Expanding Possibilities for Foreign Gene Expression by Cucumber Green Mottle Mosaic Virus Genome-Based Bipartite Vector System
Abstract
1. Introduction
2. Materials and Methodology
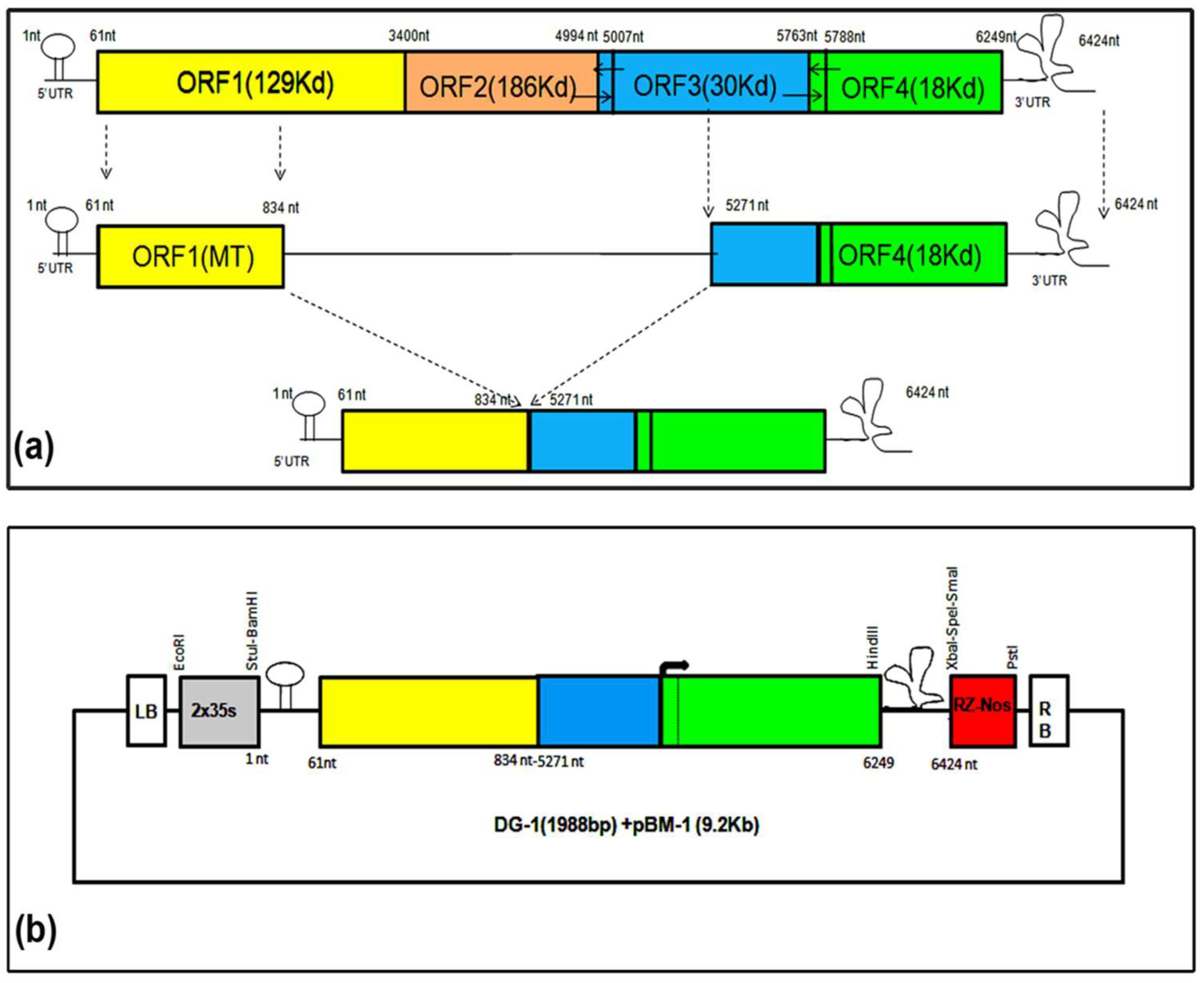
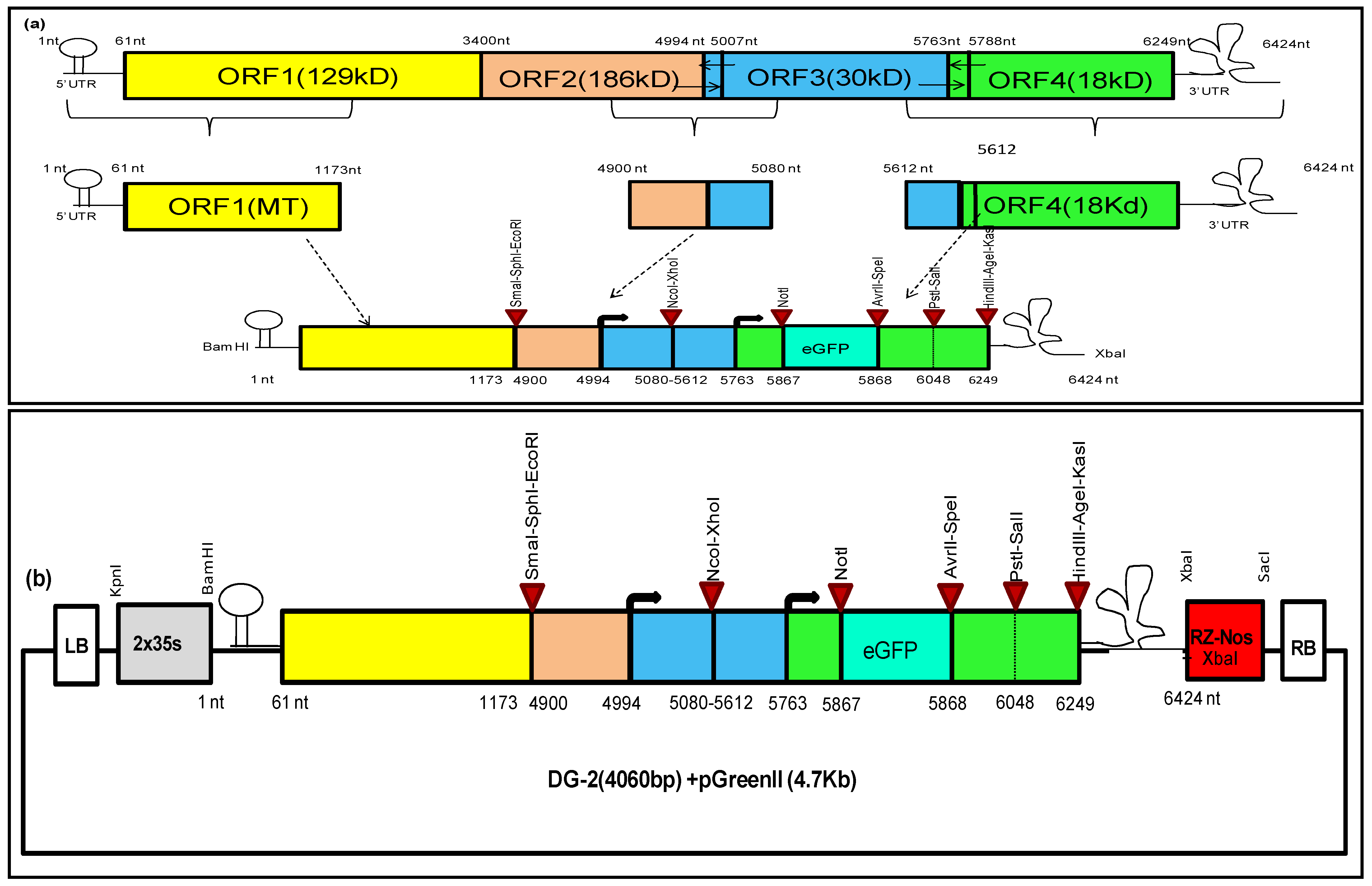
2.1. Designing of Deconstructed Virus Genomes and Analysing Their RNA Secondary Structures
2.2. Development of Deconstructed Virus Genomes
2.3. Plant Inoculation of Deconstructed Virus Genomes along with Wild-Type Virus Strains
2.4. Infectivity Assay of Wild-Type CGMMV
2.5. Trans-Replication Assay of Deconstructed Genomes of CGMMV in Different Hosts
2.6. Movement Assay of Deconstructed Genomes of CGMMV
2.7. Vector Function Assay of Deconstructed Genomes within CGMMV-Infected N. benthamiana
2.7.1. Gene-Silencing Assay of Deconstructed Genome-1-Expressing NbPDS Gene Sequence
Insertion of NbPDS Gene into DG-1 Construct, Agro-Transformation, and Plant Infiltration
Functional Validation of pDG(PDS)-1 Constructs
Northern Blot Analysis for the Confirmation of the Deletion RNA Replicon
2.7.2. In Planta Expression of Foreign Protein Using Deconstructed Genome-2
Detection of GFP Fluorescence through Confocal Microscopy
Western Blot Assay for the Confirmation of GFP Expression
2.8. Quantitative Estimation of Replication Rate of Deconstructed Genomes
3. Results
3.1. Design and Development of Deconstructed Genomes
3.2. Trans-Replication and In Planta Systemic Movement of Deconstructed Genomes of CGMMV
3.2.1. Trans-Replication and In Planta Systemic Movement of DG1
3.2.2. Trans-replication and In-Planta Systemic Movement of DG2
3.3. Translational Ability of Deconstructed Genome-Based Bipartite Vector System in CGMMV-Infected N. benthamiana
3.3.1. Functionality of Deconstructed Virus Genome-Based Silencing Vector
Development of Deconstructed Virus Genome-Based Silencing Vector
Phenotypic Silencing of NbPDS Gene Using pDG(PDS)-1 Construct in CGMMV-Infected N. benthamiana
Detection of Replication and Movement of DG(PDS)-1 in CGMMV-Infected N. benthamiana
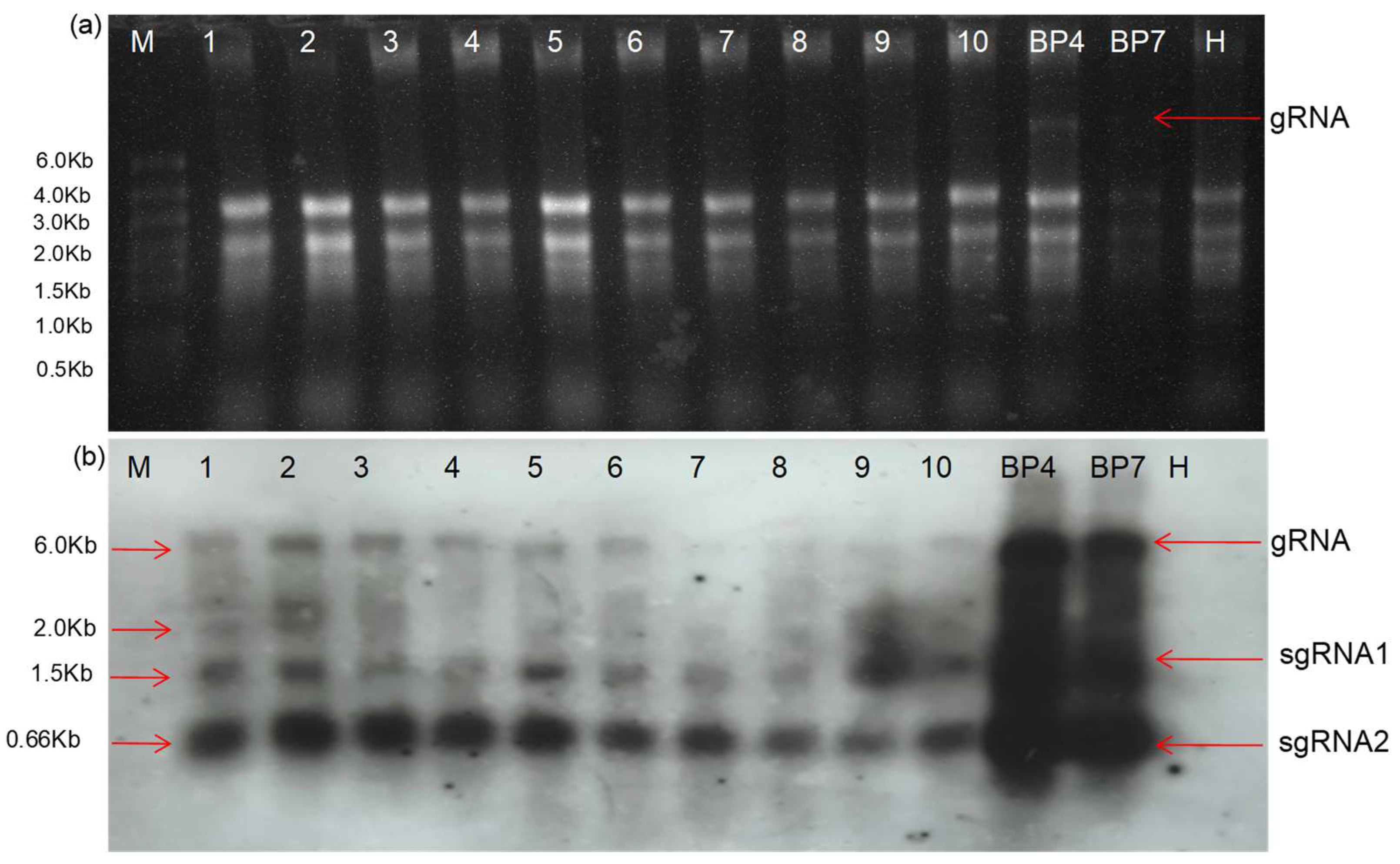
Subgenomic RNAs of Deconstructed Genome DG(PDS)-1
3.3.2. Functionality of Deconstructed Virus Genome-Based Protein Expression Vector
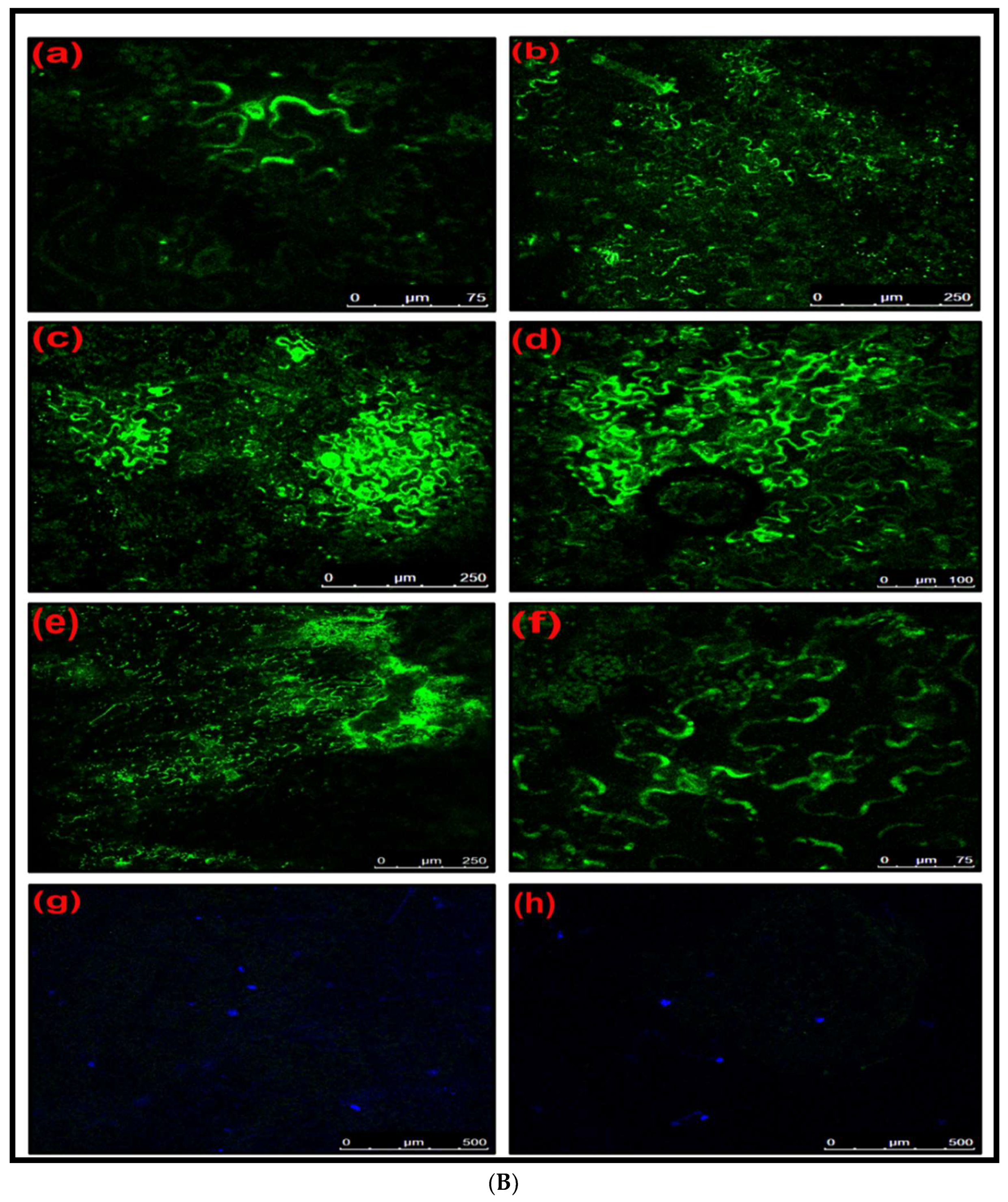
Protein Expression Ability of Deconstructed Genome 2 (DG2)
Confirmation of Protein Expression Ability of Deconstructed Genome 2
3.4. Quantification of Replication Rate of the Deconstructed Genomes
3.5. Genome Sequence Analysis of DG-1 and DG-2
3.5.1. Genome Sequence Analysis of DG-1
3.5.2. Genome Sequence Analysis of DG-2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Budzyńska, D.; Zwart, M.P.; Hasiów-Jaroszewska, B. Defective RNA particles of plant viruses-origin, structure and role in pathogenesis. Viruses 2022, 14, 2814. [Google Scholar] [CrossRef] [PubMed]
- Minicka, J.; Taberska, A.; Zarzyńska-Nowak, A.; Kubska, K.; Budzyńska, D.; Elena, S.F.; Hasiów-Jaroszewska, B. Genetic diversity of Tomato Black Ring Virus Satellite RNAs and their impact on virus replication. Int. J. Mol. Sci. 2022, 23, 9393. [Google Scholar] [CrossRef] [PubMed]
- Havelda, Z.; Dalmay, T.; Burgyan, J. Localization of cis-acting sequences essential for Cymbidium ringspot tombusvirus defective interfering RNA replication. J. Gen. Virol. 1995, 76, 2311–2316. [Google Scholar] [CrossRef] [PubMed]
- Damayanti, T.A.; Nagano, H.; Mise, K.; Furusawa, I.; Okuno, T. Positional effect of deletions on viability, especially on encapsidation, of Brome mosaic virus D-RNA in barley protoplasts. Virology 2002, 293, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Gleba, Y.; Marillonnet, S.; Klimyuk, V. Engineering viral expression vectors for plants: The ‘full virus’ and the ‘deconstructed virus’ strategies. Curr. Opin. Plant Biol. 2004, 7, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Gleba, Y.; Klimyuk, V.; Marillonnet, S. Magnifection—A new platform for expressing recombinant vaccines in plants. Vaccine 2005, 23, 2042–2048. [Google Scholar] [CrossRef] [PubMed]
- Dombrovsky, A.; Tran-Nguyen, L.T.T.; Jones, R.A.C. Cucumber green mottle mosaic virus: Rapidly increasing global distribution, etiology, epidemiology, and management. Annu. Rev. Phytopathol. 2017, 55, 231–256. [Google Scholar] [CrossRef] [PubMed]
- Ugaki, M.; Tomiyama, M.; Kakutani, T.; Hidaka, S.; Kiguchi, T.; Nagata, R.; Sato, T.; Motoyoshi, F.; Nishiguchi, M. The complete nucleotide sequence of cucumber green mottle mosaic virus (SH strain) genomic RNA. J. Gen. Virol. 1991, 72, 1487–1495. [Google Scholar] [CrossRef]
- Chen, H.; Ino, M.; Shimono, M.; Wagh, S.G.; Kobayashi, K.; Yaeno, T.; Yamaoka, N.; Bai, G.; Nishiguchi, M. A single amino acid substitution in the intervening region of 129K protein of cucumber green mottle mosaic virus resulted in attenuated symptoms. Phytopathology 2020, 110, 146–152. [Google Scholar] [CrossRef]
- Sztuba-Solińska, J.; Stollar, V.; Bujarski, J.J. Subgenomic messenger RNAs: Mastering regulation of (+)-strand RNA virus life cycle. Virology 2011, 412, 245–255. [Google Scholar] [CrossRef]
- Teoh, P.-G.; Ooi, A.-S.; AbuBakar, S.; Othman, R.Y. Virus-specific read-through codon preference affects infectivity of chimeric Cucumber green mottle mosaic viruses displaying a Dengue virus epitope. J. Biomed. Biotechnol. 2009, 2009, 781712. [Google Scholar] [CrossRef]
- Zheng, H.; Xiao, C.; Han, K.; Peng, J.; Lin, L.; Lu, Y.; Xie, L.; Wu, X.; Xu, P.; Li, G.; et al. Development of an agroinoculation system for full-length and GFP-tagged cDNA clones of cucumber green mottle mosaic virus. Arch. Virol. 2015, 160, 2867–2872. [Google Scholar] [CrossRef]
- Jailani, A.A.K.; Solanki, V.; Roy, A.; Sivasudha, T.; Mandal, B. A CGMMV genome-replicon vector with partial sequences of coat protein gene efficiently expresses GFP in Nicotiana benthamiana. Virus Res. 2017, 233, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Roy, G.; Weisburg, S.; Rabindran, S.; Yusibov, V. A novel two-component Tobacco mosaic virus-based vector system for high-level expression of multiple therapeutic proteins including a human monoclonal antibody in plants. Virology 2010, 405, 93–99. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Mathews, D.H.; Sabina, J.; Zuker, M.; Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999, 288, 911–940. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, A.; Jailani, A.A.K.; Roy, A.; Mukherjee, S.K.; Mandal, B. Prediction of putative regulatory elements in the subgenomic promoters of cucumber green mottle mosaic virus and their interactions with the RNA dependent RNA polymerase domain. Virus Dis. 2020, 31, 503–516. [Google Scholar] [CrossRef]
- Nayaka, S.N.; Jailani, A.A.K.; Ghosh, A.; Roy, A.; Mandal, B. Delivery of progeny virus from the infectious clone of cucumber green mottle mosaic virus and quantification of the viral load in different host plants. 3 Biotech 2023, 13, 209. [Google Scholar] [CrossRef]
- Jailani, A.A.K.; Chattopadhyay, A.; Washington, O.S.; Roy, A.; Mukherjee, S.K.; Mishra, N.S.; Bikash, M. Overlapping primer extension PCR-based cloning techniques engineer a cucumber green mottle mosaic virus genome-based VIGS vector. In Proceedings of the VIROCON: International Conference on “Evolution of Viruses and Viral Diseases”, Indian Virological Society, New Delhi, India, 18–20 February 2020; p. 112. [Google Scholar]
- Jailani, A.A.K.; Chattopadhyay, A.; Kumar, P.; Singh, O.W.; Mukherjee, S.K.; Roy, A.; Sanan-Mishra, N.; Mandal, B. Accelerated Long-Fragment Circular PCR for genetic manipulation of plant viruses in unveiling functional genomics. Viruses 2023, 15, 2332. [Google Scholar] [CrossRef]
- Weigel, D.; Glazebrook, J. Transformation of Agrobacterium using the freeze-thaw method. Cold Spring Harb. Protoc. 2006, 2006, pdb.prot4666. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Hema, R.; Anand, A.; Kang, L.; Udayakumar, M.; Mysore, K.S. A systematic study to determine the extent of gene silencing in Nicotiana benthamiana and other Solanaceae species when heterologous gene sequences are used for virus-induced gene silencing. New Phytol. 2007, 176, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Knapp, E.; Danyluk, G.M.; Achor, D.; Lewandowski, D.J. A bipartite Tobacco mosaic virus-defective RNA (dRNA) system to study the role of the N-terminal methyl transferase domain in cell-to-cell movement of dRNAs. Virology 2005, 341, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Mathews, D.H.; Turner, D.H. Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 2006, 16, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M.; Stiegler, P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981, 9, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Perrault, J. Origin and replication of defective interfering particles. Curr. Top. Microbiol. Immunol. 1981, 93, 151–207. [Google Scholar]
- López, C.B. Defective viral genomes: Critical danger signals of viral infections. J. Virol. 2014, 88, 8720–8723. [Google Scholar] [CrossRef]
- Lazzarini, R.A.; Keene, J.D.; Schubert, M. The origins of defective interfering particles of the negative-strand RNA viruses. Cell 1981, 26, 145–154. [Google Scholar] [CrossRef]
- Bosma, T.J.; Karagiannis, K.; Santana-Quintero, L.; Ilyushina, N.; Zagorodnyaya, T.; Petrovskaya, S.; Laassri, M.; Donnelly, R.P.; Rubin, S.; Simonyan, V.; et al. Identification and quantification of defective virus genomes in high throughput sequencing data using DVG-profiler, a novel post-sequence alignment processing algorithm. PLoS ONE 2019, 14, e0216944. [Google Scholar] [CrossRef]
- Huang, A.S.; Baltimore, D. Defective viral particles and viral disease processes. Nature 1970, 226, 325–327. [Google Scholar] [CrossRef]
- Tapia, K.; Kim, W.-K.; Sun, Y.; Mercado-López, X.; Dunay, E.; Wise, M.; Adu, M.; López, C.B. Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity. PLoS Pathog. 2013, 9, e1003703. [Google Scholar] [CrossRef] [PubMed]
- Rezelj, V.V.; Levi, L.I.; Vignuzzi, M. The defective component of viral populations. Curr. Opin. Virol. 2018, 33, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Aaskov, J.; Buzacott, K.; Thu, H.M.; Lowry, K.; Holmes, E.C. Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science 2006, 311, 236–238. [Google Scholar] [CrossRef] [PubMed]
- Calain, P.; Monroe, M.C.; Nichol, S.T. Ebola virus defective interfering particles and persistent infection. Virology 1999, 262, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Noppornpanth, S.; Smits, S.L.; Lien, T.X.; Poovorawan, Y.; Osterhaus, A.D.; Haagmans, B.L. Characterization of hepatitis C virus deletion mutants circulating in chronically infected patients. J. Virol. 2007, 81, 12496–12503. [Google Scholar] [CrossRef] [PubMed]
- Saira, K.; Lin, X.; DePasse, J.V.; Halpin, R.; Twaddle, A.; Stockwell, T.; Angus, B.; Cozzi-Lepri, A.; Delfino, M.; Dugan, V.; et al. Sequence analysis of in vivo defective interfering-like RNA of influenza A H1N1 pandemic virus. J. Virol. 2013, 87, 8064–8074. [Google Scholar] [CrossRef] [PubMed]
- Vasilijevic, J.; Zamarreño, N.; Oliveros, J.C.; Rodriguez-Frandsen, A.; Gómez, G.; Rodriguez, G.; Pérez-Ruiz, M.; Rey, S.; Barba, I.; Pozo, F.; et al. Reduced accumulation of defective viral genomes contributes to severe outcome in influenza virus infected patients. PLoS Pathog. 2017, 13, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Šantak, M.; Markušić, M.; Balija, M.L.; Kopač, S.K.; Jug, R.; Örvell, C.; Tomac, J.; Forčić, D. Accumulation of defective interfering viral particles in only a few passages in Vero cells attenuates mumps virus neurovirulence. Microbes Infect. 2015, 17, 228–236. [Google Scholar] [CrossRef] [PubMed]
- McLaren, L.C.; Holland, J.J. Defective interfering particles from poliovirus vaccine and vaccine reference strains. Virology 1974, 60, 579–583. [Google Scholar] [CrossRef]
- Treuhaft, M.W.; Beem, M.O. Defective interfering particles of respiratory syncytial virus. Infect. Immun. 1982, 37, 439–444. [Google Scholar] [CrossRef]
- Strahle, L.; Garcin, D.; Kolakofsky, D. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 2006, 351, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Fuller, F.J.; Marcus, P.I. Interferon induction by viruses. Sindbis virus: Defective-interfering particles temperature-sensitive for interferon induction. J. Gen. Virol. 1980, 48, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Vignuzzi, M.; López, C.B. Defective viral genomes are key drivers of the virus-host interaction. Nat. Microbiol. 2019, 4, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Pogany, J.; Romero, J.; Huang, Q.; Sgro, J.Y.; Shang, H.; Bujarski, J.J. De novo generation of defective interfering-like RNAs in broad bean mottle bromovirus. Virology 1995, 212, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Li, X.H.; Heaton, L.A.; Morris, T.J.; Simon, A.E. Turnip crinkle virus defective interfering RNAs intensify viral symptoms and are generated de novo. Proc. Natl. Acad. Sci. USA 1989, 86, 9173–9177. [Google Scholar] [CrossRef] [PubMed]
- Teycheney, P.-Y.; Marais, A.; Svanella-Dumas, L.; Dulucq, M.-J.; Candresse, T. Molecular characterization of banana virus X (BVX), a novel member of the Flexiviridae family. Arch. Virol. 2005, 150, 1715–1727. [Google Scholar] [CrossRef] [PubMed]
- Hillman, B.I.; Carrington, J.C.; Morris, T.J. A defective interfering RNA that contains a mosaic of a plant virus genome. Cell 1987, 51, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Knorr, D.A.; Mullin, R.H.; Hearne, P.Q.; Morris, T.J. De novo generation of defective interfering RNAs of tomato bushy stunt virus by high multiplicity passage. Virology 1991, 181, 193–202. [Google Scholar] [CrossRef]
- Lewandowski, D.J.; Dawson, W.O. Deletion of internal sequences results in tobacco mosaic virus defective RNAs that accumulate to high levels without interfering with replication of the helper virus. Virology 1998, 251, 427–437. [Google Scholar] [CrossRef]
- Knapp, E.; Dawson, W.O.; Lewandowski, D.J. Conundrum of the lack of defective RNAs (dRNAs) associated with tobamovirus Infections: dRNAs that can move are not replicated by the wild-type virus; dRNAs that are replicated by the wild-type virus do not move. J. Virol. 2001, 75, 5518–5525. [Google Scholar] [CrossRef][Green Version]
- Ogawa, T.; Watanabe, Y.; Meshi, T.; Okada, Y. Trans complementation of virus-encoded replicase components of tobacco mosaic virus. Virology 1991, 185, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Raffo, A.J.; Dawson, W.O. Construction of tobacco mosaic virus subgenomic replicons that are replicated and spread systemically in tobacco plants. Virology 1991, 184, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Donson, J.; Kearney, C.M.; Hilf, M.E.; Dawson, W.O. Systemic expression of a bacterial gene by a tobacco mosaic virus-based vector. Proc. Natl. Acad. Sci. USA 1991, 88, 7204–7208. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Lee, C.-C. Function and structural organization of the replication protein of Bamboo mosaic virus. Front. Microbiol. 2017, 8, 522. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Sivakumaran, K.; Kao, C. The N-terminal half of the brome mosaic virus 1a protein has RNA capping-associated activities: Specificity for GTP and S-adenosylmethionine. Virology 1999, 259, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Ahola, T.; Laakkonen, P.; Vihinen, H.; Kääriäinen, L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J. Virol. 1997, 71, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.L.; Han, Y.-T.; Chang, Y.-T.; Hsu, Y.-H.; Meng, M. Critical residues for GTP methylation and formation of the covalent m7GMP-enzyme intermediate in the capping enzyme domain of bamboo mosaic virus. J. Virol. 2004, 78, 1271–1280. [Google Scholar] [CrossRef]
- Hirashima, K.; Watanabe, Y. RNA helicase domain of tobamovirus replicase executes cell-to-cell movement possibly through collaboration with its nonconserved region. J. Virol. 2003, 77, 12357–12362. [Google Scholar] [CrossRef][Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chattopadhyay, A.; Jailani, A.A.K.; Roy, A.; Mukherjee, S.K.; Mandal, B. Expanding Possibilities for Foreign Gene Expression by Cucumber Green Mottle Mosaic Virus Genome-Based Bipartite Vector System. Plants 2024, 13, 1414. https://doi.org/10.3390/plants13101414
Chattopadhyay A, Jailani AAK, Roy A, Mukherjee SK, Mandal B. Expanding Possibilities for Foreign Gene Expression by Cucumber Green Mottle Mosaic Virus Genome-Based Bipartite Vector System. Plants. 2024; 13(10):1414. https://doi.org/10.3390/plants13101414
Chicago/Turabian StyleChattopadhyay, Anirudha, A. Abdul Kader Jailani, Anirban Roy, Sunil Kumar Mukherjee, and Bikash Mandal. 2024. "Expanding Possibilities for Foreign Gene Expression by Cucumber Green Mottle Mosaic Virus Genome-Based Bipartite Vector System" Plants 13, no. 10: 1414. https://doi.org/10.3390/plants13101414
APA StyleChattopadhyay, A., Jailani, A. A. K., Roy, A., Mukherjee, S. K., & Mandal, B. (2024). Expanding Possibilities for Foreign Gene Expression by Cucumber Green Mottle Mosaic Virus Genome-Based Bipartite Vector System. Plants, 13(10), 1414. https://doi.org/10.3390/plants13101414




