution. Plant Breed. Rev. 2011, 31, 21–187. [Google Scholar]
- Lee, S.S.; Hwang, B.H.; Kim, T.Y.; Yang, J.; Han, N.R.; Kim, J.; Kim, H.H.; Belandres, H.R. Developing stable cultivar through microspore mutagenesis in× Brassicoraphanus koranhort, inter-generic allopolyploid between Brassica rapa and Raphanus sativus. Am. J. Plant Sci. 2017, 8, 1345–1356. [Google Scholar] [CrossRef]
- Chai, A.L.; Xie, X.W.; Shi, Y.X.; Li, B.J. Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Can. J. Plant Pathol. 2014, 36 (Suppl. S1), 142–153. [Google Scholar] [CrossRef]
- Ogura, H. Studies on the new male sterility in Japanese radish with special reference to the utilization of this steri195ty towards the practical raising of hybrid seeds. Mem. Fac. Agric. Kagoshima Univ. 1968, 6, 40–75. [Google Scholar]
- Sakai, T.; Imamura, J. Intergeneric transfer of cytoplasmic male sterility between Raphanus sativus (cms line) and Brassica napus through cytoplast-protoplast fusion. Theor. Appl. Genet. 1990, 80, 421–427. [Google Scholar] [CrossRef]
- Lee, S.S.; Lee, S.A.; Yang, J.; Kim, J. Developing stable progenies of× Brassicoraphanus, an intergeneric allopolyploid between Brassica rapa and Raphanus sativus, through induced mutation using microspore culture. Theor. Appl. Genet. 2011, 122, 885–891. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Cai, Z.; Hu, T.; Liu, H.; Bao, C.; Mao, W.; Jin, W. Repetitive sequence analysis and karyotyping reveals centromere-associated DNA sequences in radish (Raphanus sativus L.). BMC Plant Biol. 2015, 15, 105. [Google Scholar] [CrossRef]
- McNaughton, I.H. Synthesis and sterility of Raphanobrassica. Euphytica 1973, 22, 70–88. [Google Scholar] [CrossRef]
- Zhang, L.; He, J.; He, H.; Wu, J.; Li, M. Genome-wide unbalanced expression bias and expression level dominance toward Brassica oleracea in artificially synthesized intergeneric hybrids of Raphanobrassica. Hortic. Res. 2021, 8, 246. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, S.; Liu, Z.; Zhao, W.; Zhang, X.; Song, J.; Jia, H.; Yang, W.; Ma, Y.; Wang, Y. Characterization and acceleration of genome shuffling and ploidy reduction in synthetic allopolyploids by genome sequencing and editing. Nucleic Acids Res. 2023, 51, 198–217. [Google Scholar] [CrossRef]
- Chèvre, A.M.; Eber, F.; Darmency, H.; Fleury, A.; Picault, H.; Letanneur, J.C.; Renard, M. Assessment of interspecific hybridization between transgenic oilseed rape and wild radish under normal agronomic conditions. Theor. Appl. Genet. 2000, 100, 1233–1239. [Google Scholar] [CrossRef]
- Mason, A.S.; Nelson, M.N.; Yan, G.; Cowling, W.A. Production of viable male unreduced gametes in Brassica interspecific hybrids is genotype specific and stimulated by cold temperatures. BMC Plant Biol. 2021, 11, 103. [Google Scholar] [CrossRef]
- Inomata, N. A cytogenetic study of the progenies of hybrids between Brassica napus and Brassica oleracea, Brassica bourgeaui, Brassica cretica and Brassica montana. Plant Breed. 2002, 121, 174–176. [Google Scholar] [CrossRef]
- Higgins, E.E.; Howell, E.C.; Armstrong, S.J.; Parkin, I.A. A major quantitative trait locus on chromosome A9, BnaPh1, controls homoeologous recombination in Brassica napus. New Phytol. 2021, 229, 3281–3293. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, H.L.; Luo, P. Production and cytogenetics of intergeneric hybrids between Brassica napus and Orychophragmus violaceus. Theor. Appl. Genet. 1995, 91, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Alix, K.; Joets, J.; Ryder, C.D.; Moore, J.; Barker, G.C.; Bailey, J.P.; King, G.J.; Heslop-Harrison, J.S. The CACTA transposon Bot1 played a major role in Brassica genome divergence and gene proliferation. Plant J. 2008, 56, 1030–1044. [Google Scholar] [CrossRef]
- Zhong, X.B.; de Jong, J.H.; Zabel, P. Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH). Chromosome Res. 1996, 4, 24–28. [Google Scholar] [CrossRef] [PubMed]
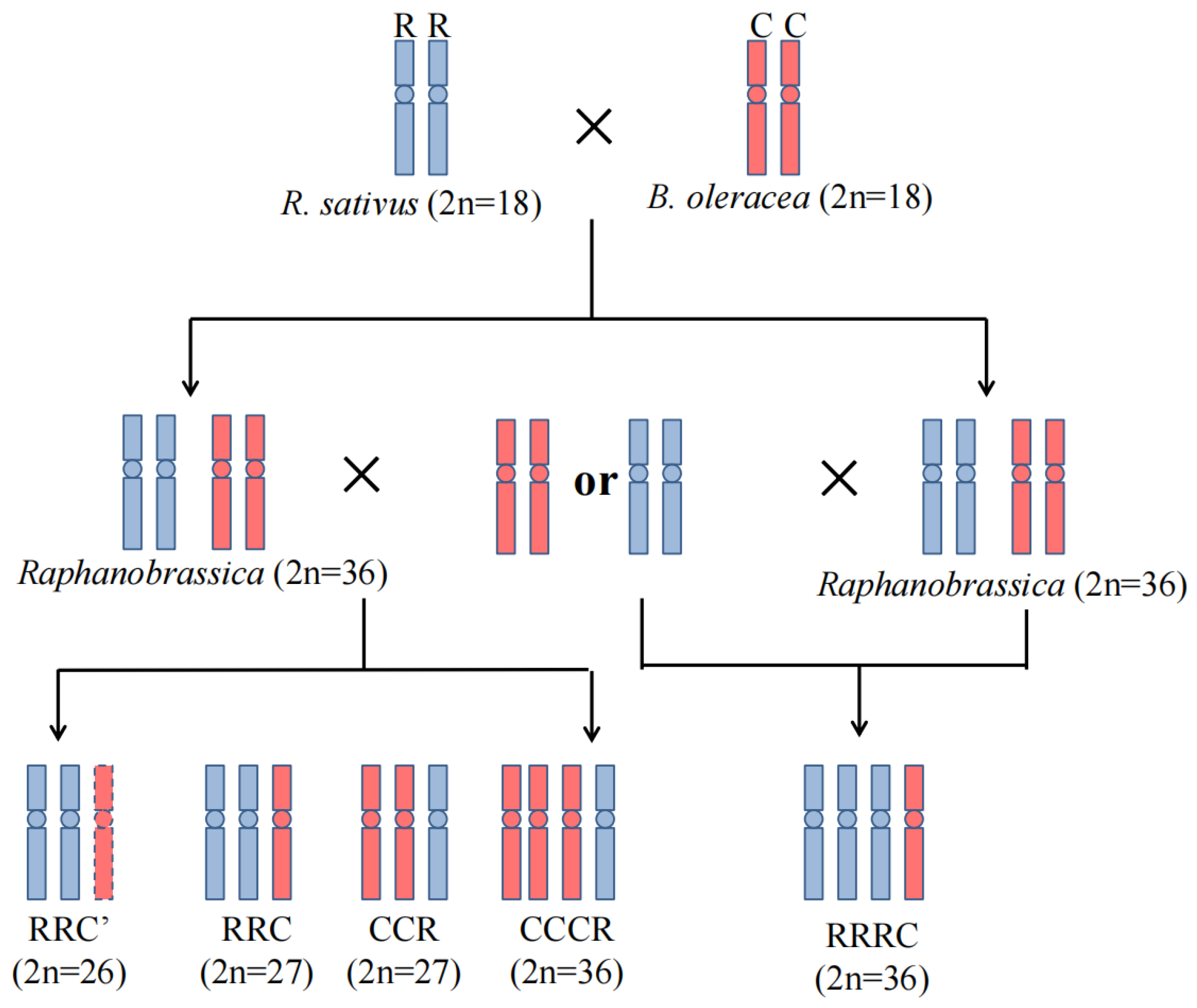


| Cross Combinations | Emasculated Flowers Number | Obtained Embryos (%) | Genome Constitution of Embryos (%) |
|---|---|---|---|
| CC × RRCC | 523 | 0 | — |
| RR × RRCC | 174 | 1 (0.57) | RRRC (0.57) |
| RRCC × CC | 432 | 3 (0.69) | 2 CCR (0.46); 1 CCCR (0.23) |
| RRCC × RR | 548 | 10 (1.82) | 2 RRC (0.36); 1 RRC’ (0.18); 7 RRCC (1.28) |
| Total | 1677 | 14 (0.83) | — |
| Hybrids Types | R Chromosome Signals | C Chromosome Signals |
|---|---|---|
| RRC’ | 18 | 8 |
| RRC | 18 | 9 |
| RRRC | 27 | 9 |
| CCR | 9 | 18 |
| CCCR | 9 | 27 |
| Hybrids Types | Total PMCs | Homoeologous Pairing in PMCs | Ratio | Allosyndesis Types | Autosyndesis Types |
|---|---|---|---|---|---|
| RRCb | 49 | 35 | 71.43% | R-C; R-R-C; C-C-R; R-C-R; C-R-C; R-C-C-R; C-C-C-R; R-R-R-C; | C-C |
| RRRCa | 17 | 15 | 88.23% | R-C; R-R-C; C-C-R; R-R-C-R; C-C-C-R; C-C-R-R; C-C-C-C-R; | C-C |
| CCRc | 31 | 11 | 35.48% | R-C; C-C-R; C-C-R-R; R-C-C-R; C-C-C-C-R; | C-C-C; R-R-R; R-R |
| CCCRb | 49 | 34 | 69.39% | R-C; R-R-C; C-C-R; R-C-R; C-R-C; C-C-C-R; C-C-R-C; C-C-C-R-R; | R-R-R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Lei, S.; Fang, S.; Tai, N.; Yu, W.; Yang, Z.; Gu, L.; Wang, H.; Du, X.; Zhu, B.; et al. Identification, Characterization, and Cytological Analysis of Several Unexpected Hybrids Derived from Reciprocal Crosses between Raphanobrassica and Its Diploid Parents. Plants 2023, 12, 1875. https://doi.org/10.3390/plants12091875
Yu J, Lei S, Fang S, Tai N, Yu W, Yang Z, Gu L, Wang H, Du X, Zhu B, et al. Identification, Characterization, and Cytological Analysis of Several Unexpected Hybrids Derived from Reciprocal Crosses between Raphanobrassica and Its Diploid Parents. Plants. 2023; 12(9):1875. https://doi.org/10.3390/plants12091875
Chicago/Turabian StyleYu, Jie, Shaolin Lei, Shiting Fang, Niufang Tai, Wei Yu, Ziwei Yang, Lei Gu, Hongcheng Wang, Xuye Du, Bin Zhu, and et al. 2023. "Identification, Characterization, and Cytological Analysis of Several Unexpected Hybrids Derived from Reciprocal Crosses between Raphanobrassica and Its Diploid Parents" Plants 12, no. 9: 1875. https://doi.org/10.3390/plants12091875
APA StyleYu, J., Lei, S., Fang, S., Tai, N., Yu, W., Yang, Z., Gu, L., Wang, H., Du, X., Zhu, B., & Cai, M. (2023). Identification, Characterization, and Cytological Analysis of Several Unexpected Hybrids Derived from Reciprocal Crosses between Raphanobrassica and Its Diploid Parents. Plants, 12(9), 1875. https://doi.org/10.3390/plants12091875







