Morphological and Molecular Identification of Phytophthora capsici Isolates with Differential Pathogenicity in Sechium edule
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Pathogen Isolation
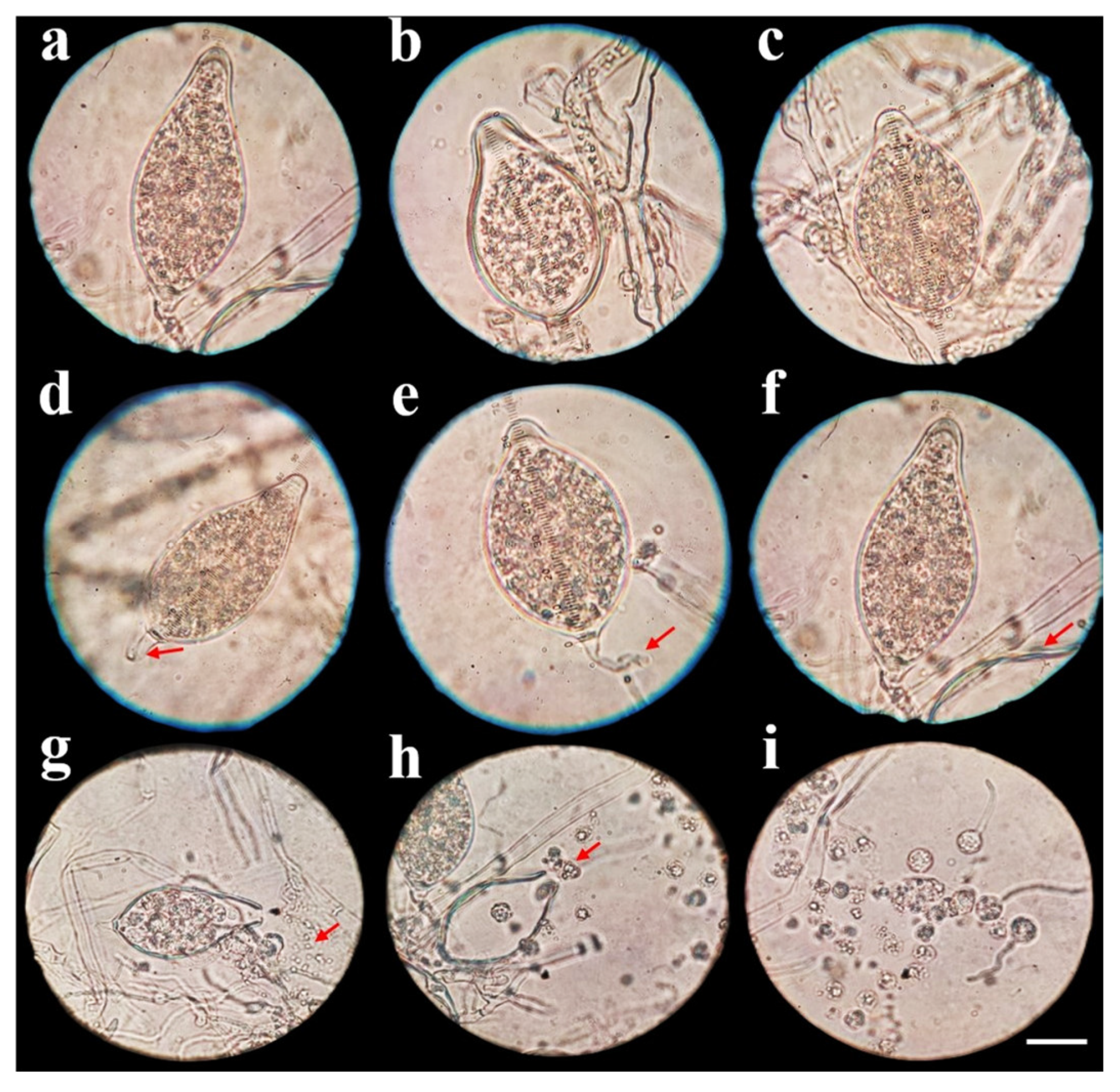
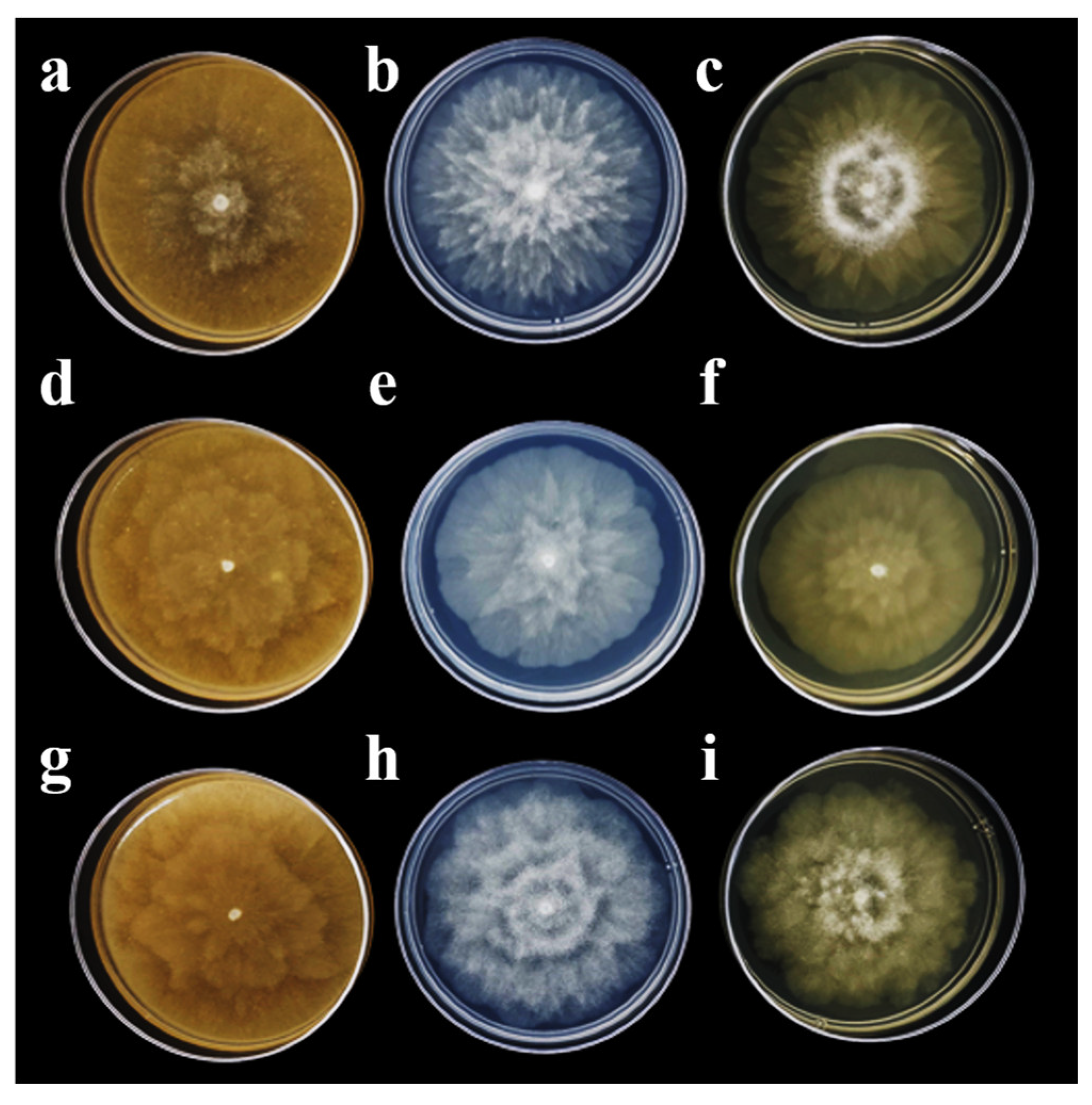
2.3. Morphological Characterization
2.4. Molecular Identification
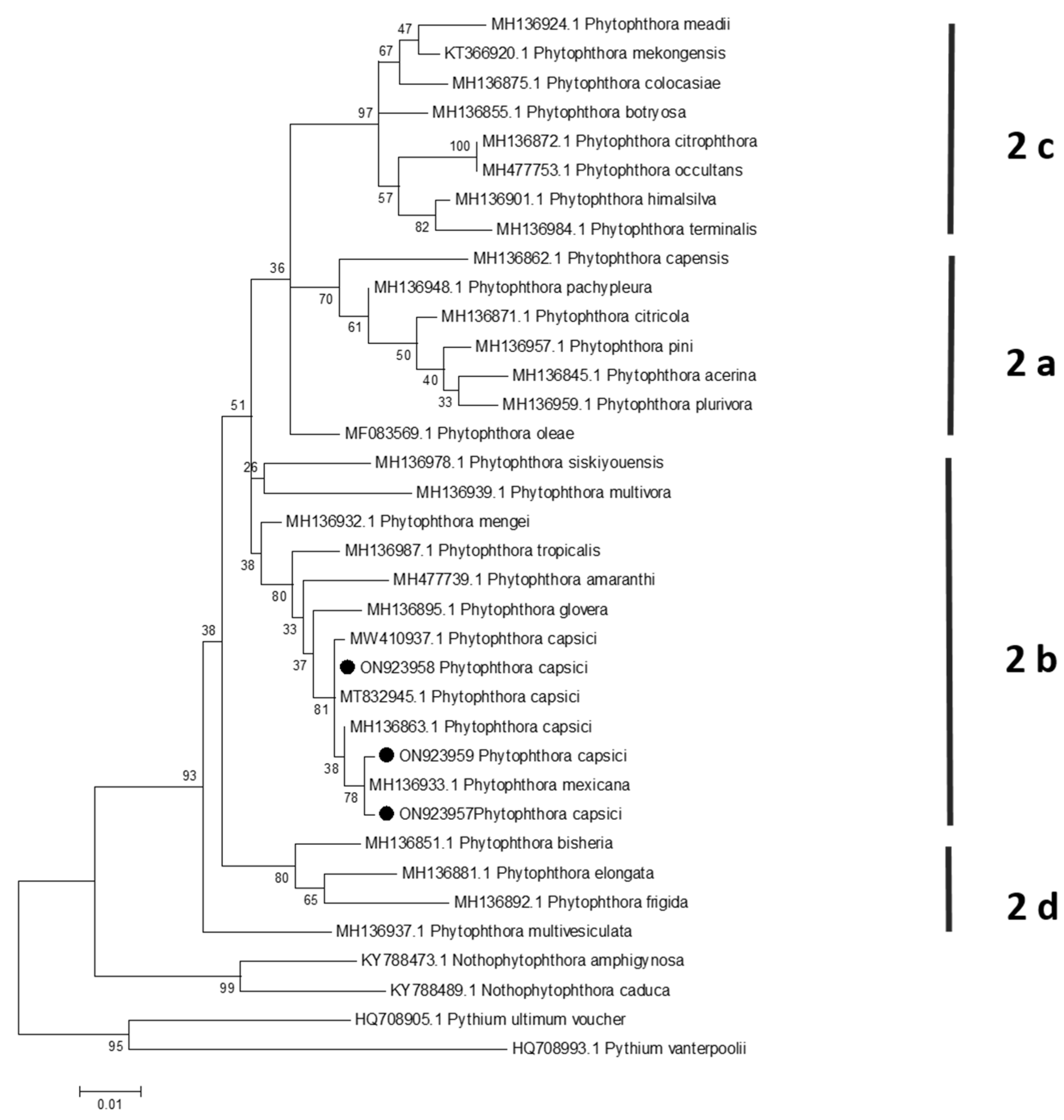
2.5. Phylogenetic Analysis
2.6. Mating Type
2.7. Pathogenicity Test
2.8. Sensitivity to Metalaxyl
2.9. Data Analysis
3. Results
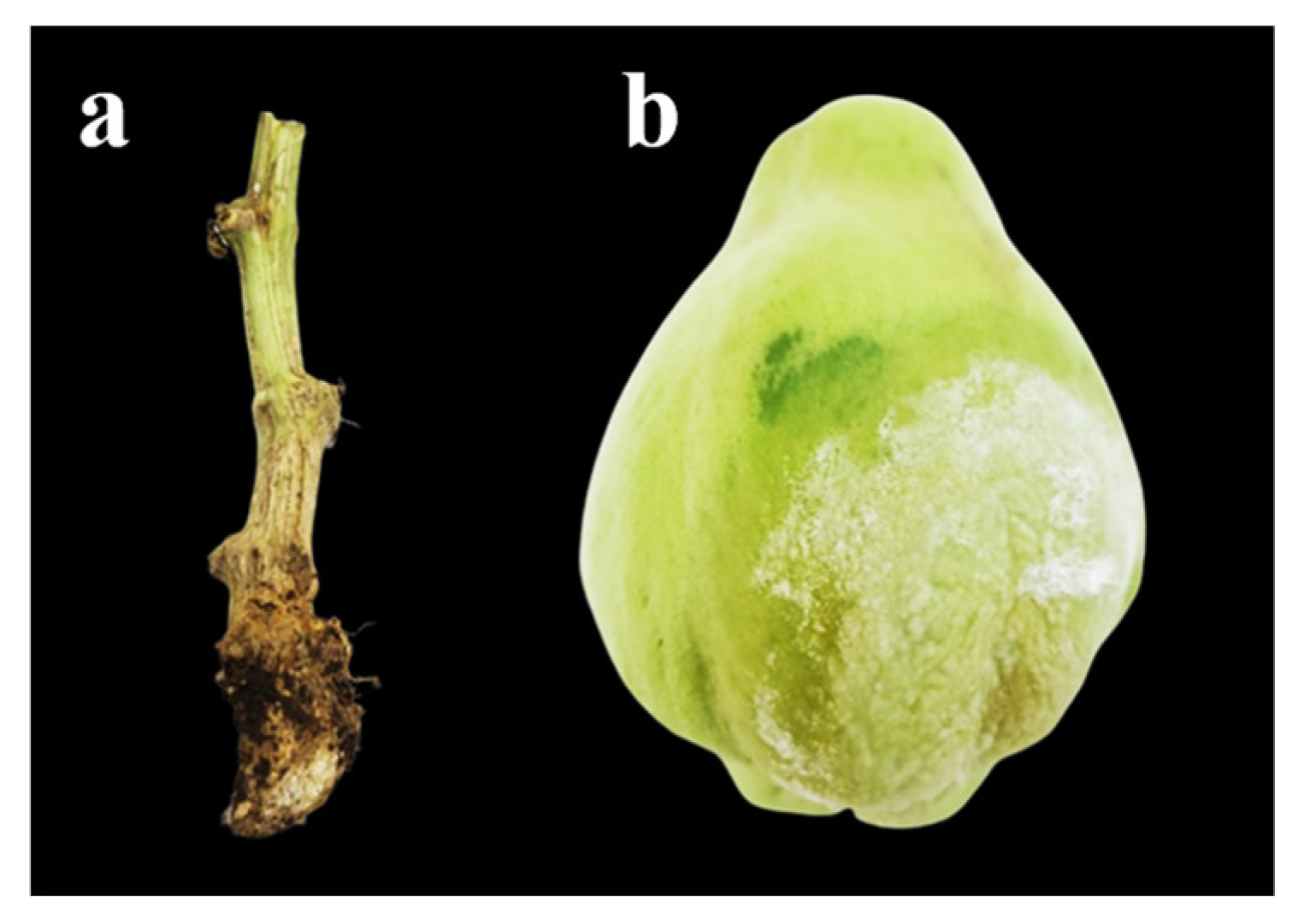
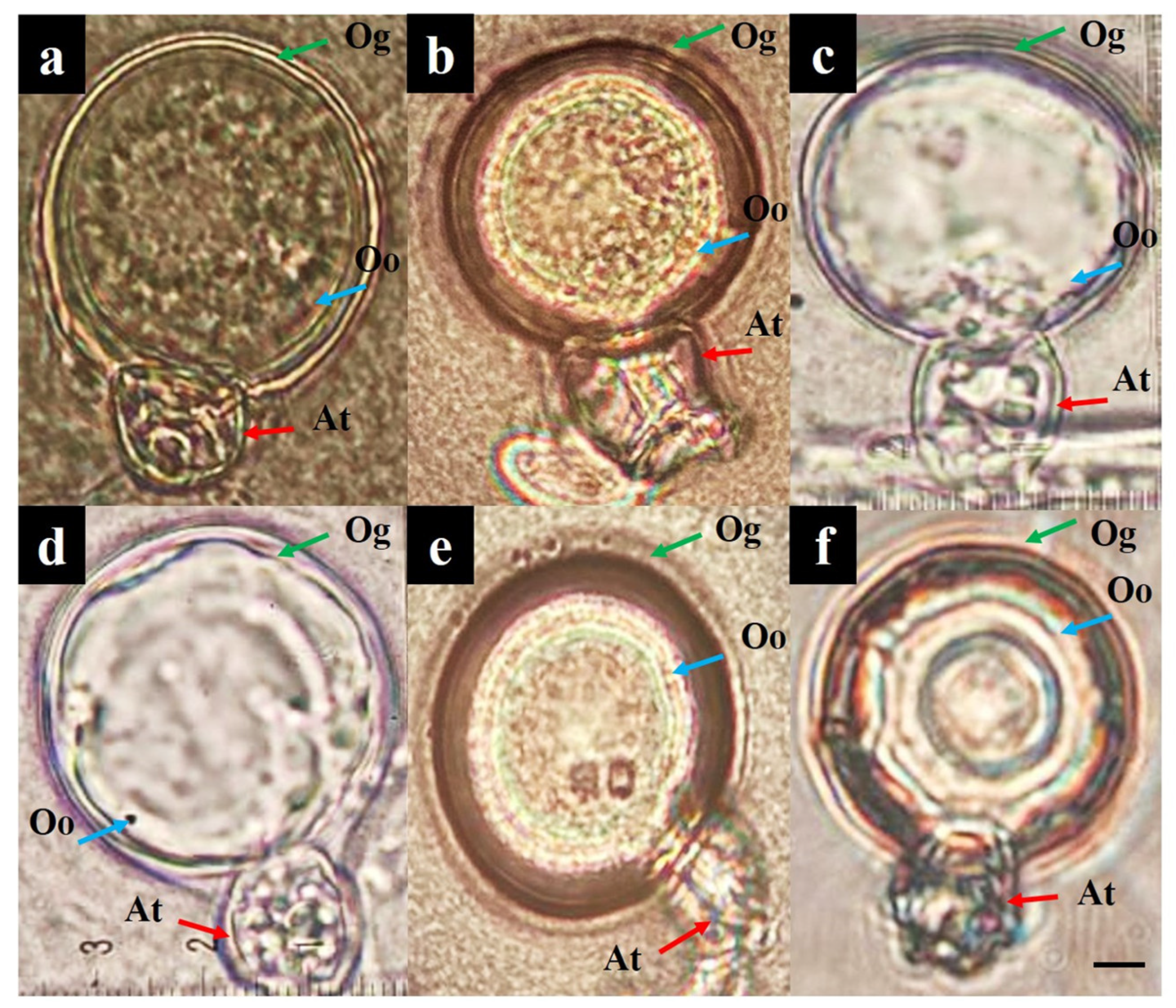
3.1. Pathogen Isolation and Morphological Characterization
3.2. Molecular Identification
3.3. Phylogenetic Analysis
3.4. Mating Type
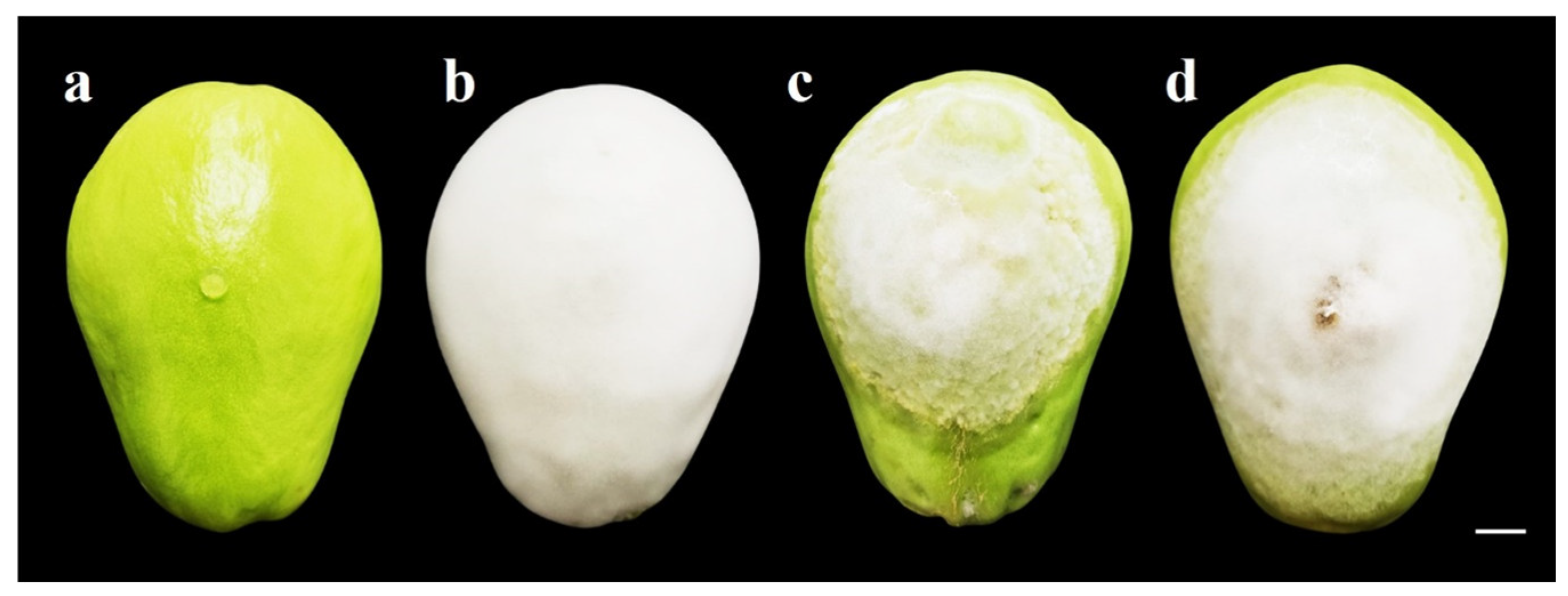
3.5. Pathogenicity Test
3.6. Sensitivity to Metalaxyl
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lamour, K.H.; Stam, R.; Jupe, J.; Huitema, E. The oomycete broad-host-range pathogen Phytophthora capsici. Mol. Plant Pathol. 2012, 13, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Bassani, I.; Larousse, M.; Tran, Q.D.; Attard, A.; Galiana, E. Phytophthora zoospores: From perception of environmental signals to inoculum formation on the host-root surface. Comput. Struct. Biotechnol. J. 2020, 18, 766–3773. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, S.; Furzer, O.; Jones, J.D.G.; Judelson, H.S.; Ali, G.S.; Dalio, R.J.D.; Roy, S.G.; Schena, L.; Zambounis, A.; Panabières, F.; et al. The top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 2015, 16, 413–434. [Google Scholar] [CrossRef] [PubMed]
- Abad, Z.G.; Burgess, T.I.; Redford, A.J.; Bienapfl, J.C.; Srivastava, S.; Mathew, R.; Jennings, K. IDphy: An international online resource for molecular and morphological identification of Phytophthora. Plant Dis. 2023, 107, 987–998. [Google Scholar] [CrossRef] [PubMed]
- Jupe, J.; Stam, R.; Howden, A.J.M.; Morris, J.A.; Zhang, R.; Hedley, P.E.; Huitema, E. Phytophthora capsici-tomato interaction features dramatic shifts in gene expresión associated with a hemi-biotrophic lifestyle. Genome Biol. 2013, 14, R63. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.N.; Abad, Z.G.; Balci, Y.; Ivors, K. Identification and detection of Phytophthora: Reviewing our progress, and identifying our needs. Plant Dis. 2012, 96, 1080–1103. [Google Scholar] [CrossRef] [PubMed]
- Majid, M.U.; Awan, M.F.; Fatima, K.; Tahir, M.S.; Ali, Q.; Rashid, B.; Husnain, T. Phytophthora capsici on chilli pepper (Capsicum annuum L.) and its management through genetic and bio-control: A review. Zemdirb.-Agric. 2016, 103, 419–430. [Google Scholar] [CrossRef]
- Kreutzer, W.A. A Phytophthora rot of cucumber fruit. Phytopathology 1937, 27, 955. [Google Scholar]
- Wiant, J.S.; Tucker, C.M. A rot of winter queen watermelon caused by Phytophthora capsici. J. Agric. Res. 1940, 60, 73–88. [Google Scholar]
- Olguín-Hernández, G.; Valdovinos-Ponce, G.; Cadena-Íñiguez, J.; Arévalo-Galarza, M.L. Etiology of Chayote (Sechium edule) wilting plants in the state of Veracruz. Rev. Mex. Fitopatol. 2013, 31, 161–169. [Google Scholar]
- Andrade-Luna, M.I.; Espinosa-Victoria, D.; Gómez-Rodríguez, O.; Cadena-Iñiguez, J.; Arévalo-Galarza, M.L.; Trejo-Téllez, L.I.; Delgadillo-Martínez, J. Severity of a Phytophthora capsici isolate in chayote Sechium edule plants at growth chamber level. Rev. Mex. Fitopatol. 2017, 35, 40–57. [Google Scholar] [CrossRef]
- Anandaraj, M.; Mathew, S.K.; Eapen, S.J.; Cissin, J.; Rosana, B.; Suseela, B.R. Morphological and molecular intervention in identifying Phytophthora spp. causing leaf and nut fall in nutmeg (Myristica fragrans Houtt.). Eur. J. Plant Pathol. 2020, 156, 373–386. [Google Scholar] [CrossRef]
- Das, A.K.; Nerkar, S.; Kumar, A.; Bawage, S. Detection, identification and characterization of Phytophthora spp. infecting citrus in India. J. Plant Pathol. 2016, 98, 55–69. [Google Scholar] [CrossRef]
- Chunthawodtiporn, J.; Hill, T.; Stoffel, K.; Van Deynze, A. Genetic analysis of resistance to multiple isolates of Phytophthora capsici and linkage to horticultural traits in bell pepper. HortScience 2019, 54, 1143–1148. [Google Scholar] [CrossRef]
- Gobena, D.; Roig, J.; Galmarini, C.; Hulvey, J.; Lamour, K. Genetic diversity of Phytophthora capsici isolates from pepper and pumpkin in Argentina. Mycologia 2012, 104, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Granke, L.L.; Windstam, S.T.; Hoch, H.C.; Smart, C.D.; Hausbeck, M.K. Dispersal and movement mechanisms of Phytophthora capsici sporangia. Phytopathology 2009, 99, 1258–1264. [Google Scholar] [CrossRef]
- Granke, L.L.; Quesada-Ocampo, L.M.; Hausbeck, M.K. Differences in virulence of Phytophthora capsici isolates from a worldwide collection on host fruits. Eur. J. Plant Pathol. 2012, 132, 281–296. [Google Scholar] [CrossRef]
- Sanogo, S.; Lamour, K.; Kousik, C.S.; Lozada, D.N.; Parada-Rojas, C.H.; Quesada-Ocampo, L.M.; Miller, S.A. Phytophthora capsici, 100 years later: Research mile markers from 1922 to 2022. Phytopathology 2023, 113, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Tena, A.; Rodríguez-Alvarado, G.; Luna-Ruíz, J.D.J.; Arreola-Romero, V.; Arriaga-Solorio, K.L.; Gómez-Dorantes, N.; Fernández-Pavia, P. Tolerance to virulence phenotypes of Phytophthora capsici in pasilla pepper cultivars. HortScience 2021, 56, 1239–1243. [Google Scholar] [CrossRef]
- Castro-Rocha, A.; Shrestha, S.; Lyon, B.; Grimaldo-Pantoja, G.L.; Flores-Marges, J.P.; Valero-Galván, J.; Lamour, K. An initial assessment of genetic diversity for Phytophthora capsici in northern and central Mexico. Mycol. Prog. 2016, 15, 1–12. [Google Scholar] [CrossRef]
- Johanson, A. Detection of banana leaf spot pathogens by PCR. Bull. OEPP 1995, 25, 99–107. [Google Scholar] [CrossRef]
- Robideau, G.P.; De Cock, A.W.A.M.; Coffey, M.D.; Voglmayr, H.; Brouwer, H.; Bala, K.; Chitty, D.W.; Désaulniers, N.; Eggertson, Q.A.; Gachon, C.M.M.; et al. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Resour. 2011, 11, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Tyler, B.M.; Hong, C. An expanded phylogeny for the genus Phytophthora. IMA Fungus 2017, 8, 355–384. [Google Scholar] [CrossRef] [PubMed]
- Coomber, A.; Saville, A.; Carbone, I.; Ristaino, J.B. An open-access T-BAS phylogeny for emerging Phytophthora species. PLoS ONE 2023, 18, e0283540. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Tena, A. Análisis Fenotípico y Genómico de Aislados de Phytophthora capsici del centro de México, y su Interacción con Cultivares de Chile (Capsicum annuum). Doctoral’s Thesis, Universidad Michoacana de San Nicolás de Hidalgo, Morelia, Mexico, 2020. [Google Scholar]
- Reyes-Tena, A.; Rodríguez-Alvarado, G.; Fernández-Pavía, S.P.; Pedraza-Santos, M.E.; Larsen, J.; Vázquez-Marrufo, G. Morphological characterization of Phytophthora capsici isolates from Jalisco and Michoacán. Mexico. Rev. Mex. Fitopatol. 2021, 39, 75–93. [Google Scholar] [CrossRef]
- Li, P.; Liu, D.; Guo, M.; Pan, Y.; Chen, F.; Zhang, H.; Gao, Z. A PCR-based Assay for Distinguishing between A1 and A2 Mating Types of Phytophthora capsici. J. Am. Soc. Hortic. 2017, 142, 260–264. [Google Scholar] [CrossRef]
- Lamour, K.H.; Hausbeck, M.K. Mefenoxam insensitivity and the sexual stage of Phytophthora capsici in Michigan cucurbit fields. Phytopathology 2000, 90, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Zar, J.H. Biostatistical Analysis, 5th ed.; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2010; 665p. [Google Scholar]
- Barchenger, D.W.; Lamour, K.H.; Bosland, P.W. Challenges and strategies for breeding resistance in Capsicum annuum to the multifarious pathogen, Phytophthora capsici. Front. Plant Sci. 2018, 9, 628. [Google Scholar] [CrossRef]
- Kreutzer, W.A.; Bodine, E.W.; Durrell, L.W. Cucurbit disease and rot of tomato fruit caused by Phytophthora capsici. Phytopathology 1940, 30, 972–976. [Google Scholar]
- Cheng, W.; Lin, M.; Qiu, M.; Kong, L.; Xu, Y.; Li, Y.; Wang, Y.; Ye, W.; Dong, S.; He, S.; et al. Chitin synthase is involved in vegetative growth, asexual reproduction and pathogenesis of Phytophthora capsici and Phytophthora sojae. Environ. Microbiol. 2019, 21, 4537–4547. [Google Scholar] [CrossRef] [PubMed]
- Aragaki, M.; Uchida, J.Y. Morphological distinctions between Phytophthora capsici and P. tropicalis sp. nov. Mycologia 2001, 93, 137–145. [Google Scholar] [CrossRef]
- Pons-Hernández, J.L.; Guerrero-Aguilar, B.Z.; González-Chavira, M.M.; González-Pérez, E.; Villalobos-Reyes, S.; Muñoz-Sánchez, C.I. Phenotypic variability of Phytophthora capsici isolates in Guanajuato. Mex. J. Agric. Sci. 2020, 11, 1891–1901. [Google Scholar] [CrossRef]
- Soto-Plancarte, A.; Fernández-Pavía, S.P.; Rodríguez-Alvarado, G.; López-Pérez, L.; Fernández-Pavía, Y.L.; Pedraza-Santos, M.E.; Álvarez-Vargas, J.E. Phytophthora capsici and P. drechsleri mating types A1 and A2 coexist in ornamental nursery plants. Rev. Mex. Fitopatol. 2018, 36, 298–307. [Google Scholar] [CrossRef]
- Schlink, K. Identification and characterization of differentially expressed genes from Fagus sylvatica roots after infection with Phytophthora citricola. Plant Cell Rep. 2009, 28, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Meszka, B.; Michalecka, M. Identification of Phytophthora spp. isolated from plants and soil samples on strawberry plantations in Poland. J. Plant Dis. Prot. 2016, 123, 29–36. [Google Scholar] [CrossRef]
- Beketova, M.P.; Sokolova, E.A.; Malyuchenko, O.P.; Alekseev, Y.I.; Kuznetsova, M.A.; Kozlovsky, B.E.; Rogozina, E.V.; Khavkin, E.E. On molecular identification of Phytophthora infestans genotypes. Russ. Agric. Sci. 2014, 40, 435–438. [Google Scholar] [CrossRef]
- Cooke, D.E.L.; Duncan, J.M.; Williams, N.A.; Hagenaar de-Weerdt, M.; Bonants, P.J.M. Identification of Phytophthora species on the basis of restriction enzyme fragment analysis of the internal transcribed spacer regions of ribosomal RNA*. Bull. OEPP 2000, 30, 519–523. [Google Scholar] [CrossRef]
- Rodríguez-Alvarado, G.; Díaz-Celaya, M.; Grünwald, N.J.; Fieland, V.; Garay-Serrano, E.; Fernández-Pavía, S.P. Phytophthora palmivora agente causal de la pudrición de fruto de papaya (Carica papaya) en Chiapas, México. Biotecnol. Sustentabilidad 2020, 5, 37–47. [Google Scholar] [CrossRef]
- Yang, X.; Hong, C. Differential usefulness of nine commonly used genetic markers for identifying Phytophthora species. Front. Microbiol. 2018, 9, 2334. [Google Scholar] [CrossRef]
- Afandi, A.; Masanto; Wibowo, A.; Afandi; Loekito, S.; Subandiyah, S.; Hieno, A.; Otsubo, K.; Koji, K. Molecular identification of oomycetes related to horticultural crops in Southern Sumatera and Java, Indonesia. J. Trop. Plant Pests Dis. 2022, 22, 90–99. [Google Scholar] [CrossRef]
- Silva-Rojas, H.V.; Fernández-Pavía, S.P.; Góngora-Canul, C.; Macías-López, B.C.; Ávila-Quezada, G.D. Spatio-Temporal Distribution of Pepper (Capsicum annuum L.) Wilt in Chihuahua and Identification of the Causal Agent Phytophthora capsici Leo. Rev. Mex. Fitopatol. 2009, 27, 134–147. [Google Scholar]
- Díaz-Nájera, J.F.; Vargas Hernández, M.; Leyva Mir, S.G.; Ayvar Serna, S.; Michel Aceves, A.C.; Alvarado Gómez, O.G.; Morphological and molecular identification of Phytophthora capsici L. in pipiana pumpkin and its greenhouse management. Rev. Chapingo Ser. Hortic. 2015, 21, 157–168. [Google Scholar] [CrossRef]
- Vogel, G.; Gore, M.A.; Smart, C.D. Genome-wide association study in New York Phytophthora capsici isolates reveals loci involved in mating type and mefenoxam sensitivity. Phytopathology 2021, 111, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Cardenas-Rodríguez, J.; Yáñez Juárez, M.G.; López Orona, C.A.; Ayala Tafoya, F.; López Urquidez, G.A.; Romero Gómez, S.J. Phytophthora species associated with irrigation water in the Culiacán Valley. Rev. Mex. Cienc. Agric. 2021, 12, 473–483. [Google Scholar] [CrossRef]
- Chepsergon, J.; Motaung, T.E.; Bellieny-Rabelo, D.; Moleleki, L.N. Organize, don’t agonize: Strategic success of Phytophthora species. Microorganisms 2020, 8, 917. [Google Scholar] [CrossRef]
- Jia, Y.J.; Feng, B.Z.; Sun, W.X.; Zhang, X.G. Polygalacturonase, pectate lyase and pectin methylesterase activity in pathogenic strains of Phytophthora capsici incubated under different conditions. J. Phytopathol. 2009, 157, 585–591. [Google Scholar] [CrossRef]
- Qi, R.D.; Ding, J.C.; Gao, Z.M.; Ni, C.G.; Jiang, J.J.; Li, P. Resistance of Phytophthora capsici isolates to metalaxyl in Anhui Province. Acta Phytopathol. 2008, 35, 245–250. [Google Scholar]
- Lamour, K.H.; Hausbeck, M.K. The dynamics of mefenoxam insensitivity in a recombining population of Phytophthora capsici characterized with amplified fragment length polymorphism markers. Phytopathology 2001, 91, 553–557. [Google Scholar] [CrossRef]


| Isolate Code | Location | Coordinates 1 | Sporangium Length (µm) 2 | Sporangium Width (µm) 2 | Papilla Length (µm) 2 | Sporangia Form |
|---|---|---|---|---|---|---|
| A1-C | Tozongo, Coscomatepec, Mexico | −97.060556, 19.100556, 1660 | 76.43 ± 1.41 a | 43.85 ± 0.36 a | 6.65 ± 0.12 a | Papillate (ellipsoid piriform ovoid) |
| A2-H | Tenejapa, Huatusco, Mexico | −97.004167, 19.130556, 1360 | 73.20 ± 1.22 a | 42.01 ± 0.22 b | 6.33 ± 0.10 a | Papillate (ellipsoid piriform ovoid) |
| A3-O | Rincon Grande, Orizaba, Mexico | −97.078874, 18.837815, 1230 | 73.91 ± 1.45 a | 42.21 ± 0.25 b | 6.38 ± 0.11 a | Papillate (ellipsoid piriform ovoid) |
| Isolate Code | Primers | Amplicon Size (pb) | GenBank-Accession Number of Isolates from This Study | Similarity with P. capsici Sequences in GenBank |
|---|---|---|---|---|
| A1-C | ITS 5/ITS 4 | 772 | ON886223 | 100% (MG865467.1) |
| A2-H | 648 | ON898572 | 100% (MF322868.1) | |
| A3-O | 689 | ON898571 | 100% (MH025884.1) | |
| A1-C | COI F/COI R | 646 | ON923957 | 100% (NC_063804.1) |
| A2-H | 642 | ON923958 | 100% (MT832945.1) | |
| A3-O | 693 | ON923959 | 99.86% (MN369544.1) |
| Crosses of Isolates and Strains | Mating Type | Oogonia Diameter (µm) 1 | Oospore Diameter (µm) 1 |
|---|---|---|---|
| A1-C x A2-H | A2 x A1 | 38.2 ± 1.5 | 31.5 ± 0.3 |
| A2-H x A3-O | A1 x A2 | 37.9 ± 0.8 | 31.2 ± 1.1 |
| CPV-259 x A1-C | A1 x A2 | 38.0 ± 0.5 | 31.1 ± 1.3 |
| CPV-259 x A3-O | A1 x A2 | 37.6 ± 0.7 | 30.9 ± 0.4 |
| CPV-276 x A1-C | A1 x A2 | 38.1 ± 1.2 | 31.4 ± 0.6 |
| CPV-276 x A3-O | A1 x A2 | 37.7 ± 0.9 | 31.0 ± 1.0 |
| Isolate | Lesion Length (cm) 1 | Lesion Width (cm) 1 | Mycelium Length (cm) 1 | Mycelium Width (cm) 1 |
|---|---|---|---|---|
| A1-C | 12.81 ± 0.07 a | 9.80 ± 0.06 a | 11.85 ± 0.06 a | 8.90 ± 0.03 a |
| A2-H | 8.84 ± 0.04 c | 5.79 ± 0.07 c | 7.88 ± 0.03 c | 4.80 ± 0.06 c |
| A3-O | 9.79 ± 0.05 b | 6.89 ± 0.04 b | 8.82 ± 0.05 b | 5.78 ± 0.06 b |
| Isolate | Length/Breadth Ratio (cm) | Sensitivity to Metalaxyl (%) a | Sensitivity Scale a |
|---|---|---|---|
| A1-C | 2.85 ± 0.01 | 34.62 | IS |
| A2-H | 1.82 ± 0.01 | 21.98 | S |
| A3-O | 1.86 ± 0.02 | 25.88 | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soto-Contreras, A.; Caamal-Chan, M.G.; Ramírez-Mosqueda, M.A.; Murguía-González, J.; Núñez-Pastrana, R. Morphological and Molecular Identification of Phytophthora capsici Isolates with Differential Pathogenicity in Sechium edule. Plants 2024, 13, 1602. https://doi.org/10.3390/plants13121602
Soto-Contreras A, Caamal-Chan MG, Ramírez-Mosqueda MA, Murguía-González J, Núñez-Pastrana R. Morphological and Molecular Identification of Phytophthora capsici Isolates with Differential Pathogenicity in Sechium edule. Plants. 2024; 13(12):1602. https://doi.org/10.3390/plants13121602
Chicago/Turabian StyleSoto-Contreras, Anell, María G. Caamal-Chan, Marco A. Ramírez-Mosqueda, Joaquín Murguía-González, and Rosalía Núñez-Pastrana. 2024. "Morphological and Molecular Identification of Phytophthora capsici Isolates with Differential Pathogenicity in Sechium edule" Plants 13, no. 12: 1602. https://doi.org/10.3390/plants13121602





