The Ability of Different Tea Tree Germplasm Resources in South China to Aggregate Rhizosphere Soil Characteristic Fungi Affects Tea Quality
Abstract
1. Introduction
2. Results and Discussion
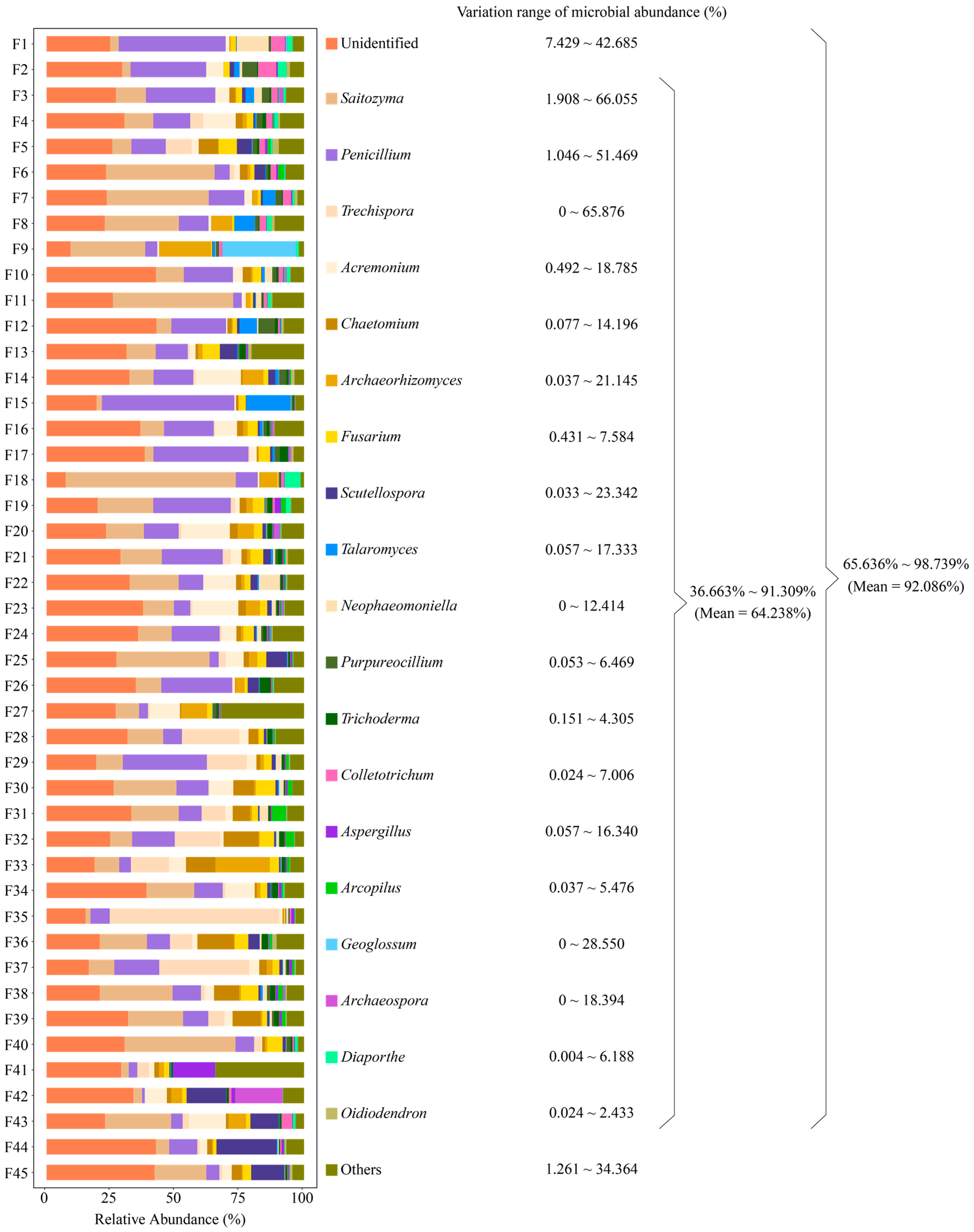
2.1. Basic Information on the Rhizosphere Soil Fungal Community of Tea Tree
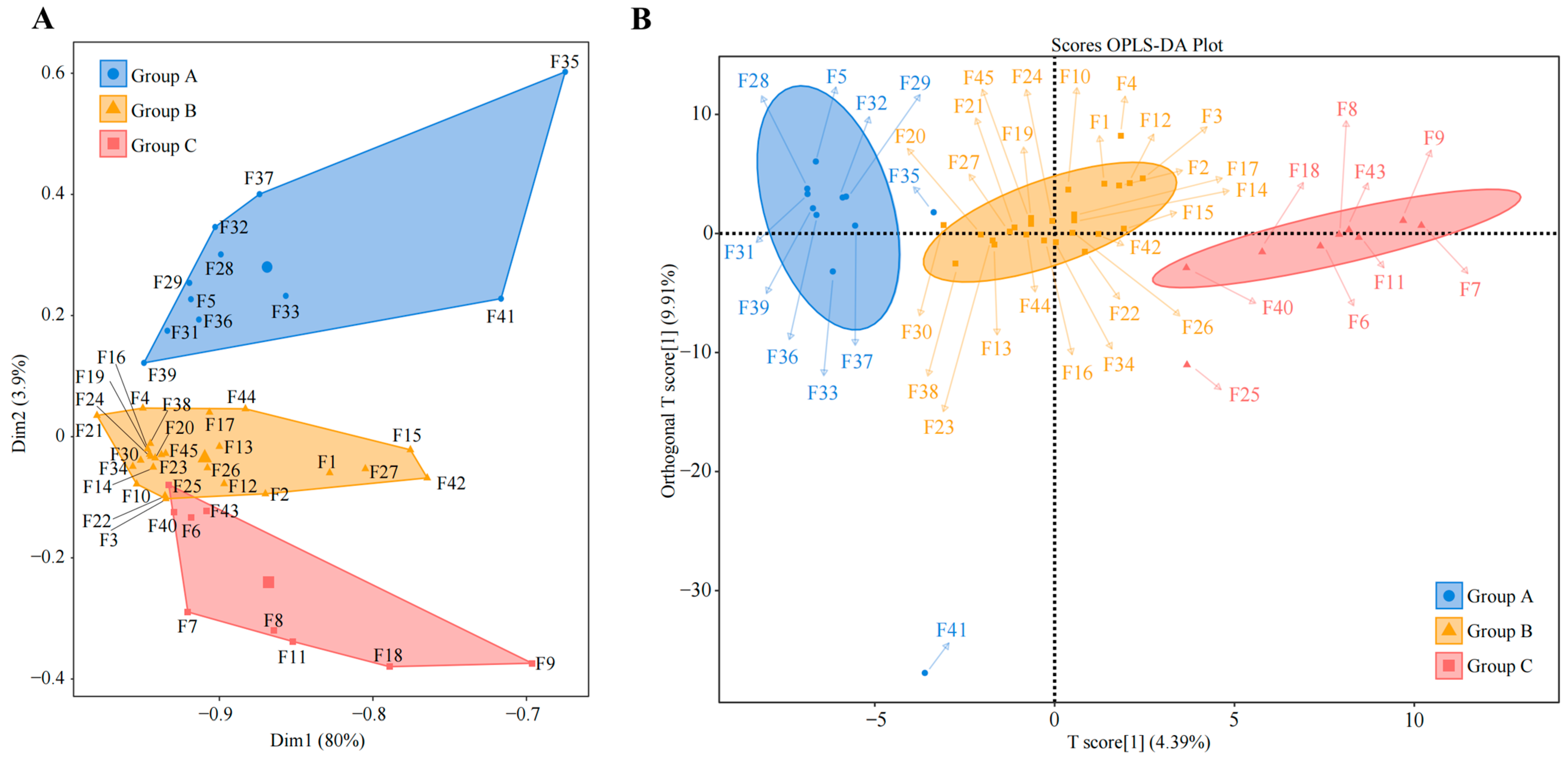
2.2. Tea Germplasm Resources Recruited and Aggregated Similar Dominant Fungal Populations
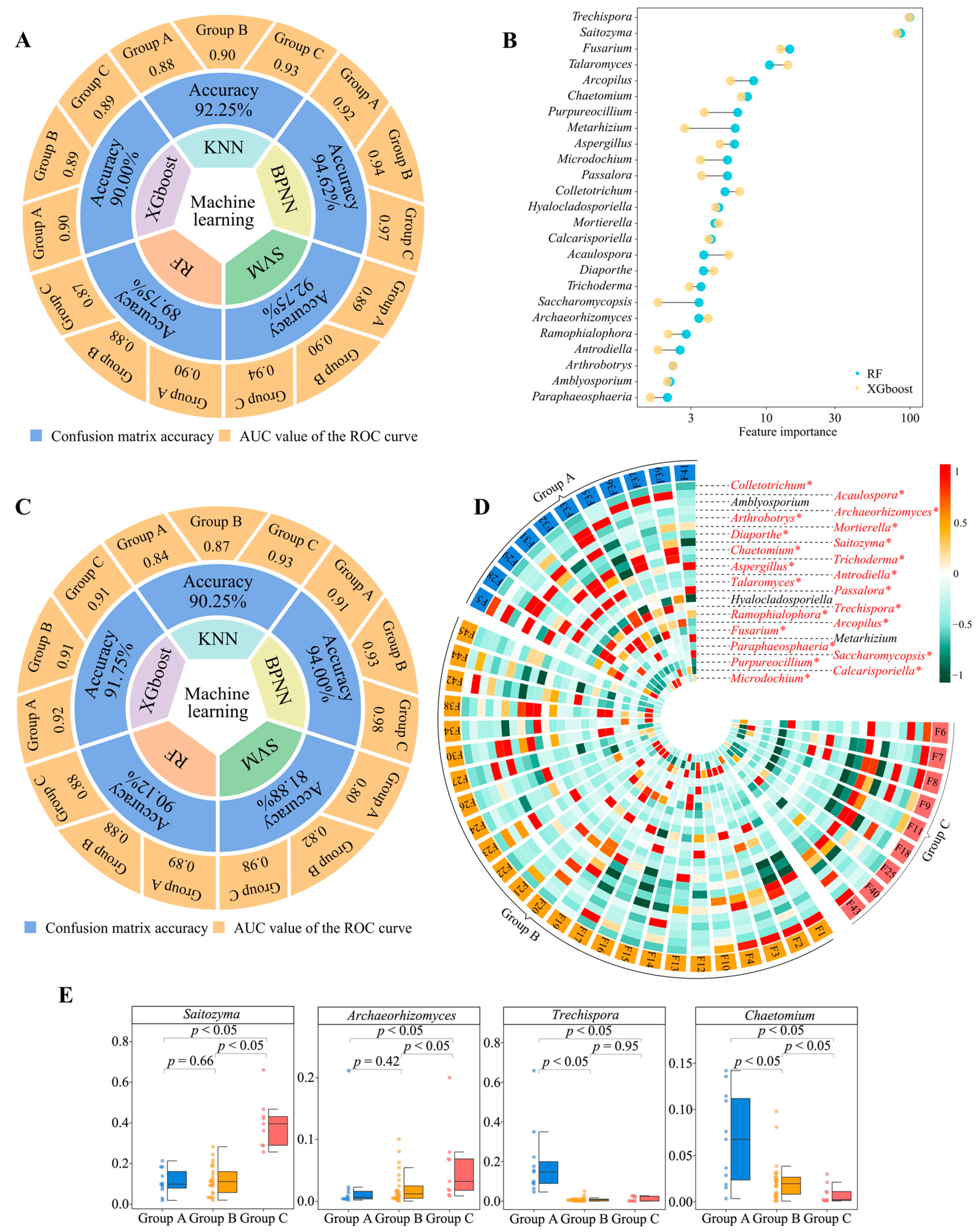
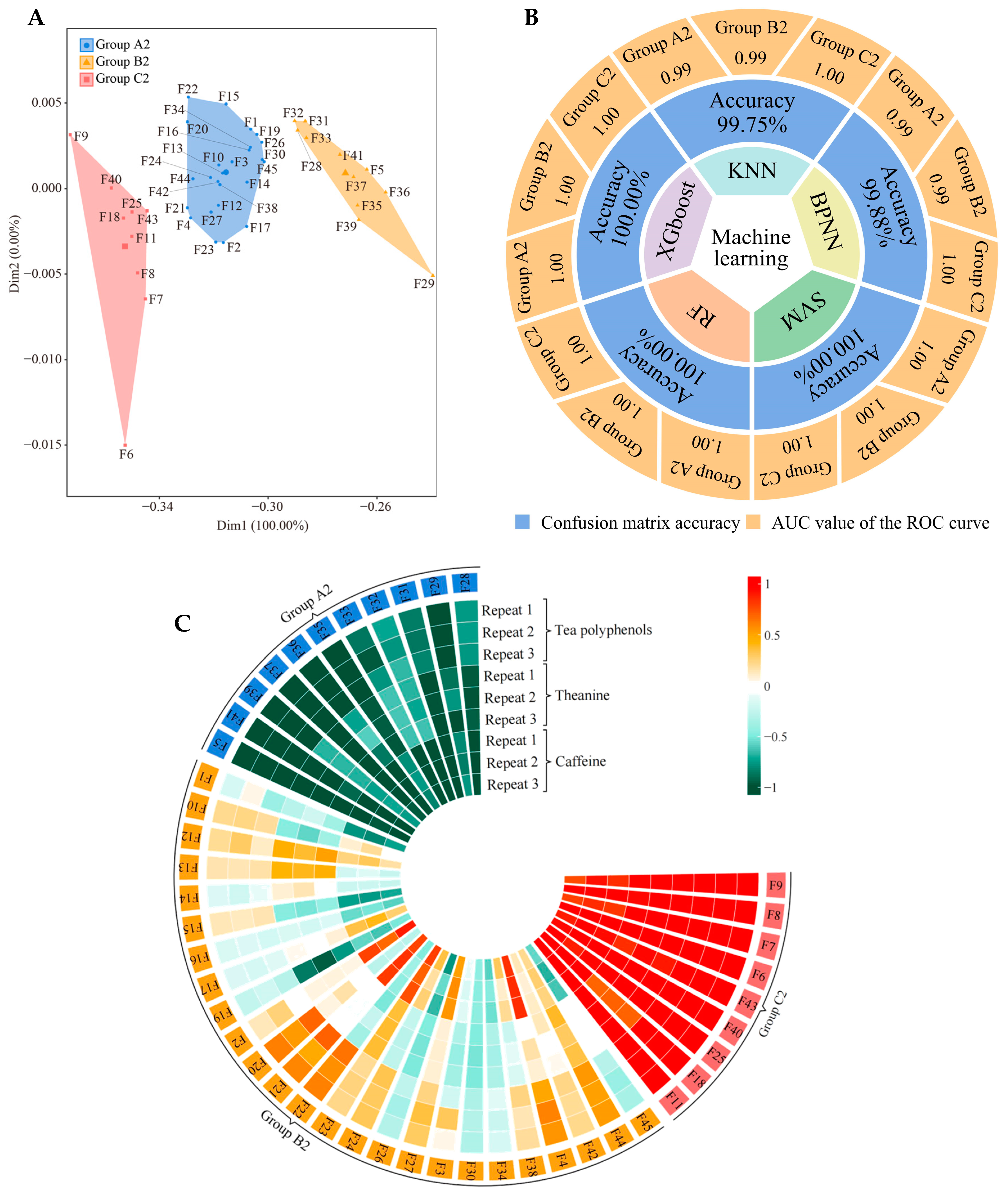
2.3. Screening and Validation of Characteristic Fungi
2.4. Soil Available Nutrient Content
2.5. Tea Quality Index Content Analysis
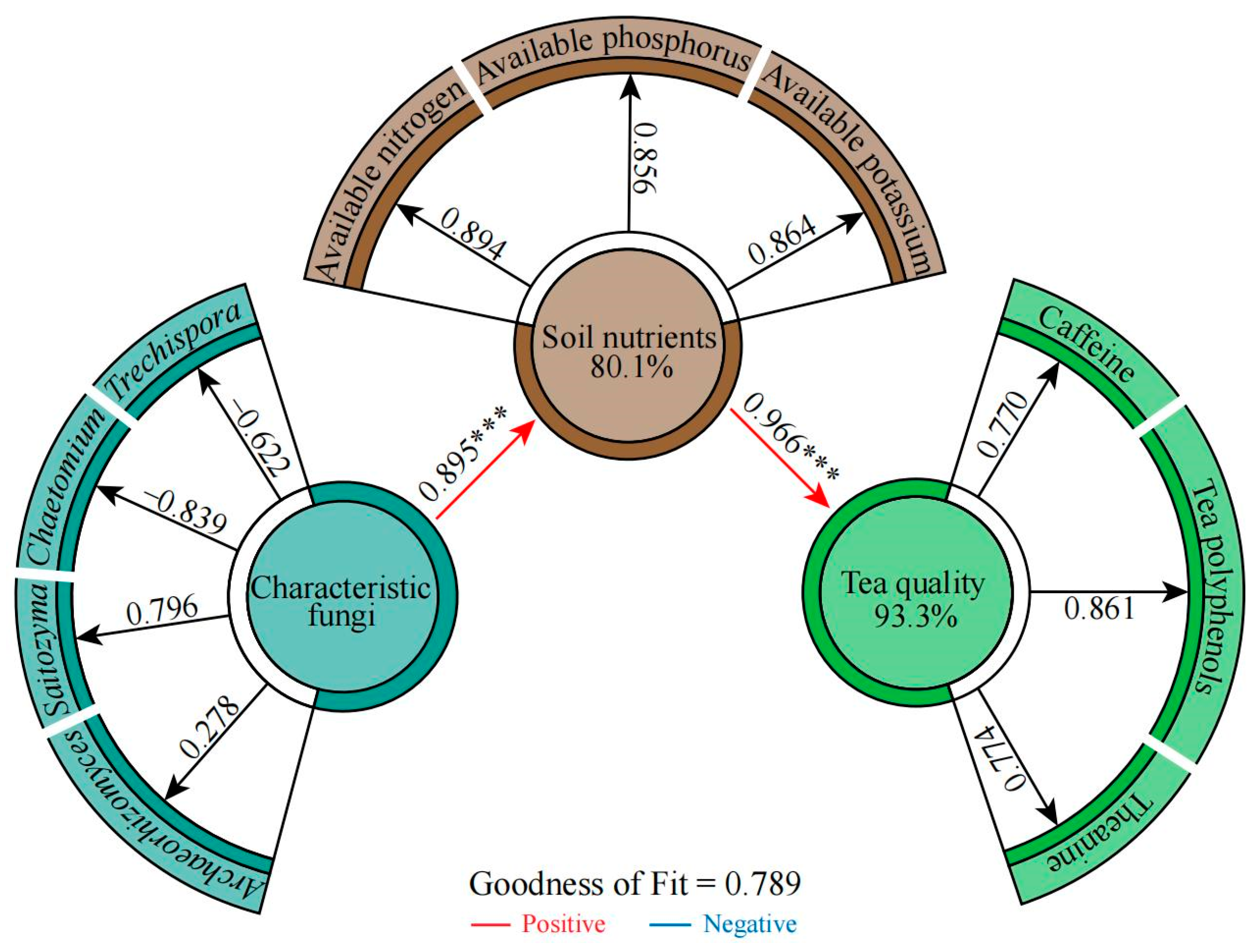
2.6. Correlation Analysis among Soil Characteristic Fungi, Available Nutrients, and Tea Quality Indexes
3. Materials and Methods
3.1. Sample Collection
3.2. Fungal ITS Amplicon Sequencing
3.3. Bioinformatics Analysis
3.4. Construction and Evaluation of Machine and Deep Learning Models
3.5. qRT-PCR Analysis of Soil Characteristic Fungi
3.6. Determination of Soil Available Nutrient Content
3.7. Determination of Tea Quality Index Content
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Galviz, Y.C.; Ribeiro, R.V.; Souza, G.M. Yes, plants do have memory. Theor. Exp. Plant Phys. 2020, 32, 195–202. [Google Scholar] [CrossRef]
- Karban, R. Plant behaviour and communication. Ecol. Lett. 2008, 11, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Padisak, J. Seasonal succession of phytoplankton in a large shallow lake (Balaton, Hungary)—A dynamic approach to ecological memory, its possible role and mechanisms. J. Ecol. 1992, 80, 217–230. [Google Scholar] [CrossRef]
- Leopold, A.C. Smart plants: Memory and communication without brains. Plant Signal. Behav. 2014, 9, e972268. [Google Scholar] [CrossRef]
- Richards, L.A.; Dyer, L.A.; Forister, M.L.; Smilanich, A.M.; Dodson, C.D.; Leonard, M.D.; Jeffrey, C.S. Phytochemical diversity drives plant–insect community diversity. Proc. Natl. Acad. Sci. USA 2015, 112, 10973–10978. [Google Scholar] [CrossRef] [PubMed]
- Das, N.; Mishra, S.K.; Bishayee, A.; Ali, E.S.; Bishayee, A. The phytochemical, biological, and medicinal attributes of phytoecdysteroids: An updated review. Acta Pharm. Sin. B 2021, 11, 1740–1766. [Google Scholar] [CrossRef] [PubMed]
- Karban, R. Plant communication. Annu. Rev. Ecol. Evol. Syst. 2021, 52, 1–24. [Google Scholar] [CrossRef]
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Sadhukhan, A.; Prasad, S.S.; Mitra, J.; Siddiqui, N.; Sahoo, L.; Kobayashi, Y.; Koyama, H. How do plants remember drought? Planta 2022, 256, 7. [Google Scholar] [CrossRef]
- Bhar, A.; Chakraborty, A.; Roy, A. Plant responses to biotic stress: Old memories matter. Plants 2021, 11, 84. [Google Scholar] [CrossRef]
- Wilpiszeski, R.L.; Aufrecht, J.A.; Retterer, S.T.; Sullivan, M.B.; Graham, D.E.; Pierce, E.M.; Zablocki, O.D.; Palumbo, A.V.; Elias, D.A. Soil aggregate microbial communities: Towards understanding microbiome interactions at biologically relevant scales. Appl. Environ. Microb. 2019, 85, e00324-19. [Google Scholar] [CrossRef]
- Timmis, K.; Ramos, J.L. The soil crisis: The need to treat as a global health problem and the pivotal role of microbes in prophylaxis and therapy. Microb. Biotechnol. 2021, 14, 769–797. [Google Scholar] [CrossRef]
- Sokol, N.W.; Slessarev, E.; Marschmann, G.L.; Nicolas, A.; Blazewicz, S.J.; Brodie, E.L.; Firestone, M.K.; Foley, M.M.; Hestrin, R.; Hungate, B.A.; et al. Life and death in the soil microbiome: How ecological processes influence biogeochemistry. Nat. Rev. Microbiol. 2022, 20, 415–430. [Google Scholar] [CrossRef]
- Fields, B.; Friman, V.P. Microbial eco-evolutionary dynamics in the plant rhizosphere. Curr. Opin. Microbiol. 2022, 68, 102153. [Google Scholar] [CrossRef] [PubMed]
- Angulo, V.; Beriot, N.; Garcia-Hernandez, E.; Li, E.; Masteling, R.; Lau, J.A. Plant–microbe eco-evolutionary dynamics in a changing world. New Phytol. 2022, 234, 1919–1928. [Google Scholar] [CrossRef]
- Trivedi, P.; Batista, B.D.; Bazany, K.E.; Singh, B.K. Plant–microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 2022, 234, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.G.; Song, G.C.; Ryu, C.M. Inheritance of seed and rhizosphere microbial communities through plant–soil feedback and soil memory. Environ. Microbiol. Rep. 2019, 11, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Jacques, C.; Salon, C.; Barnard, R.L.; Vernoud, V.; Prudent, M. Drought stress memory at the plant cycle level: A review. Plants 2021, 10, 1873. [Google Scholar] [CrossRef]
- Canarini, A.; Schmidt, H.; Fuchslueger, L.; Martin, V.; Herbold, C.W.; Zezula, D.; Gündler, P.; Hasibeder, R.; Jecmenica, M.; Bahn, M.; et al. Ecological memory of recurrent drought modifies soil processes via changes in soil microbial community. Nat. Commun. 2021, 12, 5308. [Google Scholar] [CrossRef]
- Kannenberg, S.A.; Schwalm, C.R.; Anderegg, W.R.L. Ghosts of the past: How drought legacy effects shape forest functioning and carbon cycling. Ecol. Lett. 2020, 23, 891–901. [Google Scholar] [CrossRef]
- Kottawa-Arachchi, J.D.; Gunasekare, M.K.; Ranatunga, M.A. Biochemical diversity of global tea [Camellia sinensis (L.) O. Kuntze] germplasm and its exploitation: A review. Genet. Resour. Crop Evol. 2019, 66, 259–273. [Google Scholar] [CrossRef]
- He, H.Z.; Lin, W.X.; Min, Q.W.; Fan, S.S.; Chen, Z.D. Discussion on the conservation and utilization of ancient tea tree resources in southern Fujian. J. Ecol. Rural Environ. 2022, 38, 1508–1513. [Google Scholar]
- Ahmed, S.; Griffin, T.S.; Kraner, D.; Schaffner, M.K.; Sharma, D.; Hazel, M.; Leitch, A.R.; Orians, C.M.; Han, W.; Stepp, J.R.; et al. Environmental factors variably impact tea secondary metabolites in the context of climate change. Front. Plant Sci. 2019, 10, 939. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Geng, Y.; Liang, T. Optimization of reduced chemical fertilizer use in tea gardens based on the assessment of related environmental and economic benefits. Sci. Total Environ. 2020, 713, 136439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yi, X.; Gao, X.; Wang, M.; Shao, C.; Lv, Z.; Chen, J.; Liu, Z.; Shen, C. Physiological and biochemical responses of tea seedlings (Camellia sinensis) to simulated acid rain conditions. Ecotox. Environ. Saf. 2020, 192, 110315. [Google Scholar] [CrossRef] [PubMed]
- Kc, Y.; Parajuli, A.; Khatri, B.B.; Shiwakoti, L.D. Phytochemicals and quality of green and black teas from different clones of tea plant. J. Food Qual. 2020, 2020, 8874271. [Google Scholar] [CrossRef]
- Li, J.W.; Li, H.; Liu, Z.W.; Wang, Y.X.; Chen, Y.; Yang, N.; Hu, Z.H.; Li, T.; Zhuang, J. Molecular markers in tea plant (Camellia sinensis): Applications to evolution, genetic identification, and molecular breeding. Plant Physiol. Biochem. 2023, 198, 107704. [Google Scholar] [CrossRef]
- Xia, E.H.; Tong, W.; Wu, Q.; Wei, S.; Zhao, J.; Zhang, Z.Z.; Wei, C.L.; Wan, X.C. Tea plant genomics: Achievements, challenges and perspectives. Hortic. Res. 2020, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, H.; Chang, Y.; Ma, C.; Wang, L.; Hao, X.; Li, A.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 4447. [Google Scholar] [CrossRef]
- Yu, X.; Xiao, J.; Chen, S.; Yu, Y.; Ma, J.; Lin, Y.; Li, R.; Lin, J.; Fu, Z.; Zhou, Q.; et al. Metabolite signatures of diverse Camellia sinensis tea populations. Nat. Commun. 2020, 11, 5586. [Google Scholar] [CrossRef]
- Zou, D.; Yin, X.L.; Gu, H.W.; Peng, Z.X.; Ding, B.M.; Li, Z.S.; Hu, X.C.; Long, W.J.; Fu, H.Y.; She, Y.B. Insight into the effect of cultivar and altitude on the identification of EnshiYulu tea grade in untargeted metabolomics analysis. Food Chem. 2024, 436, 137768. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive Earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef] [PubMed]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; Da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef]
- Vieira, S.; Sikorski, J.; Dietz, S.; Herz, K.; Schrumpf, M.; Bruelheide, H.; Scheel, D.; Friedrich, M.W.; Overmann, J. Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J. 2020, 14, 463–475. [Google Scholar] [CrossRef]
- Huang, X.F.; Chaparro, J.M.; Reardon, K.F.; Zhang, R.; Shen, Q.; Vivanco, J.M. Rhizosphere interactions: Root exudates, microbes, and microbial communities. Botany 2014, 92, 267–275. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Zhou, Y.; Coventry, D.R.; Gupta, V.V.; Fuentes, D.; Merchant, A.; Kaiser, B.N.; Li, J.S.; Wei, Y.L.; Liu, H.; Wang, Y.Y.; et al. The preceding root system drives the composition and function of the rhizosphere microbiome. Genome Boil. 2020, 21, 89. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Zverev, A.O.; Kichko, A.A.; Pinaev, A.G.; Provorov, N.A.; Andronov, E.E. Diversity indices of plant communities and their rhizosphere microbiomes: An attempt to find the connection. Microorganisms 2021, 9, 2339. [Google Scholar] [CrossRef]
- Raimi, A.R.; Ezeokoli, O.T.; Adeleke, R.A. Soil nutrient management influences diversity, community association and functional structure of rhizosphere bacteriome under vegetable crop production. Front. Microbiol. 2023, 14, 1229873. [Google Scholar] [CrossRef]
- Prashar, P.; Kapoor, N.; Sachdeva, S. Rhizosphere: Its structure, bacterial diversity and significance. Rev. Environ. Sci. Bio/Technol. 2014, 13, 63–77. [Google Scholar] [CrossRef]
- Hartmann, A.; Schmid, M.; Tuinen, D.V.; Berg, G. Plant-driven selection of microbes. Plant Soil 2009, 321, 235–257. [Google Scholar] [CrossRef]
- Lambers, H.; Mougel, C.; Jaillard, B.; Hinsinger, P. Plant-microbe-soil interactions in the rhizosphere: An evolutionary perspective. Plant Soil 2009, 321, 83–115. [Google Scholar] [CrossRef]
- Zhang, K.; Adams, J.M.; Shi, Y.; Yang, T.; Sun, R.; He, D.; Ni, Y.; Chu, H. Environment and geographic distance differ in relative importance for determining fungal community of rhizosphere and bulk soil. Environ. Microbiol. 2017, 19, 3649–3659. [Google Scholar] [CrossRef]
- Bickford, W.A.; Zak, D.R.; Kowalski, K.P.; Goldberg, D.E. Differences in rhizosphere microbial communities between native and non-native Phragmites australis may depend on stand density. Ecol. Evol. 2020, 10, 11739–11751. [Google Scholar] [CrossRef]
- Qiao, Q.; Wang, F.; Zhang, J.; Chen, Y.; Zhang, C.; Liu, G.; Zhang, H.; Ma, C.; Zhang, J. The variation in the rhizosphere microbiome of cotton with soil type, genotype and developmental stage. Sci. Rep. 2017, 7, 3940. [Google Scholar] [CrossRef]
- Dong, Z.Y.; Rao, M.P.N.; Liao, T.J.; Li, L.; Liu, Y.H.; Xiao, M.; Mohamad, O.A.A.; Tian, Y.Y.; Li, W.J. Diversity and function of rhizosphere microorganisms between wild and cultivated medicinal plant Glycyrrhiza uralensis Fisch under different soil conditions. Arch. Microbiol. 2021, 203, 3657–3665. [Google Scholar] [CrossRef]
- Feng, H.; Fu, R.; Hou, X.; Lv, Y.; Zhang, N.; Liu, Y.; Xu, Z.; Miao, Y.; Krell, T.; Shen, Q.; et al. Chemotaxis of beneficial rhizobacteria to root exudates: The first step towards root–microbe rhizosphere interactions. Int. J. Mol. Sci. 2021, 22, 6655. [Google Scholar] [CrossRef]
- Zhao, M.; Zhao, J.; Yuan, J.; Hale, L.; Wen, T.; Huang, Q.; Vivanco, J.; Zhou, J.; Kowalchuk, G.; Shen, Q. Root exudates drive soil-microbe-nutrient feedbacks in response to plant growth. Plant Cell Environ. 2021, 44, 613–628. [Google Scholar] [CrossRef]
- Luo, X.; Liu, K.; Shen, Y.; Yao, G.; Yang, W.; Mortimer, P.E.; Gui, H. Fungal community composition and diversity vary with soil horizons in a subtropical forest. Front. Microbiol. 2021, 12, 650440. [Google Scholar] [CrossRef]
- He, M.; Zhong, X.; Xia, Y.; Xu, L.; Zeng, Q.; Yang, L.; Fan, Y. Long-term nitrogen addition exerts minor effects on microbial community but alters sensitive microbial species in a subtropical natural forest. Forests 2023, 14, 928. [Google Scholar] [CrossRef]
- Rodriguez-Mena, S.; Camacho, M.; de los Santos, B.; Miranda, L.; Camacho-Sanchez, M. Microbiota modulation in blueberry rhizosphere by biocontrol bacteria. Microbiol. Res. 2022, 13, 809–824. [Google Scholar] [CrossRef]
- Chen, J.Z.; Huang, X.L.; Liu, J.M.; Sun, Q.W. Dominant microbial community in rhizosphere affecting active components of Cinnamomum migao at different ripening stages and their response to soil properties. J. Plant Nutr. Fertil. 2021, 27, 1779–1791. [Google Scholar]
- Cao, G.; Wu, L.; Qu, X.; Dai, L.; Ye, Y.; Xu, S.; Wu, C.; Chen, Y. Correlation between changes in soil properties and microbial diversity driven by different management in artificial Chinese fir (Cunninghamia lanceolata (Lamb.) Hook.) plantations. Forests 2023, 14, 877. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Q.; Zhong, Z.; Huang, Z.; Bian, F.; Yang, C.; Wen, X. Changes in soil organic carbon fractions and fungal communities, subsequent to different management practices in Moso bamboo plantations. J. Fungi 2022, 8, 640. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Y.; Wei, F.; Zhao, L.; Zhou, J.; Qi, G.; Ma, Z.; Zhu, H.; Feng, H.; Feng, Z. Applications of Chaetomium globosum CEF-082 improve soil health and mitigate the continuous cropping obstacles for Gossypium hirsutum. Ind. Crop. Prod. 2023, 197, 116586. [Google Scholar] [CrossRef]
- Kang, H.J.; Chai, A.; Lin, Z.H.; Shi, Y.X.; Xie, X.W.; Li, L.; Fan, T.F.; Xiang, S.; Xie, J.M.; Li, B.J. Deciphering differences in microbial community diversity between clubroot-diseased and healthy soils. Microorganisms 2024, 12, 251. [Google Scholar] [CrossRef]
- Liu, R.; Han, C.; Kang, Y.; Jiang, Y.B.; Deng, H.; Liu, K.L.; Zhang, J.B.; Zhong, W.H. Active microbial taxa preferentially assimilate inorganic nitrogen in acidic upland soils using a 15N-DNA-SIP approach. Soil Biol. Biochem. 2024, 188, 109226. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Zhang, J.; Xia, S.; Qin, H.; Feng, C.; Bie, S.; Wang, X. Influence of incorporating decomposition agent in oil rapeseed return practice on fungal community, soil fertility and the cotton yield. J. Biobased Mater. Bioenergy 2022, 16, 686–695. [Google Scholar] [CrossRef]
- Yang, X.; Ma, L.; Ji, L.; Shi, Y.; Yi, X.; Yang, Q.; Ni, K.; Ruan, J. Long-term nitrogen fertilization indirectly affects soil fungi community structure by changing soil and pruned litter in a subtropical tea (Camellia sinensis L.) plantation in China. Plant Soil 2019, 444, 409–426. [Google Scholar] [CrossRef]
- Qian, Z.; Zhuang, S.; Gao, J.; Tang, L. Can aeration improve bamboo soil fertility of soil below bamboo and fungal diversity under mulching conditions? Land Degrad. Dev. 2022, 33, 2353–2365. [Google Scholar] [CrossRef]
- Ma, Y.H.; Fu, S.L.; Zhang, X.P.; Zhao, K.; Chen, H.Y. Intercropping improves soil nutrient availability, soil enzyme activity and tea quantity and quality. Appl. Soil Ecol. 2017, 119, 171–178. [Google Scholar] [CrossRef]
- Duan, Y.; Shen, J.; Zhang, X.; Wen, B.; Ma, Y.; Wang, Y.; Fang, W.; Zhu, X. Effects of soybean–tea intercropping on soil-available nutrients and tea quality. Acta Physiol. Plant 2021, 41, 140. [Google Scholar] [CrossRef]
- Farooq, T.H.; Kumar, U.; Mo, J.; Shakoor, A.; Wang, J.; Rashid, M.H.U.; Tufail, M.A.; Chen, X.; Yan, W. Intercropping of peanut–tea enhances soil enzymatic activity and soil nutrient status at different soil profiles in subtropical southern China. Plants 2021, 10, 881. [Google Scholar] [CrossRef]
- Jia, X.; Wang, Y.; Zhang, Q.; Lin, S.; Zhang, Y.; Du, M.; Chen, M.; Ye, J.; Wu, Z.; Wang, H. Reasonable deep application of sheep manure fertilizer to alleviate soil acidification to improve tea yield and quality. Front. Plant Sci. 2023, 14, 1179960. [Google Scholar] [CrossRef]
- Tang, S.; Pan, W.K.; Tang, R.; Ma, Q.X.; Zhou, J.J.; Zheng, N.; Wang, J.; Sun, T.; Wu, L.H. Effects of balanced and unbalanced fertilisation on tea quality, yield, and soil bacterial community. Appl. Soil Ecol. 2022, 175, 104442. [Google Scholar] [CrossRef]
- Wen, B.; Li, R.; Zhao, X.; Ren, S.; Chang, Y.; Zhang, K.; Wang, S.; Guo, G.; Zhu, X. A quadratic regression model to quantify plantation soil factors that affect tea quality. Agriculture 2021, 11, 1225. [Google Scholar] [CrossRef]
- Ye, J.; Wang, Y.; Wang, Y.; Hong, L.; Jia, X.; Kang, J.; Lin, S.; Wu, Z.; Wang, H. Improvement of soil acidification in tea plantations by long-term use of organic fertilizers and its effect on tea yield and quality. Front. Plant Sci. 2022, 13, 1055900. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Q.; Li, J.; Lin, S.; Jia, X.; Zhang, Q.; Ye, J.; Wang, H.; Wu, Z. Study on the effect of pH on rhizosphere soil fertility and the aroma quality of tea trees and their tnteractions. Agriculture 2023, 13, 1739. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, Y.; Wang, Y.; Lin, S.; Chen, M.; Cheng, P.; Ye, J.; Miao, P.; Jia, L.; Wang, H. Effects of pruning on tea tree growth, tea quality, and rhizosphere soil microbial community. Microbiol. Spectr. 2023, 11, e01601-23. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End read mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Ye, J.; McGinnis, S.; Madden, T.L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res. 2006, 34, W6–W9. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, X.; Fan, W.; Shen, L.; Zhang, Z.; Liu, X.; Du, J.; Li, H.; Chen, Y.; Li, H. Fast density peak clustering for large scale data based on kNN. Knowl.-Based Syst. 2020, 187, 104824. [Google Scholar] [CrossRef]
- Cervantes, J.; Garcia-Lamont, F.; Rodríguez-Mazahua, L.; Lopez, A. A comprehensive survey on support vector machine classification: Applications, challenges and trends. Neurocomputing 2020, 408, 189–215. [Google Scholar] [CrossRef]
- Zhang, J.; Qu, S. Optimization of backpropagation neural network under the adaptive genetic algorithm. Complexity 2021, 2021, 1718234. [Google Scholar] [CrossRef]
- Boateng, E.Y.; Otoo, J.; Abaye, D.A. Basic tenets of classification algorithms K-nearest-neighbor, support vector machine, random forest and neural network: A review. J. Data Anal. Inform. Process. 2020, 8, 341–357. [Google Scholar] [CrossRef]
- Ai, F.F.; Bin, J.; Zhang, Z.; Huang, J.H.; Wang, J.B.; Liang, Y.Z.; Yu, L.; Yang, Z.Y. Application of random forests to select premium quality vegetable oils by their fatty acid composition. Food Chem. 2014, 143, 472–478. [Google Scholar] [CrossRef]
- Sagi, O.; Rokach, L. Approximating XGBoost with an interpretable decision tree. Inform. Sci. 2021, 572, 522–542. [Google Scholar] [CrossRef]
- Pan, S.; Zhang, X.; Xu, W.; Yin, J.; Gu, H.; Yu, X. Rapid On-site identification of geographical origin and storage age of tangerine peel by Near-infrared spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 271, 120936. [Google Scholar] [CrossRef]
- Ganaie, M.A.; Hu, M.; Malik, A.K.; Tanveer, M.; Suganthan, P.N. Ensemble deep learning: A review. Eng. Appl. Artif. Intel. 2022, 115, 105151. [Google Scholar] [CrossRef]
- Zhao, Y.R.; Yu, K.Q.; Li, X.; He, Y. Detection of fungus in fectionon petals of rapeseed (Brassica napus L.) using NIR hyperspectral imaging. Sci. Rep. 2016, 6, 38878. [Google Scholar] [CrossRef]
- Valavi, R.; Guillera-Arroita, G.; Lahoz-Monfort, J.J.; Elith, J. Predictive performance of presence-only species distribution models: A benchmark study with reproducible code. Ecol. Monogr. 2022, 92, e01486. [Google Scholar] [CrossRef]
- Jia, X.; Zhang, Q.; Wang, Y.; Zhang, Y.; Li, M.; Cheng, P.; Chen, M.; Lin, S.; Zou, J.; Ye, J.; et al. Changes of physiological characteristics, element accumulation and hormone metabolism of tea leaves in response to soil pH. Front. Plant Sci. 2023, 14, 1266026. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Q.; Wang, Y.; Lin, S.; Chen, M.; Cheng, P.; Wang, Y.; Du, M.; Jia, X.; Wang, H.; et al. Effects of magnesium on transcriptome and physicochemical index of tea leaves. Plants 2023, 12, 1810. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, X.; Lin, S.; Zhang, Q.; Wang, Y.; Hong, L.; Li, M.; Zhang, S.; Wang, T.; Jia, M.; Luo, Y.; et al. The Ability of Different Tea Tree Germplasm Resources in South China to Aggregate Rhizosphere Soil Characteristic Fungi Affects Tea Quality. Plants 2024, 13, 2029. https://doi.org/10.3390/plants13152029
Jia X, Lin S, Zhang Q, Wang Y, Hong L, Li M, Zhang S, Wang T, Jia M, Luo Y, et al. The Ability of Different Tea Tree Germplasm Resources in South China to Aggregate Rhizosphere Soil Characteristic Fungi Affects Tea Quality. Plants. 2024; 13(15):2029. https://doi.org/10.3390/plants13152029
Chicago/Turabian StyleJia, Xiaoli, Shaoxiong Lin, Qi Zhang, Yuhua Wang, Lei Hong, Mingzhe Li, Shuqi Zhang, Tingting Wang, Miao Jia, Yangxin Luo, and et al. 2024. "The Ability of Different Tea Tree Germplasm Resources in South China to Aggregate Rhizosphere Soil Characteristic Fungi Affects Tea Quality" Plants 13, no. 15: 2029. https://doi.org/10.3390/plants13152029
APA StyleJia, X., Lin, S., Zhang, Q., Wang, Y., Hong, L., Li, M., Zhang, S., Wang, T., Jia, M., Luo, Y., Ye, J., & Wang, H. (2024). The Ability of Different Tea Tree Germplasm Resources in South China to Aggregate Rhizosphere Soil Characteristic Fungi Affects Tea Quality. Plants, 13(15), 2029. https://doi.org/10.3390/plants13152029






