Drought Responses in Poaceae: Exploring the Core Components of the ABA Signaling Pathway in Setaria italica and Setaria viridis
Abstract
:1. Introduction
2. Results
2.1. The Core ABA Signaling Genes of S. italica and S. viridis Are Highly Conserved
2.2. Phylogenetic Analyses Offer a Valuable Tool for Associating Setaria spp. Genes to Other Homologous Genes with Well-Defined Functions
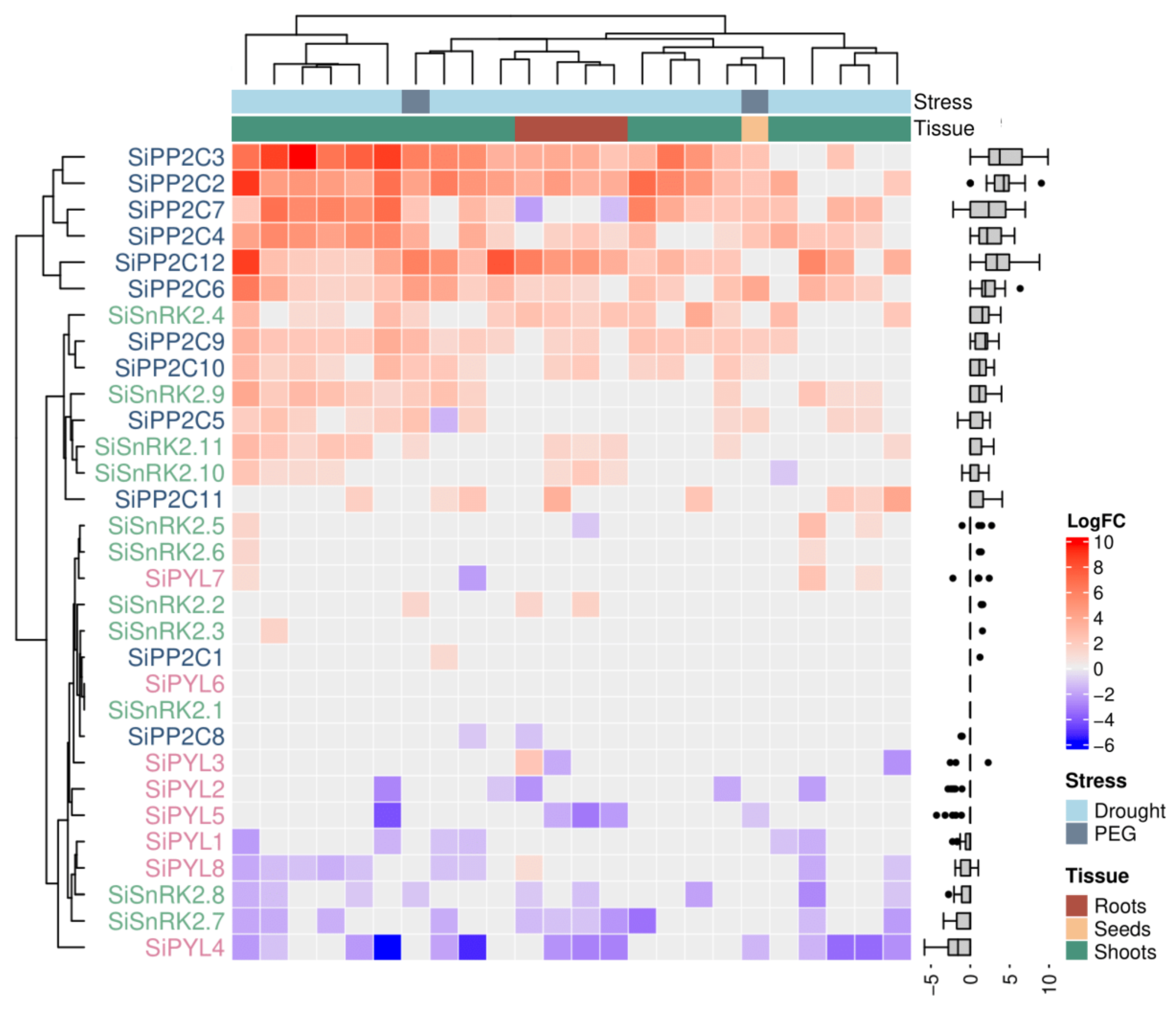
2.3. Meta-Analysis of Gene Expression Supports a Diversity of Roles of ABA Core Components during the Response to Water Stress
3. Discussion
4. Materials and Methods
4.1. Mining and Gene Structure Analyses
4.2. Phylogenetic Reconstructions
4.3. Meta-Analysis of Gene Expression Profile
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Olds, C.L.; Glennon, E.K.K.; Luckhart, S. Abscisic Acid: New Perspectives on an Ancient Universal Stress Signaling Molecule. Microbes Infect. 2018, 20, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Hartung, W. The Evolution of Abscisic Acid (ABA) and ABA Function in Lower Plants, Fungi and Lichen. Funct. Plant Biol. 2010, 37, 806–812. [Google Scholar] [CrossRef]
- Trivedi, D.K.; Gill, S.S.; Tuteja, N. Abscisic Acid (ABA): Biosynthesis, Regulation, and Role in Abiotic Stress Tolerance. In Abiotic Stress Response in Plants; Tuteja, N., Gill, S.S., Eds.; Wiley-VCH Verlag GmbH & Co.: Weinheim, Germany, 2016; pp. 311–322. [Google Scholar] [CrossRef]
- Duarte, K.E.; de Souza, W.R.; Santiago, T.R.; Sampaio, B.L.; Ribeiro, A.P.; Cotta, M.G.; da Cunha, B.A.D.B.; Marraccini, P.R.R.; Kobayashi, A.K.; Molinari, H.B.C. Identification and Characterization of Core Abscisic Acid (ABA) Signaling Components and Their Gene Expression Profile in Response to Abiotic Stresses in Setaria viridis. Sci. Rep. 2019, 9, 4028. [Google Scholar] [CrossRef] [PubMed]
- Addicott, F.T. Abscisic Acid: Discovery, and Exploration of Properties. In Discoveries in Plant Biology; Kung, S.-D., Yang, S.-F., Eds.; World Scientific Publishing: Singapore, 1998; Volume I, pp. 33–46. [Google Scholar] [CrossRef]
- Shen, X.; Nan, H.; Jiang, Y.; Zhou, Y.; Pan, X. Genome-Wide Identification, Expression and Interaction Analysis of GmSnRK2 and Type A PP2C Genes in Response to Abscisic Acid Treatment and Drought Stress in Soybean Plant. Int. J. Mol. Sci. 2022, 23, 13166. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R. Abscisic Acid Synthesis and Response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic Acid Dynamics, Signaling, and Functions in Plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Hu, B.; Cao, J.; Ge, K.; Li, L. The Site of Water Stress Governs the Pattern of ABA Synthesis and Transport in Peanut. Sci. Rep. 2016, 6, 32143. [Google Scholar] [CrossRef]
- Fidler, J.; Graska, J.; Gietler, M.; Nykiel, M.; Prabucka, B.; Rybarczyk-Płońska, A.; Muszyńska, E.; Morkunas, I.; Labudda, M. PYR/PYL/RCAR Receptors Play a Vital Role in the Abscisic-Acid-Dependent Responses of Plants to External or Internal Stimuli. Cells 2022, 11, 1352. [Google Scholar] [CrossRef]
- Kim, J.S.; Mizoi, J.; Yoshida, T.; Fujita, Y.; Nakajima, J.; Ohori, T.; Todaka, D.; Nakashima, K.; Hirayama, T.; Shinozaki, K.; et al. An ABRE Promoter Sequence Is Involved in Osmotic Stress-Responsive Expression of the DREB2A Gene, Which Encodes a Transcription Factor Regulating Drought-Inducible Genes in Arabidopsis. Plant Cell Physiol. 2011, 52, 2136–2146. [Google Scholar] [CrossRef]
- Gómez-Porras, J.L.; Riaño-Pachón, D.; Dreyer, I.; Mayer, J.E.; Mueller-Roeber, B. Genome-Wide Analysis of ABA-Responsive Elements ABRE and CE3 Reveals Divergent Patterns in Arabidopsis and Rice. BMC Genom. 2007, 8, 260. [Google Scholar] [CrossRef]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between Two Cis-acting Elements, ABRE and DRE in ABA-dependent Expression of Arabidopsis Rd29A Gene in Response to Dehydration and High-Salinity Stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Yin, P.; Li, W.; Wang, L.; Yan, C.; Lin, Z.; Wu, J.Z.; Wang, J.; Yan, S.F.; Yan, N. The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins. Mol. Cell 2011, 42, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chan, Z.; Xing, L.; Liu, X.; Hou, Y.J.; Chinnusamy, V.; Wang, P.; Duan, C.; Zhu, J.K. The Unique Mode of Action of a Divergent Member of the ABA-Receptor Protein Family in ABA and Stress Signaling. Cell Res. 2013, 23, 1380–1395. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.J.; Wan, Y.; Liu, W.; Xie, S.; et al. Arabidopsis Duodecuple Mutant of PYL ABA Receptors Reveals PYL Repression of ABA-Independent SnRK2 Activity. Cell Rep. 2018, 23, 3340–3351.e5. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernández, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR Receptors Play a Major Role in Quantitative Regulation of Stomatal Aperture and Transcriptional Response to Abscisic Acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Grill, E.; Meskiene, I.; Schweighofer, A. Type 2C Protein Phosphatases in Plants. FEBS J. 2012, 280, 681–693. [Google Scholar] [CrossRef]
- Saha, J.; Chatterjee, C.; Sengupta, A.; Gupta, K.; Gupta, B. Genome-Wide Analysis and Evolutionary Study of Sucrose Non-Fermenting 1-Related Protein Kinase 2 (SnRK2) Gene Family Members in Arabidopsis and Oryza. Comput. Biol. Chem. 2013, 49, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Yamamoto, S.; Minami, H.; Kagaya, Y.; Hattori, T. Differential Activation of the Rice Sucrose Nonfermenting1-Related Protein Kinase2 Family by Hyperosmotic Stress and Abscisic Acid. Plant Cell 2004, 16, 1163–1177. [Google Scholar] [CrossRef]
- Suguiyama, V.F.; Rodriguez, J.D.P.; dos Santos, T.C.N.; Lira, B.S.; de Haro, L.A.; Silva, J.P.N.; Borba, E.L.; Purgatto, E.; da Silva, E.A.; Bellora, N.; et al. Regulatory Mechanisms behind the Phenotypic Plasticity Associated with Setaria Italica Water Deficit Tolerance. Plant Mol. Biol. 2022, 109, 761–780. [Google Scholar] [CrossRef]
- Sun, C.; Gao, X.; Chen, X.; Fu, J.; Zhang, Y. Metabolic and Growth Responses of Maize to Successive Drought and Re-Watering Cycles. Agric. Water Manag. 2016, 172, 62–73. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, J.; Lei, T.; He, B.; Wu, Z.; Liu, M.; Mo, X.; Geng, G.; Li, X.; Zhou, H.; et al. Temporal-Spatial Characteristics of Severe Drought Events and Their Impact on Agriculture on a Global Scale. Quat. Int. 2014, 349, 10–21. [Google Scholar] [CrossRef]
- Dekker, J. The Foxtail (Setaria) Species-Group. Weed Sci. 2003, 51, 641–656. [Google Scholar] [CrossRef]
- Diao, X.; Jia, G. Origin and Domestication of Foxtail Millet. In Genetics and Genomics of Setaria; Doust, A.N., Diao, X., Eds.; Springer International Publishing: Beijing, China, 2017; pp. 61–72. ISBN 9783319451053. [Google Scholar]
- Lata, C.; Gupta, S.; Prasad, M. Foxtail Millet: A Model Crop for Genetic and Genomic Studies in Bioenergy Grasses. Crit. Rev. Biotechnol. 2012, 33, 328–343. [Google Scholar] [CrossRef]
- Ruiz-Partida, R.; Rosario, S.M.; Lozano-Juste, J. An Update on Crop ABA Receptors. Plants 2021, 10, 1087. [Google Scholar] [CrossRef] [PubMed]
- Mega, R.; Abe, F.; Kim, J.S.; Tsuboi, Y.; Tanaka, K.; Kobayashi, H.; Sakata, Y.; Hanada, K.; Tsujimoto, H.; Kikuchi, J.; et al. Tuning Water-Use Efficiency and Drought Tolerance in Wheat Using Abscisic Acid Receptors. Nat. Plants 2019, 5, 153–159. [Google Scholar] [CrossRef]
- Yadav, S.K.; Venkata, V.; Kumar, S.; Verma, R.K.; Yadav, P.; Saroha, A.; Wankhede, D.P.; Chaudhary, B.; Chinnusamy, V. Genome-Wide Identification and Characterization of ABA Receptor PYL Gene Family in Rice. BMC Genom. 2020, 21, 676. [Google Scholar] [CrossRef]
- Quadrana, L.; Almeida, J.; Asís, R.; Duffy, T.; Dominguez, P.G.; Bermúdez, L.; Conti, G.; Corrêa Da Silva, J.V.; Peralta, I.E.; Colot, V.; et al. Natural Occurring Epialleles Determine Vitamin e Accumulation in Tomato Fruits. Nat. Commun. 2014, 5, 4027. [Google Scholar] [CrossRef]
- n.d. Google Scholar. Available online: https://scholar.google.com/ (accessed on 1 October 2020).
- National Library of Medicine of PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 1 October 2020).
- Wang, R.-L.; Wendel, J.F.; Dekker, J.H. Weedy Adaptation in Setaria spp. i. Isozyme Analysis of Genetic Diversity and Population Genetic Structure in Setaria viridis. Am. J. Bot. 1995, 82, 308–317. [Google Scholar] [CrossRef]
- D’Ennequin, M.L.T.; Panaud, O.; Toupance, B.; Sarr, A. Assessment of Genetic Relationships between Setaria italica and Its Wild Relative S. viridis Using AFLP Markers. Theor. Appl. Genet. 2000, 100, 1061–1066. [Google Scholar] [CrossRef]
- Wang, C.; Chen, J.; Zhi, H.; Yang, L.; Li, W.; Wang, Y.; Li, H.; Zhao, B.; Chen, M.; Diao, X. Population Genetics of Foxtail Millet and Its Wild Ancestor. BMC Genet. 2010, 11, 90. [Google Scholar] [CrossRef]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA Perception and Signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Dalal, M.; Inupakutika, M. Transcriptional Regulation of ABA Core Signaling Component Genes in Sorghum (Sorghum bicolor L. Moench). Mol. Breed. 2014, 34, 1517–1525. [Google Scholar] [CrossRef]
- Rubio, S.; Rodrigues, A.; Saez, A.; Dizon, M.B.; Galle, A.; Kim, T.H.; Santiago, J.; Flexas, J.; Schroeder, J.I.; Rodriguez, P.L. Triple Loss of Function of Protein Phosphatases Type 2C Leads to Partial Constitutive Response to Endogenous Abscisic Acid. Plant Physiol. 2009, 150, 1345–1355. [Google Scholar] [CrossRef]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 Protein Kinases Are the Main Positive Regulators of Abscisic Acid Signaling in Response to Water Stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef]
- Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Rodrigues, A.; Pizzio, G.A.; Rodriguez, P.L. Selective Inhibition of Clade A Phosphatases Type 2C by PYR/PYL/RCAR Abscisic Acid Receptors. Plant Physiol. 2012, 158, 970–980. [Google Scholar] [CrossRef]
- Bhaskara, G.B.; Nguyen, T.T.; Verslues, P.E. Unique Drought Resistance Functions of the Highly ABA-Induced Clade a Protein Phosphatase 2Cs. Plant Physiol. 2012, 160, 379–395. [Google Scholar] [CrossRef]
- Sun, L.; Wang, Y.P.; Chen, P.; Ren, J.; Ji, K.; Li, Q.; Li, P.; Dai, S.J.; Leng, P. Transcriptional Regulation of SlPYL, SlPP2C, and SlSnRK2 Gene Families Encoding ABA Signal Core Components during Tomato Fruit Development and Drought Stress. J. Exp. Bot. 2011, 62, 5659–5669. [Google Scholar] [CrossRef]
- Boneh, U.; Biton, I.; Zheng, C.; Schwartz, A.; Ben-Ari, G. Characterization of Potential ABA Receptors in Vitis vinifera. Plant Cell Rep. 2012, 31, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.M.; Liu, X.D.; Waseem, M.; Guang-Qian, Y.; Alabdallah, N.M.; Jahan, M.S.; Fang, X.W. ABA Activated SnRK2 Kinases: An Emerging Role in Plant Growth and Physiology. Plant Signal. Behav. 2022, 17, e2071024. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, M.; Umezawa, T.; Nakashima, K.; Kidokoro, S.; Takasaki, H.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two Closely Related Subclass II SnRK2 Protein Kinases Cooperatively Regulate Drought-Inducible Gene Expression. Plant Cell Physiol. 2010, 51, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, W.; Mao, X.; Jing, R.; Jia, H. Differential Activation of the Wheat SnRK2 Family by Abiotic Stresses. Front. Plant Sci. 2016, 7, 420. [Google Scholar] [CrossRef]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of ABA in Integrating Plant Responses to Drought and Salt Stresses. Field Crop. Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Santiago, J.; Dupeux, F.; Round, A.; Antoni, R.; Park, S.Y.; Jamin, M.; Cutler, S.R.; Rodriguez, P.L.; Márquez, J.A. The Abscisic Acid Receptor PYR1 in Complex with Abscisic Acid. Nature 2009, 462, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Umezawa, T. The PP2c-SnRK2 Complex: The Central Regulator of an Abscisic Acid Signaling Pathway. Plant Signal. Behav. 2010, 5, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, D.; Dudek, S.; Ritchie, M.D.; Pendergrass, S.A. Visualizing Genomic Information across Chromosomes with PhenoGram. BioData Min. 2013, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van De Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Kohany, O.; Gentles, A.J.; Hankus, L.; Jurka, J. Annotation, Submission and Screening of Repetitive Elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform. 2006, 7, 474. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.3.1. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 February 2024).
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Posit Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2023. [Google Scholar]
- Qi, X.; Xie, S.; Liu, Y.; Yi, F.; Yu, J. Genome-Wide Annotation of Genes and Noncoding RNAs of Foxtail Millet in Response to Simulated Drought Stress by Deep Sequencing. Plant Mol. Biol. 2013, 83, 459–473. [Google Scholar] [CrossRef]
- Tang, S.; Li, L.; Wang, Y.; Chen, Q.; Zhang, W.; Jia, G.; Zhi, H.; Zhao, B.; Diao, X. Genotype-Specific Physiological and Transcriptomic Responses to Drought Stress in Setaria italica (an Emerging Model for Panicoideae Grasses). Sci. Rep. 2017, 7, 10009. [Google Scholar] [CrossRef]
- Qin, L.; Chen, E.; Li, F.; Yu, X.; Liu, Z.; Yang, Y.; Wang, R.; Zhang, H.; Wang, H.; Liu, B.; et al. Genome-Wide Gene Expression Profiles Analysis Reveal Novel Insights into Drought Stress in Foxtail Millet (Setaria italica L.). Int. J. Mol. Sci. 2020, 21, 8520. [Google Scholar] [CrossRef]
- Yu, A.; Zhao, J.; Wang, Z.; Cheng, K.; Zhang, P.; Tian, G.; Liu, X.; Guo, E.; Du, Y.; Wang, Y. Transcriptome and Metabolite Analysis Reveal the Drought Tolerance of Foxtail Millet Significantly Correlated with Phenylpropanoids-Related Pathways during Germination Process under PEG Stress. BMC Plant Biol. 2020, 20, 274. [Google Scholar] [CrossRef]
- Guo, Y.; Hao, D.; Wang, X.; Wang, H.; Wu, Z.; Yang, P.; Zhang, B. Comparative Transcriptomics Reveals Key Genes Contributing to the Differences in Drought Tolerance among Three Cultivars of Foxtail Millet (Setaria italica). Plant Growth Regul. 2023, 99, 45–64. [Google Scholar] [CrossRef]
- Zhang, R.; Zhi, H.; Li, Y.; Guo, E.; Feng, G.; Tang, S.; Guo, W.; Zhang, L.; Jia, G.; Diao, X. Response of Multiple Tissues to Drought Revealed by a Weighted Gene Co-Expression Network Analysis in Foxtail Millet [Setaria italica (L.) P. Beauv.]. Front. Plant Sci. 2022, 12, 746166. [Google Scholar] [CrossRef]
- Cui, X.; Wang, B.; Chen, Z.; Guo, J.; Zhang, T.; Zhang, W.; Shi, L. Comprehensive Physiological, Transcriptomic, and Metabolomic Analysis of the Key Metabolic Pathways in Millet Seedling Adaptation to Drought Stress. Physiol. Plant. 2023, 175, e14122. [Google Scholar] [CrossRef] [PubMed]
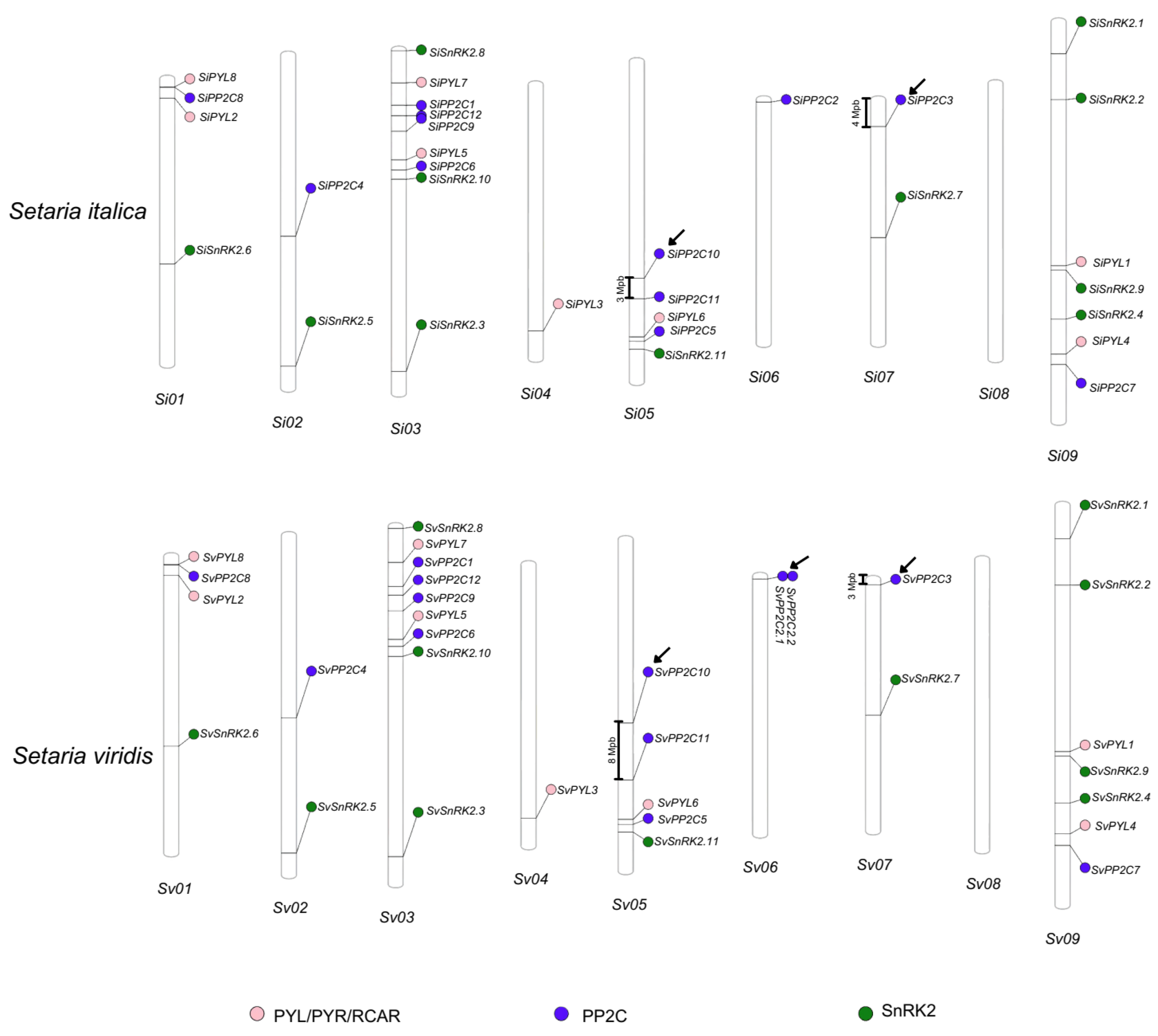
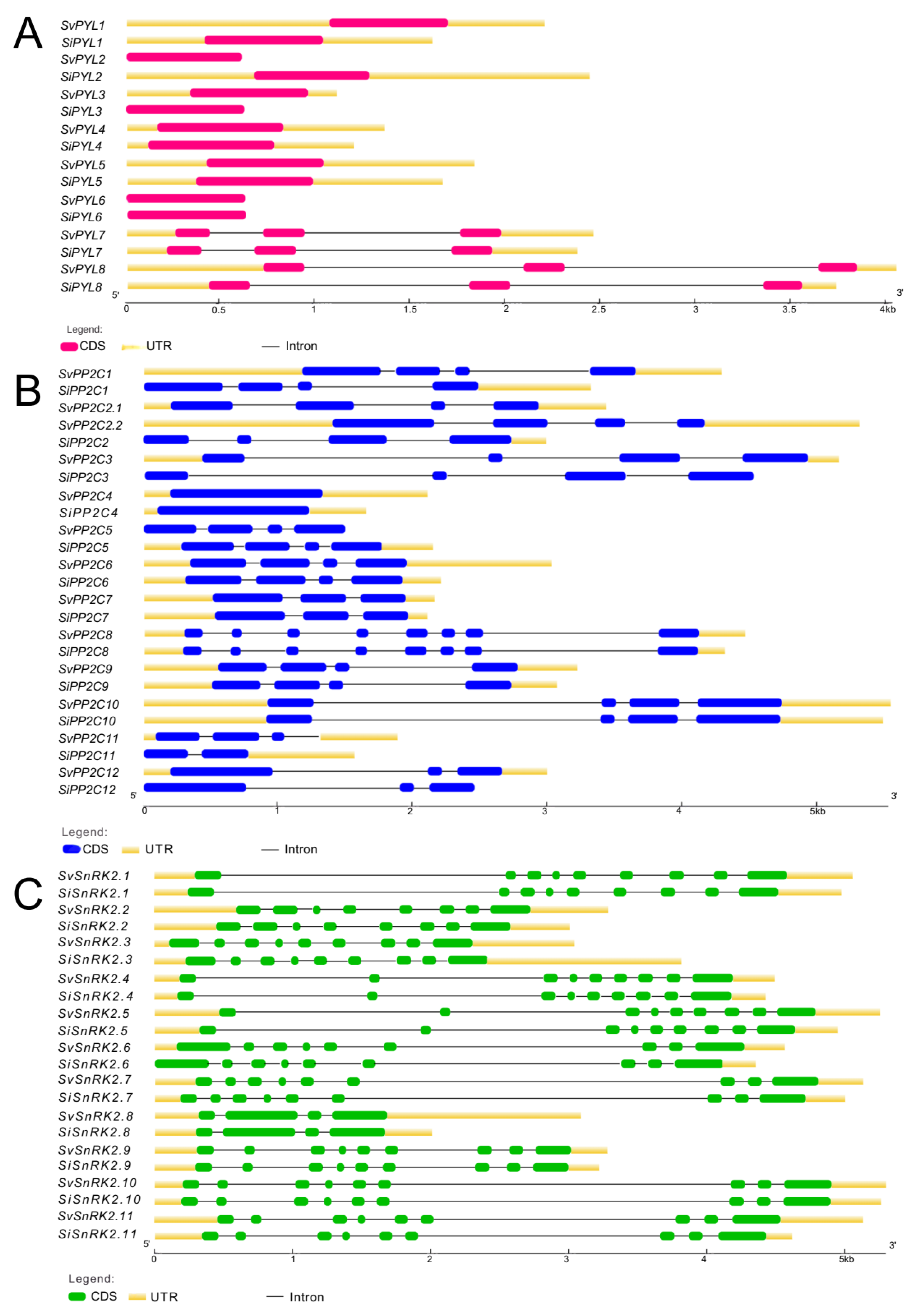
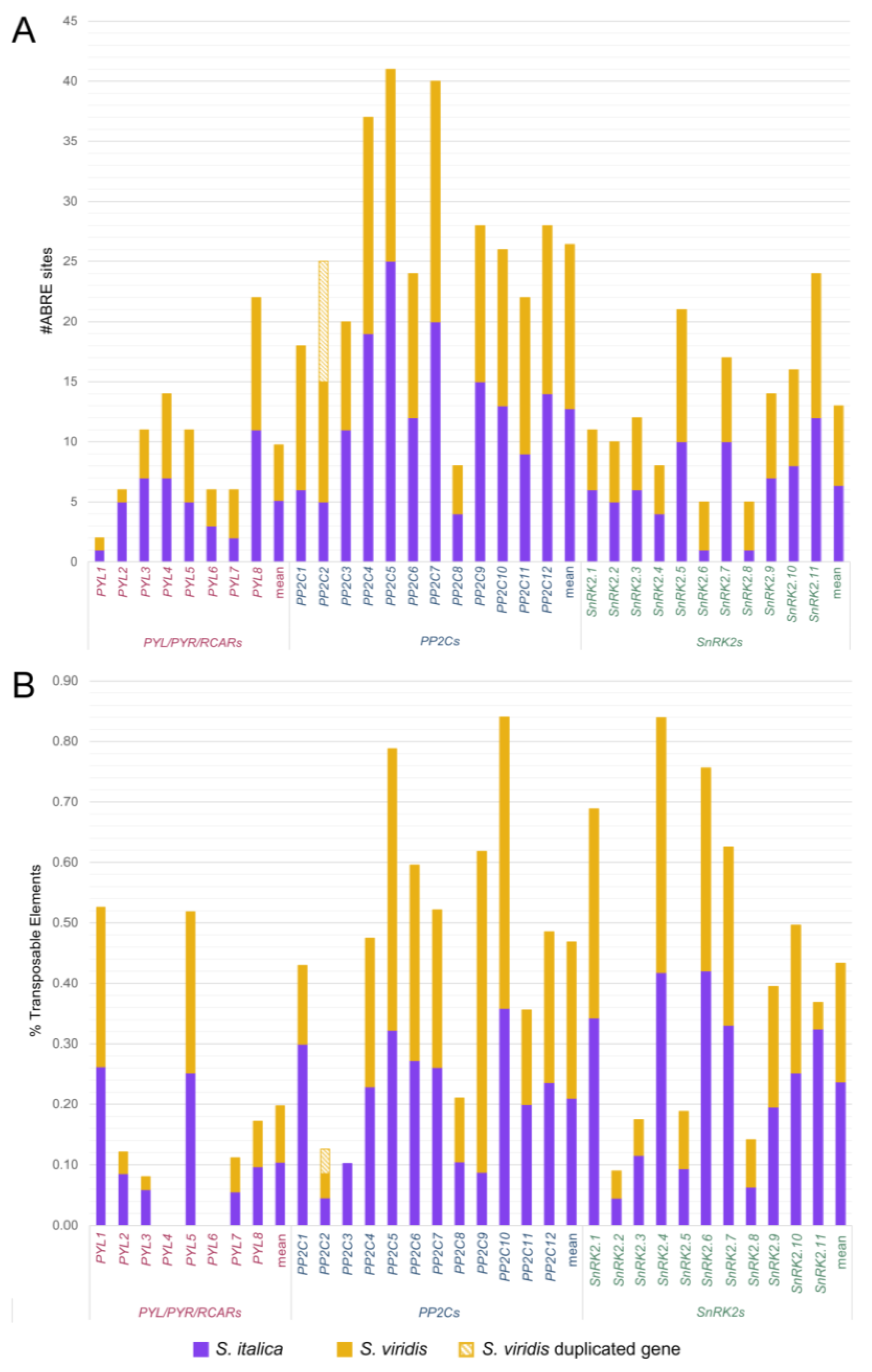
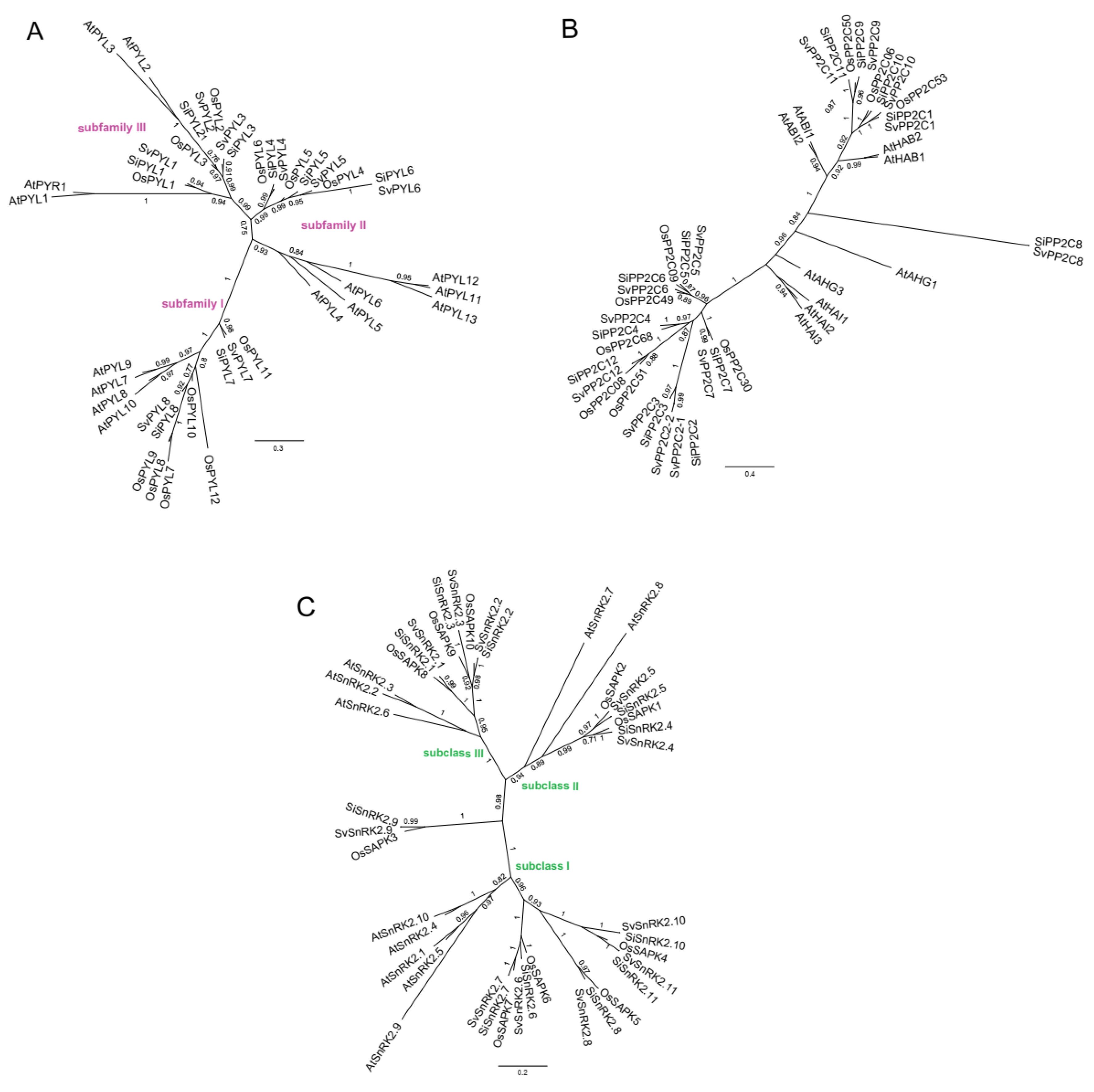
| Loci | Scaffold | Transcription Sites | Protein Length | PFAM Domain * | Phytozome Gene Identifier | |
|---|---|---|---|---|---|---|
| Start | Stop | |||||
| SiPYL1 | 9 | 36003871 | 36005478 | 207 | 42–192 | Seita.9G311900 |
| SiPYL2 | 1 | 2860240 | 2862675 | 201 | 38–183 | Seita.1G030500 |
| SiPYL3 | 4 | 36338581 | 36339202 | 206 | 47–198 | Seita.4G239500 |
| SiPYL4 | 9 | 48993015 | 48994206 | 220 | 67–213 | Seita.9G437300 |
| SiPYL5 | 3 | 16201029 | 16202684 | 204 | 53–197 | Seita.3G207900 |
| SiPYL6 | 5 | 40592574 | 40593198 | 207 | 48–195 | Seita.5G369100 |
| SiPYL7 | 3 | 4851938 | 4854302 | 205 | 46–192 | Seita.3G076200 |
| SiPYL8 | 1 | 1185260 | 1188981 | 211 | 53–198 | Seita.1G013900 |
| SvPYL1 | 9 | 34761413 | 34763538 | 207 | 48–192 | Sevir.9G318000 |
| SvPYL2 | 1 | 2710051 | 2713429 | 201 | 43–180 | Sevir.1G031000 |
| SvPYL3 | 4 | 35821353 | 35821973 | 206 | 52–198 | Sevir.4G251800 |
| SvPYL4 | 9 | 46321820 | 46323300 | 220 | 70–213 | Sevir.9G441100 |
| SvPYL5 | 3 | 15949957 | 15951675 | 204 | 56–197 | Sevir.3G213000 |
| SvPYL6 | 5 | 39467969 | 39468592 | 207 | 51–194 | Sevir.5G374800 |
| SvPYL7 | 3 | 5043188 | 5045597 | 141 | 47–131 | Sevir.3G077900 |
| SvPYL8 | 1 | 1156602 | 1160325 | 211 | 55–198 | Sevir.1G013800 |
| SiPP2C1 | 3 | 8217300 | 8220607 | 451 | 133–434 | Seita.3G121900 |
| SiPP2C2 | 6 | 375937 | 378923 | 444 | 107–433 | Seita.6G005300 |
| SiPP2C3 | 7 | 4005506 | 4010019 | 453 | 116–442 | Seita.7G021400 |
| SiPP2C4 | 2 | 26789985 | 26791627 | 374 | 76–362 | Seita.2G177500 |
| SiPP2C5 | 5 | 41297870 | 41300008 | 401 | 87–390 | Seita.5G379400 |
| SiPP2C6 | 3 | 17714334 | 17716534 | 422 | 85–411 | Seita.3G218800 |
| SiPP2C7 | 9 | 50584247 | 50586340 | 397 | 71–330 | Seita.9G460200 |
| SiPP2C8 | 1 | 1218094 | 1222395 | 358 | 105–345 | Seita.1G014100 |
| SiPP2C9 | 3 | 12049066 | 12052126 | 381 | 58–364 | Seita.3G164700 |
| SiPP2C10 | 5 | 32027849 | 32033321 | 479 | 176–462 | Seita.5G257700 |
| SiPP2C11 | 5 | 35033197 | 35034758 | 226 | 44–226 | Seita.5G294000 |
| SiPP2C12 | 3 | 9730764 | 9733216 | 399 | 89–376 | Seita.3G139000 |
| SvPP2C1 | 3 | 8481733 | 8485238 | 451 | 176–434 | Sevir.3G124100 |
| SvPP2C2.1 | 6 | 387891 | 391296 | 444 | 113–396 | Sevir.6G004900 |
| SvPP2C2.2 | 6 | 387891 | 391296 | 328 | 132–281 | Sevir.6G005000 |
| SvPP2C3 | 7 | 785451 | 790598 | 451 | 119–410 | Sevir.7G004500 |
| SvPP2C4 | 2 | 25703196 | 25705262 | 376 | 78–364 | Sevir.2G184100 |
| SvPP2C5 | 5 | 40142449 | 40144716 | 401 | 87–390 | Sevir.5G384400 |
| SvPP2C6 | 3 | 16922545 | 16924778 | 422 | 85–411 | Sevir.3G223900 |
| SvPP2C7 | 9 | 47906442 | 47908435 | 397 | 71–330 | Sevir.9G463500 |
| SvPP2C8 | 1 | 1190419 | 1194718 | 358 | 105–345 | Sevir.1G014100 |
| SvPP2C9 | 3 | 11944691 | 11947945 | 381 | 58–364 | Sevir.3G168300 |
| SvPP2C10 | 5 | 25838987 | 25843846 | 479 | 176–462 | Sevir.5G211600 |
| SvPP2C11 | 5 | 33933094 | 33934782 | 226 | 44–226 | Sevir.5G296900 |
| SvPP2C12 | 3 | 9748111 | 9750887 | 398 | 89–376 | Sevir.3G141300 |
| SiSnRK2.1 | 9 | 4716245 | 4721220 | 366 | 28–284 | Seita.9G079800 |
| SiSnRK2.2 | 9 | 11473483 | 11476494 | 362 | 23–279 | Seita.9G169200 |
| SiSnRK2.3 | 3 | 47385988 | 47389812 | 374 | 37–292 | Seita.3G369900 |
| SiSnRK2.4 | 9 | 43841286 | 43845704 | 344 | 4–260 | Seita.9G379000 |
| SiSnRK2.5 | 2 | 45956775 | 45961725 | 339 | 4–260 | Seita.2G394500 |
| SiSnRK2.6 | 1 | 27251817 | 27256180 | 454 | 94–350 | Seita.1G190000 |
| SiSnRK2.7 | 7 | 20312277 | 20317276 | 358 | 4–260 | Seita.7G100500 |
| SiSnRK2.8 | 3 | 157269 | 159276 | 380 | 4–260 | Seita.3G003200 |
| SiSnRK2.9 | 9 | 36623245 | 36626468 | 333 | 5–261 | Seita.9G318200 |
| SiSnRK2.10 | 3 | 19092667 | 19097929 | 360 | 4–260 | Seita.3G230400 |
| SiSnRK2.11 | 5 | 42440115 | 42444731 | 362 | 4–260 | Seita.5G395400 |
| SvSnRK2.1 | 9 | 4714220 | 4719224 | 366 | 28–284 | Sevir.9G078200 |
| SvSnRK2.2 | 9 | 11263105 | 11266229 | 362 | 23–279 | Sevir.9G167300 |
| SvSnRK2.3 | 3 | 46484835 | 46487553 | 375 | 37–293 | Sevir.3G387400 |
| SvSnRK2.4 | 9 | 41982274 | 41986678 | 344 | 4–260 | Sevir.9G384100 |
| SvSnRK2.5 | 2 | 44725651 | 44730688 | 339 | 4–260 | Sevir.2G405700 |
| SvSnRK2.6 | 1 | 26704997 | 26709453 | 454 | 94–350 | Sevir.1G194000 |
| SvSnRK2.7 | 7 | 19178334 | 19183417 | 358 | 4–260 | Sevir.7G108300 |
| SvSnRK2.8 | 3 | 274209 | 277259 | 379 | 4–260 | Sevir.3G004100 |
| SvSnRK2.9 | 9 | 35394567 | 35397834 | 333 | 5–261 | Sevir.9G323900 |
| SvSnRK2.10 | 3 | 18332249 | 18337532 | 360 | 4–260 | Sevir.3G235900 |
| SvSnRK2.11 | 5 | 41261960 | 41266866 | 362 | 4–260 | Sevir.5G400900 |
| Loci | Nucleotide | Amino Acids | ||||
|---|---|---|---|---|---|---|
| Identity | Substitutions | Indels * | Identity | Substitutions | Indels * | |
| SiPP2C1/SvPP2C1 | 99.7% | 3 | - | 99.7% | 1 | - |
| SiPP2C2/SvPP2C2.-1 | 99.2% | 9 | - | 99.5% | 2 | - |
| SiPP2C2/SvPP2C2.2 | 67.9% | 80 | 348 | 67.3% | 31 | 116 |
| SiPP2C3/SvPP2C3 | 99.4% | 2 | 6 | 99.5% | - | 2 |
| SiPP2C4/SvPP2C4 | 99.2% | 3 | 6 | 99.2% | 1 | 2 |
| SiPP2C5/SvPP2C5 | 99.9% | 1 | - | 100% | - | - |
| SiPP2C6/SvPP2C6 | 99.7% | 3 | - | 100% | - | - |
| SiPP2C7/SvPP2C7 | 100% | - | - | 100% | - | - |
| SiPP2C8/SvPP2C8 | 99.8% | 2 | - | 100% | - | - |
| SiPP2C9/SvPP2C9 | 99.4% | 6 | - | 99.2% | 3 | - |
| SiPP2C10/SvPP2C10 | 99.9% | 1 | - | 100% | - | - |
| SiPP2C11/SvPP2C11 | 99.7% | 2 | - | 99.1% | 2 | - |
| SiPP2C12/SvPP2C12 | 99.4% | 4 | 3 | 99.7% | - | 1 |
| SiPYL1/SvPYL1 | 100% | - | - | 100% | - | - |
| SiPYL2/SvPYL2 | 98.6% | 2 | 6 | 99.0% | - | 2 |
| SiPYL3/SvPYL3 | 99.8% | 1 | 0 | 100% | - | - |
| SiPYL4/SvPYL4 | 99.6% | 2 | - | 99.5% | 1 | - |
| SiPYL5/SvPYL5 | 99.1% | 5 | - | 100% | - | - |
| SiPYL6/SvPYL6 | 99.8% | 1 | - | 100% | - | - |
| SiPYL7/SvPYL7 | 100% | - | - | 100% | - | - |
| SiPYL8/SvPYL8 | 99.6% | 2 | - | 100% | - | - |
| SiSnRK2.1/SvSnRK2.1 | 99.9% | 1 | - | 100% | - | - |
| SiSnRK2.2/SvSnRK2.2 | 99.6% | 4 | - | 100% | - | - |
| SiSnRK2.3/SvSnRK2.3 | 99.3% | 4 | 3 | 99.4% | 1 | 1 |
| SiSnRK2.4/SvSnRK2.4 | 99.8% | 2 | - | 100% | - | - |
| SiSnRK2.5/SvSnRK2.5 | 99.7% | 3 | - | 100% | - | - |
| SiSnRK2.6/SvSnRK2.6 | 99.7% | 4 | - | 99.7% | 1 | - |
| SiSnRK2.7/SvSnRK2.7 | 100% | - | - | 100% | - | - |
| SiSnRK2.8/SvSnRK2.8 | 99.6% | 1 | 3 | 99.7% | - | 1 |
| SiSnRK2.9/SvSnRK2.9 | 99.9% | 1 | - | 99.7% | 1 | - |
| SiSnRK2.10/SvSnRK2.10 | 100% | - | - | 100% | - | - |
| SiSnRK2.11/SvSnRK2.11 | 100% | - | - | 100% | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Oliveira, I.P.; Schaaf, C.; de Setta, N. Drought Responses in Poaceae: Exploring the Core Components of the ABA Signaling Pathway in Setaria italica and Setaria viridis. Plants 2024, 13, 1451. https://doi.org/10.3390/plants13111451
de Oliveira IP, Schaaf C, de Setta N. Drought Responses in Poaceae: Exploring the Core Components of the ABA Signaling Pathway in Setaria italica and Setaria viridis. Plants. 2024; 13(11):1451. https://doi.org/10.3390/plants13111451
Chicago/Turabian Stylede Oliveira, Isabella Peres, Camila Schaaf, and Nathalia de Setta. 2024. "Drought Responses in Poaceae: Exploring the Core Components of the ABA Signaling Pathway in Setaria italica and Setaria viridis" Plants 13, no. 11: 1451. https://doi.org/10.3390/plants13111451
APA Stylede Oliveira, I. P., Schaaf, C., & de Setta, N. (2024). Drought Responses in Poaceae: Exploring the Core Components of the ABA Signaling Pathway in Setaria italica and Setaria viridis. Plants, 13(11), 1451. https://doi.org/10.3390/plants13111451





