Identification and Functional Characterization of the SaMYB113 Gene in Solanum aculeatissimum
Abstract
1. Introduction
2. Results
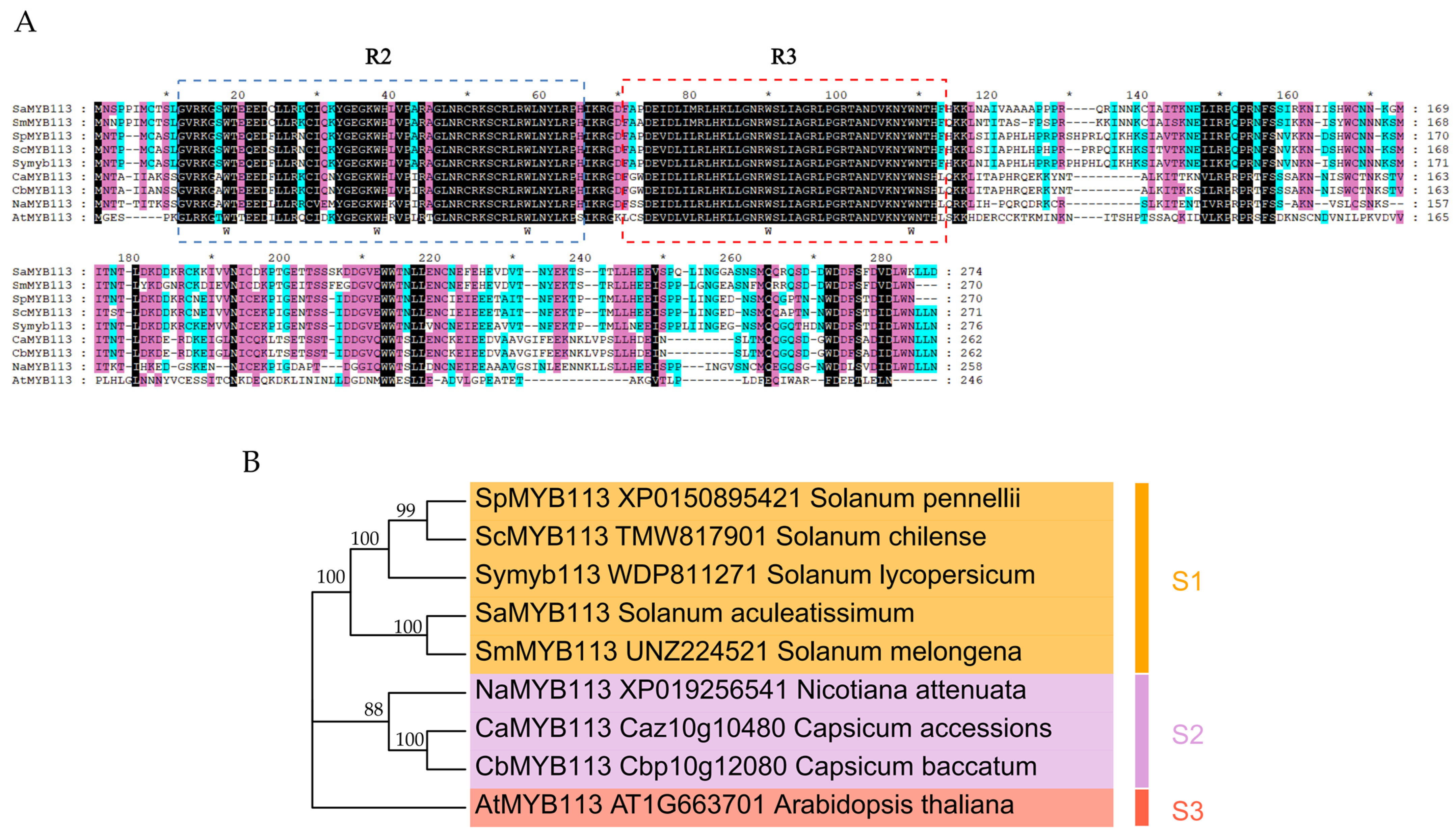
2.1. Isolation and Characterization of SaMYB113
2.2. Phylogenetic Relationship of SaMYB113 Protein
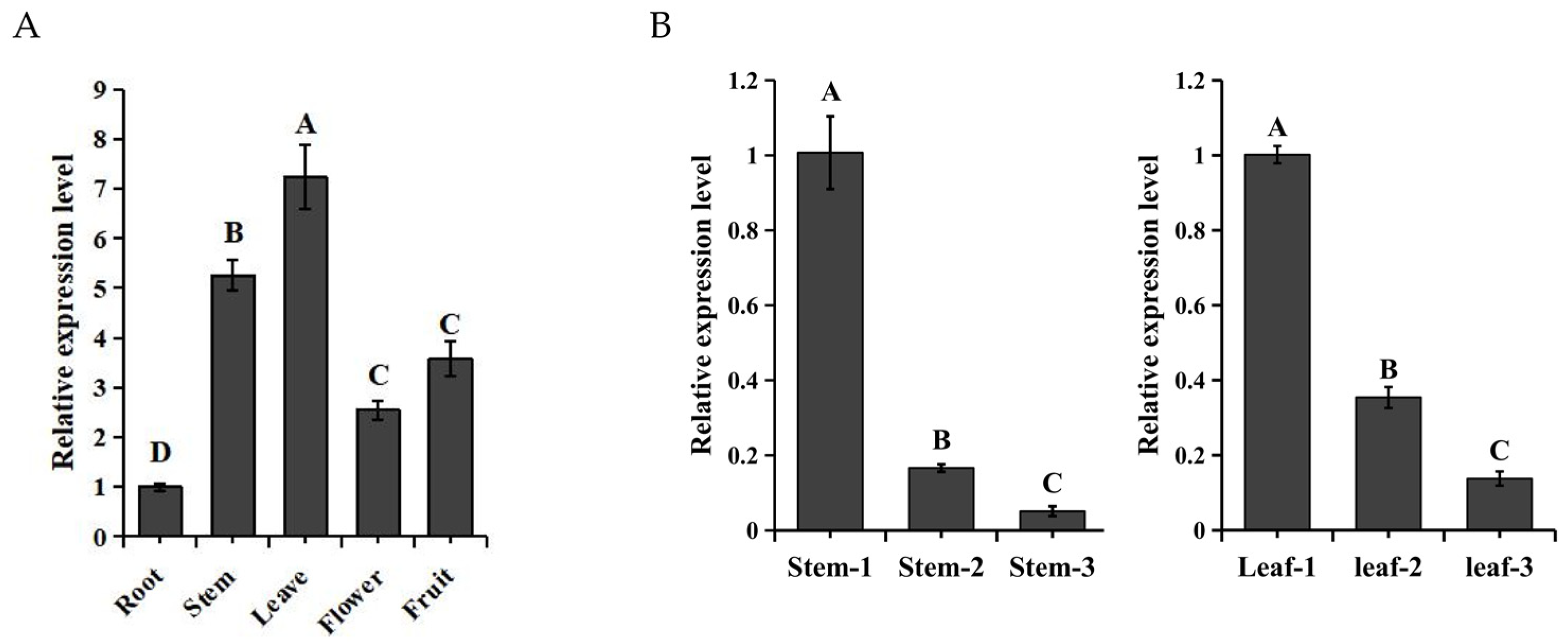
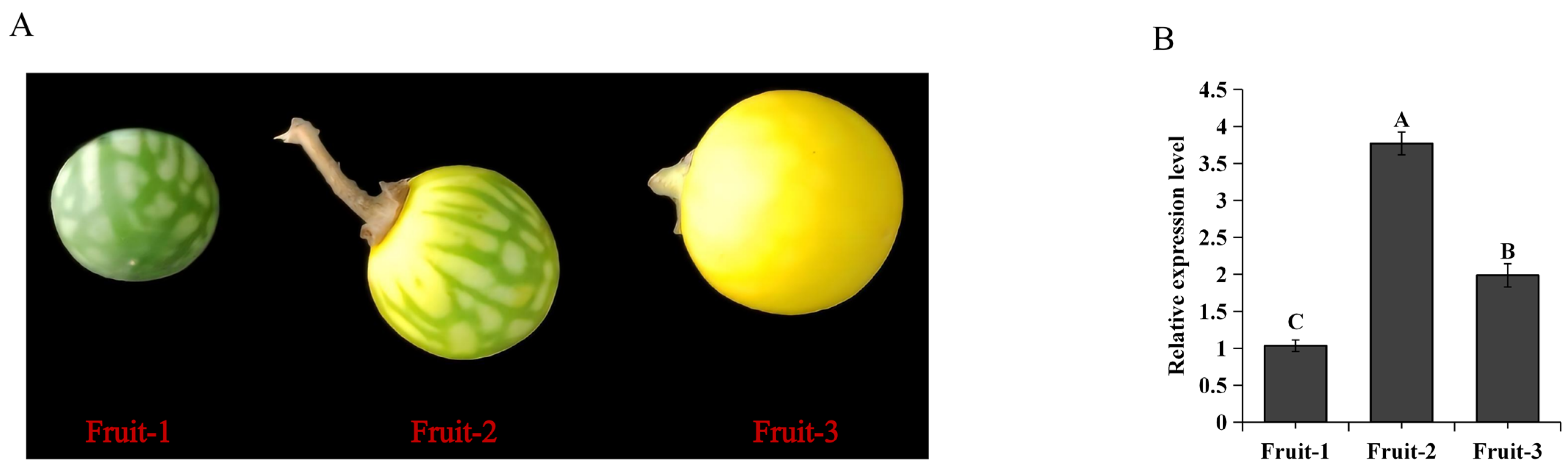
2.3. The Expression Patterns of SaMYB113
2.4. Overexpression of SaMYB113 Alters the Color of Arabidopsis Plants
2.5. Overexpression of SaMYB113 Alters the Color of Tobacco Plants
2.6. Differential Analysis of Anthocyanins in Tobacco from Different Transgenic Lines
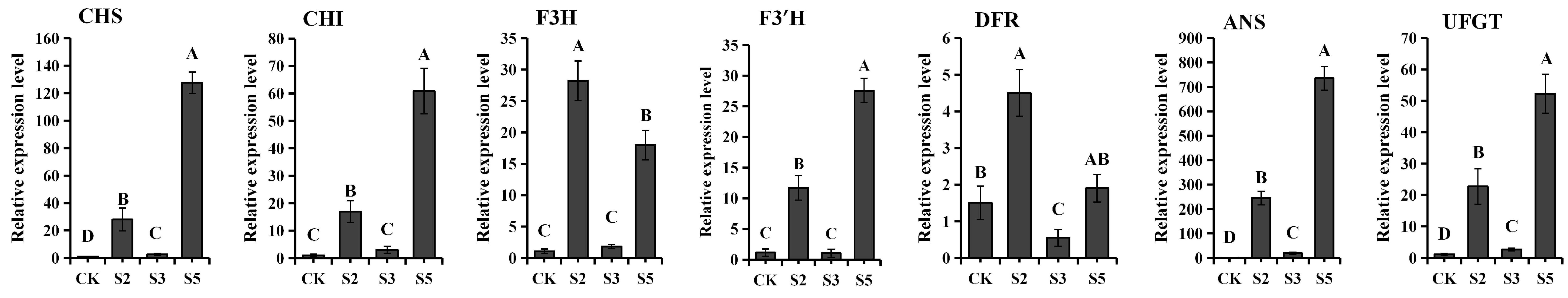
2.7. Analysis of Differences in Expression Levels of Structural Genes for Anthocyanin Synthesis in Different Tobacco Transgenic Lines
3. Discussion
4. Materials and Methods
4.1. Bioinformatics Analysis of SaMYB113
4.2. S. aculeatissimum Material, Growth Condition, and Treatment
4.3. Overexpression Vector Construction and qRT-PCR
4.4. Transgenic Arabidopsis Acquisition
4.5. Transgenic Tobacco Acquisition
4.6. Compositional Analysis of Tobacco Anthocyanins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, H.; Liu, Z.; Wu, Y.; Zheng, L.; Zhang, G. Regulatory Mechanisms of Anthocyanin Biosynthesis in Apple and Pear. Int. J. Mol. Sci. 2021, 22, 8441. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Zhang, X.; Bi, S.; You, C.; Wang, X.; Hao, Y. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, S.; Cheng, Y.; Peng, Z.; Han, J. Transcriptome profiling of anthocyanin-related genes reveals effects of light intensity on anthocyanin biosynthesis in red leaf lettuce. PeerJ 2018, 6, e4607. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Li, X.; Fu, Y.; Li, C. Environmental Stimuli and Phytohormones in Anthocyanin Biosynthesis: A Comprehensive Review. Int. J. Mol. Sci. 2023, 24, 16415. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Park, Y.; Lee, S.; Kim, D.-O. Extraction, Identification, and Health Benefits of Anthocyanins in Blackcurrants (Ribes nigrum L.). Appl. Sci. 2021, 11, 1863. [Google Scholar] [CrossRef]
- Jaakola, L.; Poole, M.; Jones, M.O.; Kämäräinen-Karppinen, T.; Koskimäki, J.J.; Hohtola, A.; Häggman, H.; Fraser, P.D.; Manning, K.; King, G.J.; et al. A SQUAMOSA MADS Box Gene Involved in the Regulation of Anthocyanin Accumulation in Bilberry Fruits. Plant Physiol. 2010, 153, 1619–1629. [Google Scholar] [CrossRef] [PubMed]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. GENETICS AND BIOCHEMISTRY OF SEED FLAVONOIDS. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Cominelli, E.; Gusmaroli, G.; Allegra, D.; Galbiati, M.; Wade, H.K.; Jenkins, G.I.; Tonelli, C. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 2008, 165, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, M.K.; Burbulis, I.E.; Winkel-Shirley, B. Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and end-products in Arabidopsis seedlings. Plant Mol. Biol. 1999, 40, 45–54. [Google Scholar] [CrossRef]
- Ilk, N.; Ding, J.; Ihnatowicz, A.; Koornneef, M.; Reymond, M. Natural variation for anthocyanin accumulation under high-light and low-temperature stress is attributable to the ENHANCER OF AG-4 2 (HUA 2) locus in combination with PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP 1) and PAP 2. New Phytol. 2015, 206, 422–435. [Google Scholar] [CrossRef]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Matus, J.T.; Aquea, F.; Arce-Johnson, P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 2008, 8, 83. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, H.; Zhu, H.; Huang, C.; Liu, C.; Chang, Y.; Kong, Z.; Zhou, Z.; Wang, G.; Lin, Y. Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 2019, 223, 705–721. [Google Scholar] [CrossRef]
- Zhou, L.; He, Y.; Li, J.; Liu, Y.; Chen, H. CBFs Function in Anthocyanin Biosynthesis by Interacting with MYB113 in Eggplant (Solanum melongena L.). Plant Cell Physiol. 2020, 61, 416–426. [Google Scholar] [CrossRef]
- Niu, S.-S.; Xu, C.-J.; Zhang, W.-S.; Zhang, B.; Li, X.; Lin-Wang, K.; Ferguson, I.B.; Allan, A.C.; Chen, K.-S. Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 2010, 231, 887–899. [Google Scholar] [CrossRef]
- Zhang, L.; Duan, Z.; Ma, S.; Sun, S.; Sun, M.; Xiao, Y.; Ni, N.; Irfan, M.; Chen, L.; Sun, Y. SlMYB7, an AtMYB4-Like R2R3-MYB Transcription Factor, Inhibits Anthocyanin Accumulation in Solanum lycopersicum Fruits. J. Agric. Food Chem. 2023, 71, 18758–18768. [Google Scholar] [CrossRef]
- Carpi, A.; Rahim, M.A.; Marin, A.; Armellin, M.; Brun, P.; Miotto, G.; Dal Monte, R.; Trainotti, L. Optimization of Anthocyanin Production in Tobacco Cells. Int. J. Mol. Sci. 2023, 24, 13711. [Google Scholar] [CrossRef]
- Zhu, L.; Liao, Y.; Lin, K.; Wu, W.; Duan, L.; Wang, P.; Xiao, X.; Zhang, T.; Chen, X.; Wang, J.; et al. Cytokinin promotes anthocyanin biosynthesis via regulating sugar accumulation and MYB113 expression in Eucalyptus. Tree Physiol. 2023, 44, tpad154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, J.; Li, P.; Sun, C.; Dong, W. Integrative metabolome and transcriptome analyses reveals the black fruit coloring mechanism of Crataegus maximowiczii CK Schneid. Plant Physiol. Biochem. 2023, 194, 111–121. [Google Scholar] [CrossRef]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Jarald, E.E.; Edwin, S.; Saini, V.; Deb, L.; Gupta, V.; Wate, S.; Busari, K. Anti-inflammatory and anthelmintic activities of Solanum khasianum Clarke. Nat. Prod. Res. 2008, 22, 269–274. [Google Scholar] [CrossRef]
- Yang, X.; Liu, F.; Zhang, Y.; Wang, L.; Cheng, Y.-f. Cold-responsive miRNAs and their target genes in the wild eggplant species Solanum aculeatissimum. BMC Genom. 2017, 18, 1000. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, W.; Hu, T.; Hu, H.; Wang, J.; Wei, Q.; Bao, C. Metabolomic and Transcriptomic Analyses Reveal the Effects of Grafting on Nutritional Properties in Eggplant. Foods 2023, 12, 3082. [Google Scholar] [CrossRef]
- Begum, T.; Munda, S.; Pandey, S.K.; Lal, M. Estimation of selection criteria through multi-year assessment of variability parameters, association studies and genetic diversity of Solanum khasianum CB Clarke. Sci. Hortic. 2022, 297, 110923. [Google Scholar] [CrossRef]
- Lal, M.; Munda, S.; Bhandari, S.; Saikia, S.; Begum, T.; Pandey, S.K. Molecular genetic diversity analysis using SSR marker amongst high solasodine content lines of Solanum khasianum CB Clarke, an industrially important plant. Ind. Crops Prod. 2022, 184, 115073. [Google Scholar] [CrossRef]
- Srivastava, M.; Sharma, S.; Misra, P. Elicitation Based Enhancement of Secondary Metabolites in Rauwolfia serpentina and Solanum khasianum Hairy Root Cultures. Pharmacogn. Mag. 2016, 12, S315. [Google Scholar] [CrossRef]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef]
- Campanella, J.J.; Smalley, J.V.; Dempsey, M.E. A phylogenetic examination of the primary anthocyanin production pathway of the Plantae. Bot. Stud. 2014, 55, 10. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cui, B.; Tan, Z.; Song, B.; Cao, H.; Zong, C. RNA sequencing and anthocyanin synthesis-related genes expression analyses in white-fruited Vaccinium uliginosum. BMC Genom. 2018, 19, 930. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A Comprehensive Review of Their Chemical Properties and Health Effects on Cardiovascular and Neurodegenerative Diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef] [PubMed]
- Lev-Yadun, S.; Gould, K.S. Role of Anthocyanins in Plant Defence. In Anthocyanins: Biosynthesis, Functions, and Applications; Winefield, C., Davies, K., Gould, K., Eds.; Springer: New York, NY, USA, 2009; pp. 22–28. [Google Scholar]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.K.; Roy, S. Genome-wide sequential, evolutionary, organizational and expression analyses of phenylpropanoid biosynthesis associated MYB domain transcription factors in Arabidopsis. J. Biomol. Struct. Dyn. 2018, 36, 1577–1601. [Google Scholar] [CrossRef] [PubMed]
- Schwinn, K.; Venail, J.; Shang, Y.; Mackay, S.; Alm, V.; Butelli, E.; Oyama, R.; Bailey, P.; Davies, K.; Martin, C. A Small Family of MYB-Regulatory Genes Controls Floral Pigmentation Intensity and Patterning in the Genus Antirrhinum. Plant Cell 2006, 18, 831–851. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Menconi, J.; Perata, P.; Gonzali, S. Novel R2R3 MYB transcription factors regulate anthocyanin synthesis in Aubergine tomato plants. BMC Plant Biol. 2023, 23, 148. [Google Scholar] [CrossRef]
- Albert, N.W.; Griffiths, A.G.; Cousins, G.R.; Verry, I.M.; Williams, W.M. Anthocyanin leaf markings are regulated by a family of R2R3-MYB genes in the genus Trifolium. New Phytol. 2015, 205, 882–893. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef] [PubMed]
- Rosinski, J.A.; Atchley, W.R. Molecular Evolution of the Myb Family of Transcription Factors: Evidence for Polyphyletic Origin. J. Mol. Evol. 1998, 46, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Ishimaru, M.; Hiraoka, K.; Honda, C. Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 2002, 215, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shan, X.; Zhou, L.; Gao, R.; Yang, S.; Wang, S.; Wang, L.; Gao, X. The R2R3-MYB Factor FhMYB5 From Freesia hybrida Contributes to the Regulation of Anthocyanin and Proanthocyanidin Biosynthesis. Front. Plant Sci. 2019, 9, 1935. [Google Scholar] [CrossRef] [PubMed]
- Docimo, T.; Francese, G.; Ruggiero, A.; Batelli, G.; De Palma, M.; Bassolino, L.; Toppino, L.; Rotino, G.L.; Mennella, G.; Tucci, M. Phenylpropanoids Accumulation in Eggplant Fruit: Characterization of Biosynthetic Genes and Regulation by a MYB Transcription Factor. Front. Plant Sci. 2016, 6, 1233. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ding, Z.; Ruan, M.; Yu, X.; Peng, M.; Liu, Y. Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci. Rep. 2017, 7, 16861. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Zhu, J.; Hao, Q.; Yuan, Y.-W.; Duan, Y.-W.; Men, S.; Wang, Q.; Hou, Q.; Liu, Z.-A.; Shu, Q. A Novel R2R3-MYB Transcription Factor Contributes to Petal Blotch Formation by Regulating Organ-Specific Expression of PsCHS in Tree Peony (Paeonia suffruticosa). Plant Cell Physiol. 2019, 60, 599–611. [Google Scholar] [CrossRef]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [CrossRef]
- Zhou, Y.; Mumtaz, M.A.; Zhang, Y.; Yang, Z.; Hao, Y.; Shu, H.; Zhu, J.; Bao, W.; Cheng, S.; Zhu, G. Response of anthocyanin biosynthesis to light by strand-specific transcriptome and miRNA analysis in Capsicum annuum. BMC Plant Biol. 2022, 22, 79. [Google Scholar] [CrossRef]
- Li, H.; He, K.; Zhang, Z.; Hu, Y. Molecular mechanism of phosphorous signaling inducing anthocyanin accumulation in Arabidopsis. Plant Physiol. Biochem. 2023, 196, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Henriques, R.; Lin, S.-S.; Niu, Q.-W.; Chua, N.-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Horsch, R.; Rogers, S.; Fraley, R. Transgenic Plants. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 1985; pp. 433–437. [Google Scholar]




| Gene Name | Forward Primer Sequence (5′ –> 3′) | Reverse Primer Sequence (5′ –> 3′) |
|---|---|---|
| SaMYB113 | GCTTCTAGGCAACAGGTGGT | CTTTGACGAGGAGGAGGAGC |
| CHS | ACTCCGGATGGCTAAGGACT | ACCTATAATGACCGCGGCTG |
| CHI | CTCAATCACCTGTTGGGGCA | CACTTTGCTGCAGGGGAAAC |
| F3H | GGCCAGACAAACCAGATGGA | ATGCCTTGGTTAAGGCCTCC |
| F3′H | GTTGATCCACAGGCGGAAGA | AACCAATCGAGTGCCGGAAT |
| DFR | TGTGGCAACGCCTATGGATT | GGACATCGACAGTTCCAGCA |
| ANS | AGGACCTCAAGTACCGACGA | TTCCAGCAACCTTGACACGA |
| UFGT | CACTTCACAATCCAACACCTCA | CGTCCAATAGGGGTAGTGCC |
| TUA | GATGTCAATGCTGCTGTCGC | CCAGGCACTACTGTTGGAGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, S.; Cai, Q.; Yang, Y.; Shen, H.; Sun, Z.; Li, L. Identification and Functional Characterization of the SaMYB113 Gene in Solanum aculeatissimum. Plants 2024, 13, 1570. https://doi.org/10.3390/plants13111570
Yi S, Cai Q, Yang Y, Shen H, Sun Z, Li L. Identification and Functional Characterization of the SaMYB113 Gene in Solanum aculeatissimum. Plants. 2024; 13(11):1570. https://doi.org/10.3390/plants13111570
Chicago/Turabian StyleYi, Songheng, Qihang Cai, Yanbo Yang, Hongquan Shen, Zhenghai Sun, and Liping Li. 2024. "Identification and Functional Characterization of the SaMYB113 Gene in Solanum aculeatissimum" Plants 13, no. 11: 1570. https://doi.org/10.3390/plants13111570
APA StyleYi, S., Cai, Q., Yang, Y., Shen, H., Sun, Z., & Li, L. (2024). Identification and Functional Characterization of the SaMYB113 Gene in Solanum aculeatissimum. Plants, 13(11), 1570. https://doi.org/10.3390/plants13111570





