Genome-Editing of FtsZ1 for Alteration of Starch Granule Size in Potato Tubers
Abstract
1. Introduction
2. Results
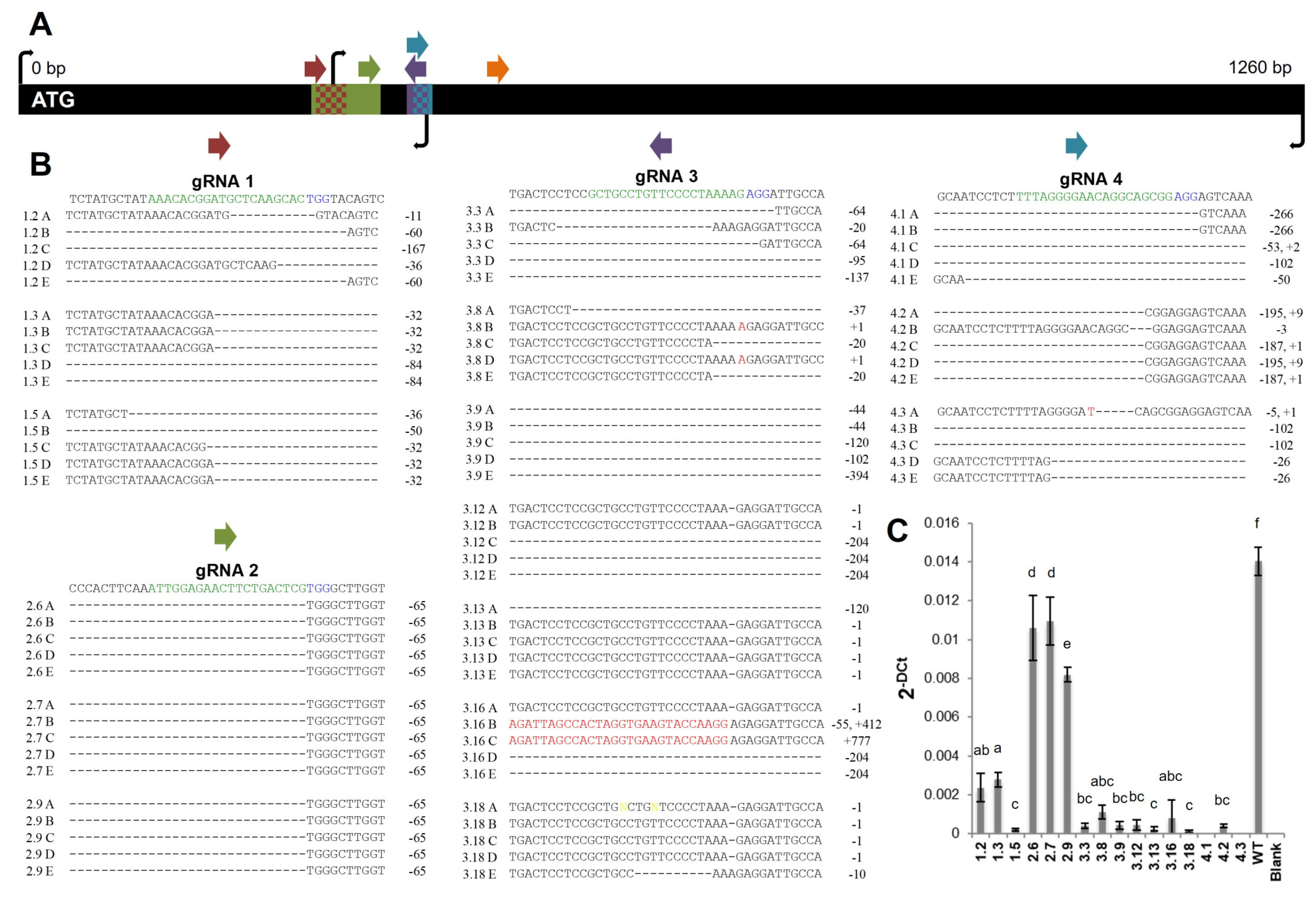
2.1. Generation of Potato Lines with Reduced FtsZ1 Expression Levels
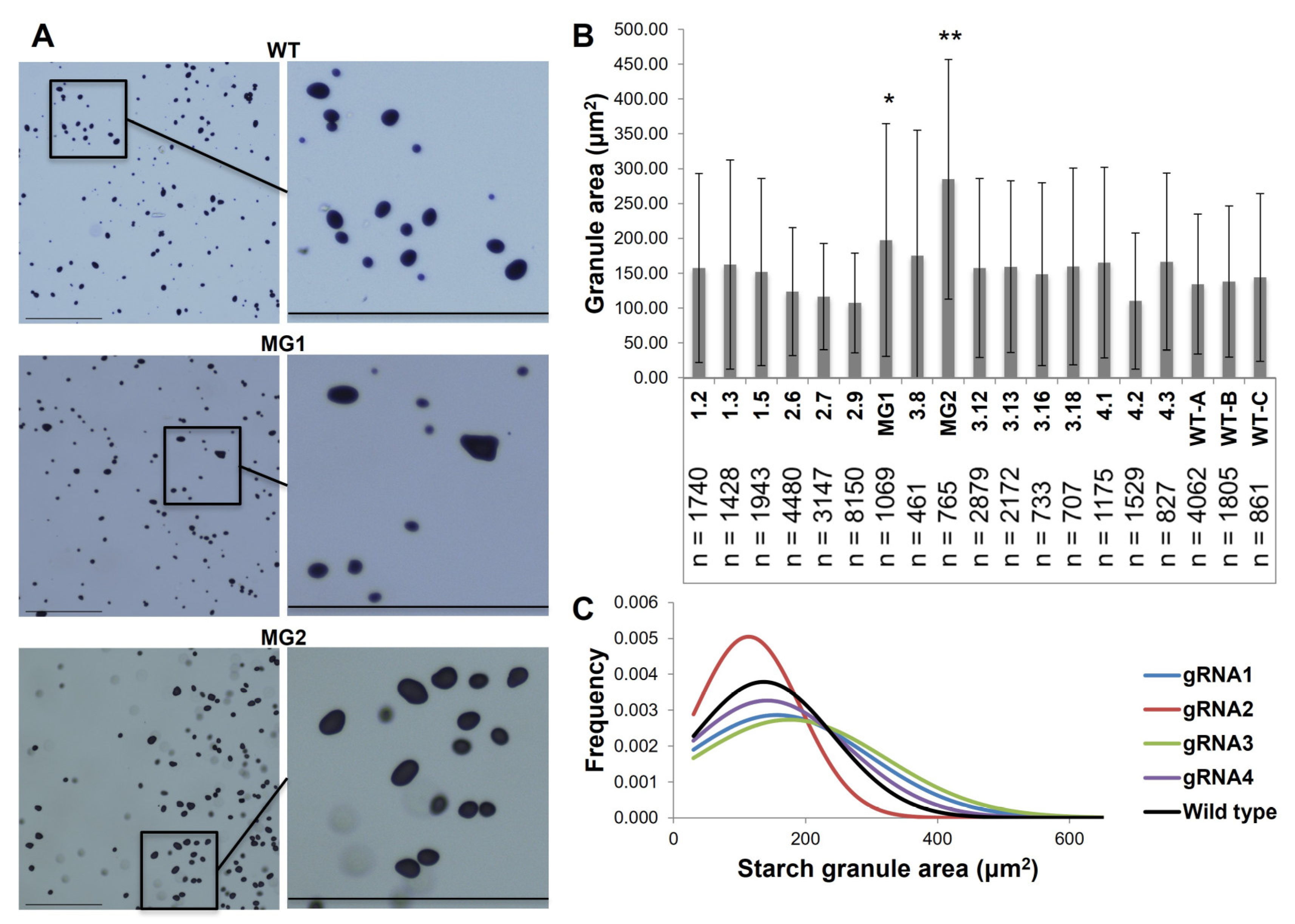
2.2. The Reduction in FtsZ1 Expression Produced Potato Tubers with Increased Starch Granule Size without Comprising Nutritional Quality
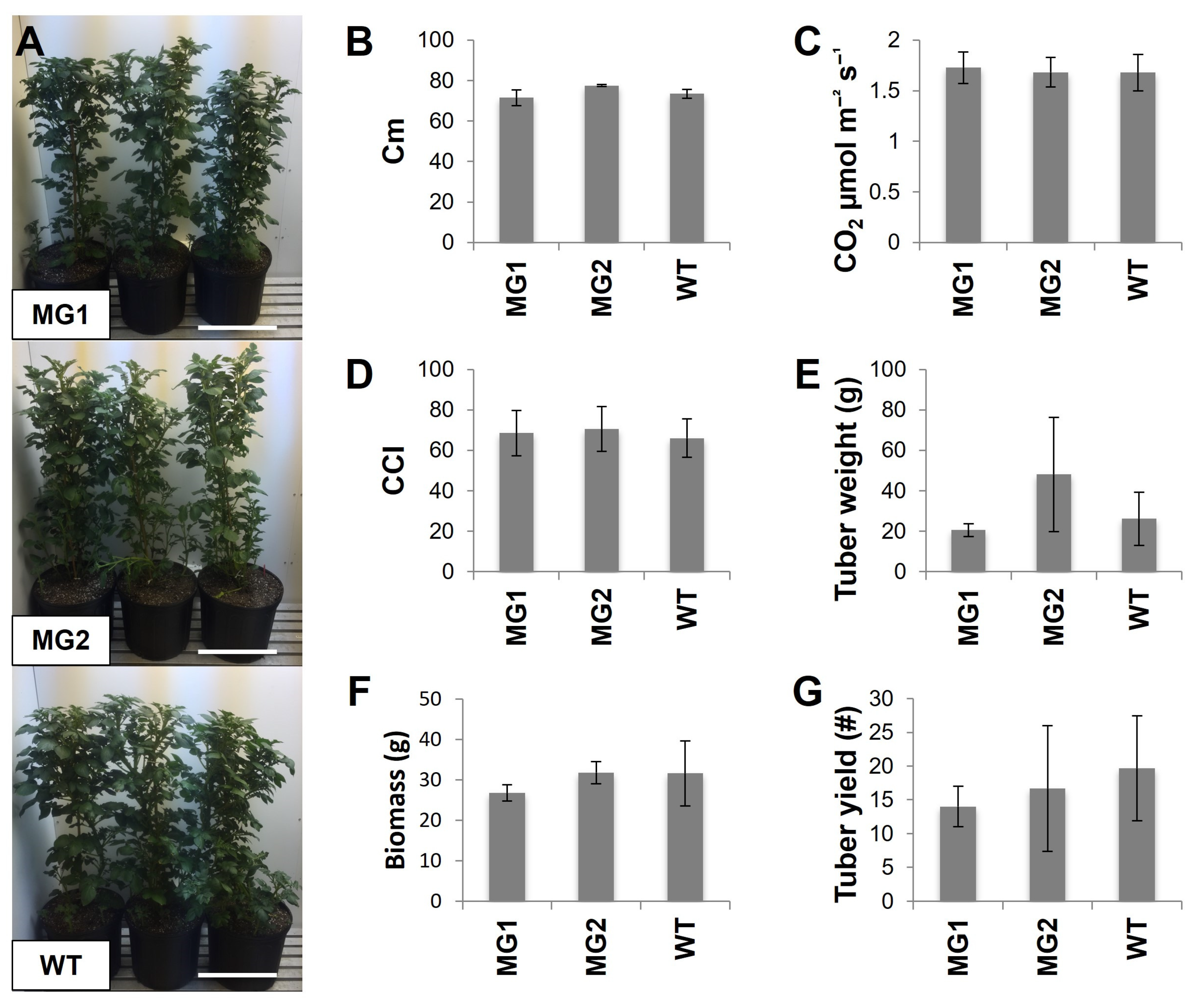
2.3. The Phenotypes and Growth Characteristics of MacroGranule1 and MacroGranule2 Plants Were Comparable to Non-Edited Wild-Type Plants
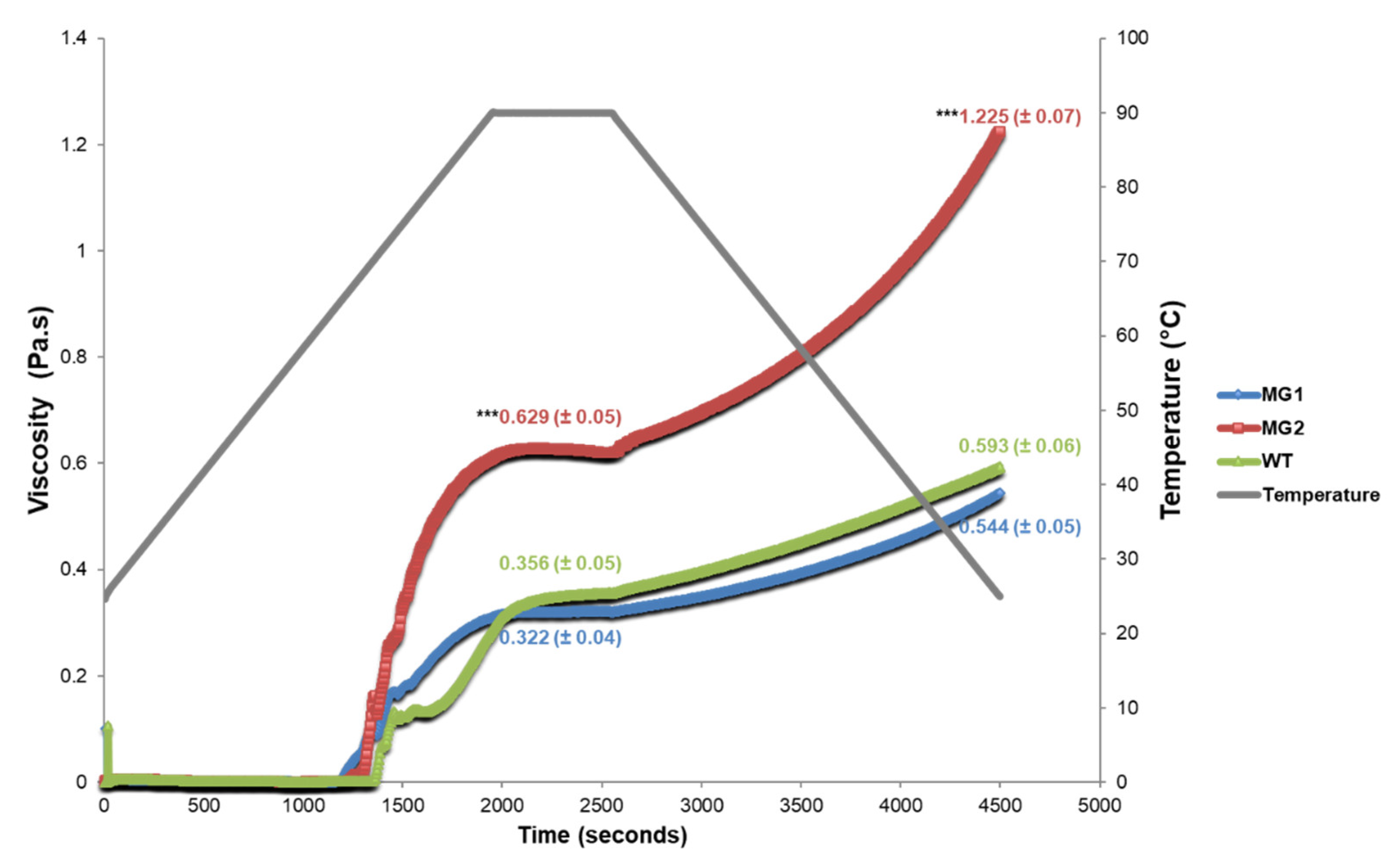
2.4. MacroGranule2 Starch Displays a Higher Viscosity Level than MacroGranule1 and Wild-Type
3. Discussion
4. Materials and Methods
4.1. FtsZ1 CDS Cloning/Sequencing
4.2. Vector Construction
4.3. Plant Transformation
4.4. Genomic DNA Extraction and Molecular Analysis
4.5. Cloning and Sequencing of Mutated Amplicons
4.6. RT PCR and q-RT PCR
4.7. Starch Granule Size Determination
4.8. Confocal Microscopy
4.9. Growth Studies
4.10. Tuber Analysis
4.11. Starch Pasting Properties
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barrell, P.J.; Meiyalaghan, S.; Jacobs, J.M.; Conner, A.J. Applications of biotechnology and genomics in potato improvement. Plant Biotechnol. J. 2013, 11, 907–920. [Google Scholar] [CrossRef]
- Lutaladio, N.C.; Castaldi, L. Potato: The hidden treasure. J. Food Compos. Anal. 2009, 22, 491–493. [Google Scholar] [CrossRef]
- Devaux, A.; Kromann, P.; Ortiz, O. Potatoes for sustainable global food security. Potato Res. 2014, 57, 185–199. [Google Scholar] [CrossRef]
- Chakraborty, R.; Kalita, P.; Sen, S. Natural starch in biomedical and food Industry: Perception and overview. Curr. Drug Discov. Technol. 2019, 16, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.P.; Cochrane, M.P.; Dale, M.F.B.; Duffus, C.M.; Lynn, A.; Morrison, I.M.; Prentice, R.D.M.; Swanston, J.S.; Tiller, S.A. Starch production and industrial use. J. Sci. Food Agric. 1999, 77, 289–311. [Google Scholar] [CrossRef]
- Kraak, A. Industrial applications of potato starch products. Ind. Crops Prod. 1992, 1, 107–112. [Google Scholar] [CrossRef]
- Burrell, M.M. Starch: The need for improved quality or quantity—An overview. J. Exp. Bot. 2003, 54, 451–456. [Google Scholar] [CrossRef]
- Jobling, S. Improving starch for food and industrial applications. Curr. Opin. Plant Biol. 2004, 7, 210–218. [Google Scholar] [CrossRef]
- Tharanathan, R.N. Starch—Value addition by modification. Crit. Rev. Food Sci. Nutr. 2005, 45, 371–384. [Google Scholar] [CrossRef]
- Davis, J.P.; Supatcharee, N.; Khandelwal, R.L.; Chibbar, R.N. Synthesis of novel starches in planta: Opportunities and challenges. Starch 2003, 55, 107–120. [Google Scholar] [CrossRef]
- Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; Wang, J.; et al. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Butler, N.M.; Atkins, P.A.; Voytas, D.F.; Douches, D.S. Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS ONE 2015, 10, e0144591. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, S.; Wang, W.; Xiong, X.; Meng, F.; Cui, X. Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 2015, 34, 1473–1476. [Google Scholar] [CrossRef]
- Andersson, M.; Turesson, H.; Nicolia, A.; Fält, A.S.; Samuelsson, M.; Hofvander, P. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 2017, 36, 117–128. [Google Scholar] [CrossRef]
- Nakayasu, M.; Akiyama, R.; Lee, H.J.; Osakabe, K.; Osakabe, Y.; Watanabe, B.; Sugimoto, Y.; Umemoto, N.; Saito, K.; Muranaka, T.; et al. Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol. Biochem. 2018, 131, 70–77. [Google Scholar] [CrossRef]
- Enciso-Rodriguez, F.; Manrique-Carpintero, N.C.; Nadakuduti, S.S.; Buell, C.R.; Zarka, D.; Douches, D. Overcoming self-incompatibility in diploid potato using CRISPR-Cas9. Front. Plant Sci. 2019, 10, 376. [Google Scholar] [CrossRef]
- Veillet, F.; Perrot, L.; Chauvin, L.; Kermarrec, M.P.; Guyon-Debast, A.; Chauvin, J.E.; Nogue, F.; Mazier, M. Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 2019, 20, 402. [Google Scholar] [CrossRef] [PubMed]
- Johansen, I.E.; Liu, Y.; Jørgensen, B.; Bennett, E.P.; Andreasson, E.; Nielsen, K.L.; Blennow, A.; Petersen, B.L. High efficacy full allelic CRISPR/Cas9 gene editing in tetraploid potato. Sci. Rep. 2019, 9, 17715. [Google Scholar] [CrossRef]
- Tuncel, A.; Corbin, K.R.; Ahn-Jarvis, J.; Harris, S.; Hawkins, E.; Smedley, M.A.; Harwood, W.; Warren, F.J.; Patron, N.J.; Smith, A.M. Cas9-mediated mutagenesis of potato starch-branching enzymes generates a range of tuber starch phenotypes. Plant Biotechnol. J. 2019, 17, 2259–2271. [Google Scholar] [CrossRef] [PubMed]
- Naeem, M.; Tetlow, I.J.; Emes, M.J. Starch synthesis in amyloplasts purified from developing potato tubers. Plant J. 2002, 11, 1095–1103. [Google Scholar] [CrossRef]
- Smith, A.M.; Denyer, K.; Martin, C. The synthesis of the starch granule. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 67–87. [Google Scholar] [CrossRef] [PubMed]
- Kram, A.M.; Oostergetel, G.T.; Van Bruggen, E. Localization of branching enzyme in potato tuber cells with the use of immunoelectron microscopy. Plant Physiol. 1993, 101, 237–243. [Google Scholar] [CrossRef]
- Ji, Q.; Oomen, R.J.; Vincken, J.P.; Bolam, D.N.; Gilbert, H.J.; Suurs, L.C.; Visser, R.G. Reduction of starch granule size by expression of an engineered tandem starch-binding domain in potato plants. Plant Biotechnol. J. 2004, 2, 251–260. [Google Scholar] [CrossRef]
- Lindeboom, N.; Chang, P.R.; Tyler, R.T. Analytical, biochemical and physicochemical aspects of starch granule size, with emphasis on small granule starches: A review. Starch 2004, 56, 89–99. [Google Scholar] [CrossRef]
- Park, S.H.; Wilson, J.D.; Seabourn, B.W. Starch granule size distribution of hard red winter and hard red spring wheat: Its effects on mixing and breadmaking quality. J. Cereal Sci. 2009, 49, 98–105. [Google Scholar] [CrossRef]
- Smith, A.M.; Martin, C. Biosynthesis and Manipulation of Plant Products, 1st ed.; Springer: Dordrecht, The Netherlands, 1993. [Google Scholar] [CrossRef]
- Stoddard, F.L. Genetics of starch granule size distribution in tetraploid and hexaploid wheats. Aust. J. Agric. Res. 2003, 54, 637–648. [Google Scholar] [CrossRef]
- Chen, Z.; Schols, H.A.; Voragen, A.G.J. Starch granule size strongly determines starch noodle processing and noodle quality. J. Food Sci. 2006, 68, 1584–1589. [Google Scholar] [CrossRef]
- Gutierrez, O.A.; Campbell, M.R.; Glover, D.V. Starch particle volume in single- and double-mutant maize endosperm genotypes involving the soft starch (h) gene. Crop. Sci. 2002, 42, 355–359. [Google Scholar] [CrossRef]
- Niu, S.; Li, X.Q.; Tang, R.; Zhang, G.; Li, X.; Cui, B.; Mikitzel, L.; Haroon, M. Starch granule sizes and degradation in sweet potatoes during storage. Postharvest Biol. Technol. 2019, 150, 137–147. [Google Scholar] [CrossRef]
- Liebers, M.; Grübler, B.; Chevalier, F.; Lerbs-Mache, S.; Merendino, L.; Blanvillain, R.; Pfannschmidt, T. Regulatory shifts in plastid transcription play a key role in morphological conversions of plastids during plant development. Front. Plant Sci. 2017, 8, 23. [Google Scholar] [CrossRef]
- Pyke, K.A. Plastid division and development. Plant Cell 1999, 11, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Osteryoung, K.W.; Pyke, K.A. Division and dynamic morphology of plastids. Annu. Rev. Plant Biol. 2014, 65, 443–472. [Google Scholar] [CrossRef]
- Vitha, S.; McAndrew, R.S.; Osteryoung, K.W. FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 2001, 153, 111–120. [Google Scholar] [CrossRef]
- Osteryoung, K.W.; Stokes, K.D.; Rutherford, S.M.; Percival, A.L.; Li, W.Y. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 1998, 10, 1991–2004. [Google Scholar] [CrossRef]
- Strepp, R.; Scholz, S.; Kruse, S.; Speth, V.; Reski, R. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA 1998, 95, 4368–4373. [Google Scholar] [CrossRef]
- Donachie, W.D. The cell cycle of Escherichia coli. Annu. Rev. Microbiol. 1993, 47, 199–230. [Google Scholar] [CrossRef]
- Margolin, W. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 2005, 6, 862–871. [Google Scholar] [CrossRef]
- TerBush, A.D.; Yoshida, Y.; Osteryoung, K.W. FtsZ in chloroplast division: Structure, function and evolution. Curr. Opin. Cell Biol. 2013, 25, 461–470. [Google Scholar] [CrossRef]
- Yoshida, Y.; Mogi, Y.; TerBush, A.D.; Osteryoung, K.W. Chloroplast FtsZ assembles into a contractible ring via tubulin-like heteropolymerization. Nat. Plants 2016, 2, 16095. [Google Scholar] [CrossRef]
- Lutkenhaus, J.F.; Wolf-Watz, H.; Donachie, W.D. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J. Bacteriol. 1980, 142, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.J.; Glynn, J.M.; Olson, B.J.; Stokes, K.D.; Osteryoung, K.W. Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol. Plant 2009, 2, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- McAndrew, R.S.; Froehlich, J.E.; Vitha, S.; Stokes, K.D.; Osteryoung, K.W. Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 2001, 127, 1656–1666. [Google Scholar] [CrossRef] [PubMed]
- Stokes, K.D.; McAndrew, R.S.; Figueroa, R.; Vitha, S.; Osteryoung, K.W. Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 2000, 124, 1668–1677. [Google Scholar] [CrossRef] [PubMed]
- de Pater, S.; Caspers, M.; Kottenhagen, M.; Meima, H.; ter Stege, R.; de Vetten, N. Manipulation of starch granule size distribution in potato tubers by modulation of plastid division. Plant Biotechnol. J. 2006, 4, 123–134. [Google Scholar] [CrossRef]
- Yun, M.S.; Kawagoe, Y. Septum formation in amyloplasts produces compound granules in the rice endosperm and is regulated by plastid division proteins. Plant Cell Physiol. 2010, 51, 1469–1479. [Google Scholar] [CrossRef]
- Occhialini, A.; Pfotenhauer, A.C.; Frazier, T.P.; Li, L.; Harbison, S.A.; Lail, A.J.; Mebane, Z.; Piatek, A.A.; Rigoulot, S.B.; Daniell, H.; et al. Generation, analysis, and transformation of macro-chloroplast potato (Solanum tuberosum) lines for chloroplast biotechnology. Sci. Rep. 2020, 10, 21144. [Google Scholar] [CrossRef]
- Christensen, D.H.; Madsen, M.H. Changes in potato starch quality during growth. Potato Res. 1996, 39, 43–50. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 2016, 7, 377. [Google Scholar] [CrossRef]
- Zhu, S.; Yu, X.; Li, Y.; Sun, Y.; Zhu, Q.; Sun, J. Highly efficient targeted gene editing in upland cotton using the CRISPR/Cas9 system. Int. J. Mol. Sci. 2018, 19, 3000. [Google Scholar] [CrossRef]
- Bánfalvi, Z.; Csákvári, E.; Villányi, V.; Kondrák, M. Generation of transgene-free PDS mutants in potato by Agrobacterium-mediated transformation. BMC Biotechnol. 2020, 20, 25. [Google Scholar] [CrossRef]
- Stupar, R.M.; Lilly, J.W.; Town, C.D.; Cheng, Z.; Kaul, S.; Buell, C.R.; Jiang, J. Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: Implication of potential sequencing errors caused by large-unit repeats. Proc. Natl. Acad. Sci. USA 2001, 98, 5099–5103. [Google Scholar] [CrossRef]
- Lough, A.N.; Roark, L.M.; Kato, A.; Ream, T.S.; Lamb, J.C.; Birchler, J.A.; Newton, K.J. Mitochondrial DNA transfer to the nucleus generates extensive insertion site variation in maize. Genetics 2008, 178, 47–55. [Google Scholar] [CrossRef]
- Michalovova, M.; Vyskot, B.; Kejnovsky, E. Analysis of plastid and mitochondrial DNA insertions in the nucleus (NUPTs and NUMTs) of six plant species: Size, relative age and chromosomal localization. Heredity 2013, 111, 314–320. [Google Scholar] [CrossRef]
- Puertas, M.J.; González-Sánchez, M. Insertions of mitochondrial DNA into the nucleus-effects and role in cell evolution. Genome 2020, 63, 365–374. [Google Scholar] [CrossRef]
- McAndrew, R.S.; Olson, B.J.; Kadirjan-Kalbach, D.K.; Chi-Ham, C.L.; Vitha, S.; Froehlich, J.E.; Osteryoung, K.W. In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem. J. 2008, 412, 367–378. [Google Scholar] [CrossRef]
- Okechukwu, P.E.; Rao, M.A. Influence of granule size on viscosity of cornstarch suspension. J. Texture Stud. 1995, 26, 501–516. [Google Scholar] [CrossRef]
- Rao, M.A.; Tattiyakul, J. Granule size and rheological behavior of heated tapioca starch dispersions. Carbohydr. Polym. 1999, 38, 123–132. [Google Scholar] [CrossRef]
- Kumar, R.; Khatkar, B.S. Thermal, pasting and morphological properties of starch granules of wheat (Triticum aestivum L.) varieties. J. Food Sci. Technol. 2017, 54, 2403–2410. [Google Scholar] [CrossRef]
- Waterschoot, J.; Gomand, S.V.; Fierens, E.; Delcour, J.A. Starch blends and their physicochemical properties. Starch 2014, 67, 1–13. [Google Scholar] [CrossRef]
- Zhu, F.; Hua, Y.; Li, G. Physicochemical properties of potato, sweet potato and quinoa starch blends. Food Hydrocoll. 2020, 100, 105278. [Google Scholar] [CrossRef]
- Waterschoot, J.; Gomand, S.V.; Willebrords, J.K.; Fierens, E.; Delcour, J.A. Pasting properties of blends of potato, rice and maize starches. Food Hydrocoll. 2014, 41, 298–308. [Google Scholar] [CrossRef]
- Chung, K.M.; Moon, T.W.; Kim, H.; Chun, J.K. Physicochemical properties of sonicated mung bean, potato, and rice starches. Cereal Chem. 2002, 79, 631–633. [Google Scholar] [CrossRef]
- Scott, S.E.; Inbar, Y.; Wirz, C.D.; Brossard, D.; Rozin, P. An overview of attitudes toward genetically engineered food. Annu. Rev. Nutr. 2018, 38, 459–479. [Google Scholar] [CrossRef] [PubMed]
- Lusk, J.L.; McFadden, B.R.; Wilson, N. Do consumers care how a genetically engineered food was created or who created it? Food Policy 2018, 78, 81–90. [Google Scholar] [CrossRef]
- Menz, J.; Modrzejewski, D.; Hartung, F.; Wilhelm, R.; Sprink, T. Genome edited crops touch the market: A view on the global development and regulatory environment. Front. Plant Sci. 2020, 11, 586027. [Google Scholar] [CrossRef]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef]
- Si, X.; Zhang, H.; Wang, Y.; Chen, K.; Gao, C. Manipulating gene translation in plants by CRISPR-Cas9-mediated genome editing of upstream open reading frames. Nat. Protoc. 2020, 15, 338–363. [Google Scholar] [CrossRef]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef]
- Curtis, M.D.; Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003, 133, 462–469. [Google Scholar] [CrossRef]
- Haeussler, M.; Schonig, K.; Eckert, H.; Eschstruth, A.; Mianne, J.; Renaud, J.B.; Schneider-Maunoury, S.; Shkumatava, A.; Teboul, L.; Kent, J.; et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016, 17, 148. [Google Scholar] [CrossRef]
- Weigel, D.; Glazebrook, J. Transformation of Agrobacterium using the freeze-thaw method. CSH Protoc. 2006, 7, pdb.prot4666. [Google Scholar] [CrossRef]
- Chronis, D.; Chen, S.; Lang, P.; Tran, T.; Thurston, D.; Wang, X. Potato transformation. Bio-Protocol 2014, 4, e1017. [Google Scholar] [CrossRef]
- Lin, M.T.; Occhialini, A.; Andralojc, P.J.; Parry, M.A.J.; Hanson, M.R. A faster rubisco with potential to increase photosynthesis in crops. Nature 2014, 513, 547–550. [Google Scholar] [CrossRef]
- Liu, Q.; Weber, E.; Currie, V.; Yada, R. Physicochemical properties of starches during potato growth. Carbohydr. Polym. 2003, 51, 213–221. [Google Scholar] [CrossRef]
- Mendez-Montealvo, G.; Wang, Y.J.; Campbell, M. Thermal and rheological properties of granular waxy maize mutant starches after β-amylase modification. Carbohydr. Polym. 2011, 83, 1106–1111. [Google Scholar] [CrossRef]
| Line/s | Phosphorus % | Phosphate % | Fat % | Nitrogen % | Protein % | Starch % | Amylose % |
|---|---|---|---|---|---|---|---|
| 1.2 | 0.11 | 0.34 | 0.36 | 0.61 | 3.81 | 59.05 (±0.89) | 29.86 (±0.50) |
| 1.3 | 0.11 | 0.34 | 0.15 | 0.61 | 3.81 | 55.44 (±1.22) | 30.28 (±0.32) |
| 1.5 | 0.10 | 0.31 | 0.32 | 0.50 | 3.13 | 59.82 (±1.74) | 29.99 (±1.18) |
| 2.6 | 0.11 | 0.33 | 0.10 | 0.50 | 3.13 | 57.79 (±2.38) | 29.10 (±0.47) |
| 2.7 | 0.12 | 0.35 | 0.16 | 0.58 | 3.63 | 57.63 (±1.96) | 25.58 (±0.265) |
| 2.9 | 0.12 | 0.36 | 0.18 | 0.54 | 3.38 | 58.95 (±1.12) | 27.64 (±0.58) |
| MG1 | 0.12 | 0.36 | 0.27 | 0.55 | 3.44 | 55.80 (±1.07) | 27.10 (±0.50) |
| 3.8 | 0.12 | 0.38 | 0.15 | 0.64 | 4.00 | 58.75 (± 0.41) | 28.89 (± 0.53) |
| MG2 | 0.13 | 0.41 | 0.10 | 0.61 | 3.81 | 57.83 (±0.63) | 26.16 (±0.41) |
| 3.12 | 0.11 | 0.34 | 0.15 | 0.55 | 3.44 | 56.89 (±0.83) | 27.04 (±0.42) |
| 3.13 | 0.11 | 0.35 | 0.10 | 0.56 | 3.50 | 56.51 (±0.37) | 30.04 (±0.56) |
| 3.16 | 0.11 | 0.33 | 0.26 | 0.54 | 3.38 | 58.76 (±0.45) | 29.12 (±0.45) |
| 3.18 | 0.11 | 0.33 | 0.30 | 0.53 | 3.31 | 57.69 (±0.73) | 29.09 (±0.40) |
| 4.1 | 0.11 | 0.33 | 0.26 | 0.58 | 3.63 | ** 50.39 (±0.84) | 27.63 (±0.38) |
| 4.2 | 0.12 | 0.38 | 0.17 | 0.71 | 4.44 | - | - |
| 4.3 | 0.13 | 0.39 | 0.29 | 0.73 | 4.56 | ** 48.74 (±0.37) | 28.33 (±0.58) |
| WT | 0.12 | 0.37 | 0.19 | 0.67 | 4.19 | 54.14 (±0.54) | 28.70 (±0.56) |
| Compiled gRNA1 | 0.11 (±0.01) | 0.33 (±0.02) | 0.28 (±0.11) | 0.57 (±0.06) | 3.58 (±0.39) | 58.10 (±2.34) | 30.04 (±0.21) |
| Compiled gRNA2 | 0.11 (±0.01) | 0.35 (±0.01) | 0.15 (±0.04) | 0.54 (±0.04) | 3.38 (±0.25) | 58.12 (±0.72) | 27.44 (±1.77) |
| Compiled gRNA3 | 0.12 (±0.01) | 0.36 (±0.03) | 0.19 (±0.08) | 0.57 (±0.04) | 3.55 (±0.25) | 57.46 (±1.12) | 28.21 (±1.43) |
| Compiled gRNA4 | 0.12 (±0.01) | 0.37 (±0.03) | 0.27 (±0.02) | 0.67 (±0.08) | 4.21 (±0.51) | 49.56 (±1.17) | 27.98 (±0.49) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pfotenhauer, A.C.; Occhialini, A.; Harbison, S.A.; Li, L.; Piatek, A.A.; Luckett, C.R.; Yang, Y.; Stewart, C.N., Jr.; Lenaghan, S.C. Genome-Editing of FtsZ1 for Alteration of Starch Granule Size in Potato Tubers. Plants 2023, 12, 1878. https://doi.org/10.3390/plants12091878
Pfotenhauer AC, Occhialini A, Harbison SA, Li L, Piatek AA, Luckett CR, Yang Y, Stewart CN Jr., Lenaghan SC. Genome-Editing of FtsZ1 for Alteration of Starch Granule Size in Potato Tubers. Plants. 2023; 12(9):1878. https://doi.org/10.3390/plants12091878
Chicago/Turabian StylePfotenhauer, Alexander C., Alessandro Occhialini, Stacee A. Harbison, Li Li, Agnieszka A. Piatek, Curtis R. Luckett, Yongil Yang, C. Neal Stewart, Jr., and Scott C. Lenaghan. 2023. "Genome-Editing of FtsZ1 for Alteration of Starch Granule Size in Potato Tubers" Plants 12, no. 9: 1878. https://doi.org/10.3390/plants12091878
APA StylePfotenhauer, A. C., Occhialini, A., Harbison, S. A., Li, L., Piatek, A. A., Luckett, C. R., Yang, Y., Stewart, C. N., Jr., & Lenaghan, S. C. (2023). Genome-Editing of FtsZ1 for Alteration of Starch Granule Size in Potato Tubers. Plants, 12(9), 1878. https://doi.org/10.3390/plants12091878









