Abstract
Seed physical dormancy (hard-seededness) is an interesting ecological phenomenon and important agronomic trait. The loss of seed coat impermeability/hard-seededness is a key target trait during the domestication of leguminous crops which allows seeds to germinate rapidly and uniformly. In this study, we examined the mutation of quantitative trait locus (QTL) genes, GmHs1-1 and GmqHS1, in 18 wild soybean (G. soja) and 23 cultivated soybean (G. max) accessions. The sequencing results indicate that a G-to-T substitution in GmqHS1 and a C-to-T substitution in GmHs1-1 occurred in all 23 cultivated soybean accessions but not in any of the 18 wild soybean accessions. The mutations in the two genes led to increased seed coat permeability in cultivated soybean. Therefore, we provide evidence that two genes, GmHs1-1 and GmqHS1, simultaneously contribute to the domestication of hard-seededness in soybeans. This finding is of great significance for genetic analysis and improved utilization of the soybean hard-seededness trait.
1. Introduction
Hard-seededness is an important trait related to soybean domestication. Compared with non-hard seeds, hard soybean seeds have higher seed vigor and longer seed lifespan [1,2,3]. Additionally, hard seeds are less prone to imbibition, delaying seed deterioration and facilitating seed preservation and transportation [2]. However, hard-seededness is not convenient for seed production in agriculture and utilization in daily life, as it not only reduces field emergence and final yield, but also affects seed consumption and processing. The breakdown of seed coat impermeability/hard-seededness is a crucial step in soybean domestication and improvement to produce high-value cultivated seeds [4]. Loss of seed coat impermeability is a key trait for many leguminous crops, which allows seeds to germinate rapidly and uniformly [5,6,7]. Permeable seed coats are also beneficial in processing seeds to produce vegetable oil and soy foods [8].
As we were interested in the seed coat permeability phenotype of legume species, two papers published in 2015 attracted our attention with their interesting results on soybean. Sun et al. [9] reported that seed coat impermeability in soybean is controlled by a major quantitative trait locus (QTL) gene, GmHs1-1, encoding a calcineurin-like protein. Soon after, another major QTL gene, qHS1, was also identified to control seed coat impermeability in soybean by fine mapping [10]. qHS1, an endo-1, 4-β-glucanase gene, promotes the accumulation of 1, 4-β-glucan in the outer layer of palisade cells, leading to the production of hard seeds. Moreover, qHS1 had the greatest effect on soybean impermeability according to another study [11]. Interestingly, the locations of these two genes are close to each other on the physical map. It is possible that these two genes function together to improve the seed coat permeability in cultivated soybean (Glycine max) [12], but each set of authors missed the gene identified by the other during their fine-mapping processes. To test the above hypothesis, we analyzed the single-nucleotide polymorphism (SNP) variation in different wild soybean (Glycine soja) and elite cultivated soybean populations. Our findings, unsurprisingly, indicated that these two genes, not one, simultaneously exist and contribute to improving seed coat permeability in cultivated soybean. Our analyses suggest that both the Hs1-1 and qHS1 genes are functional in controlling seed coat impermeability in legume species. Hs1-1 and qHS1 function together to facilitate cultivated soybean domestication for seed coat permeability.
2. Results and Discussion
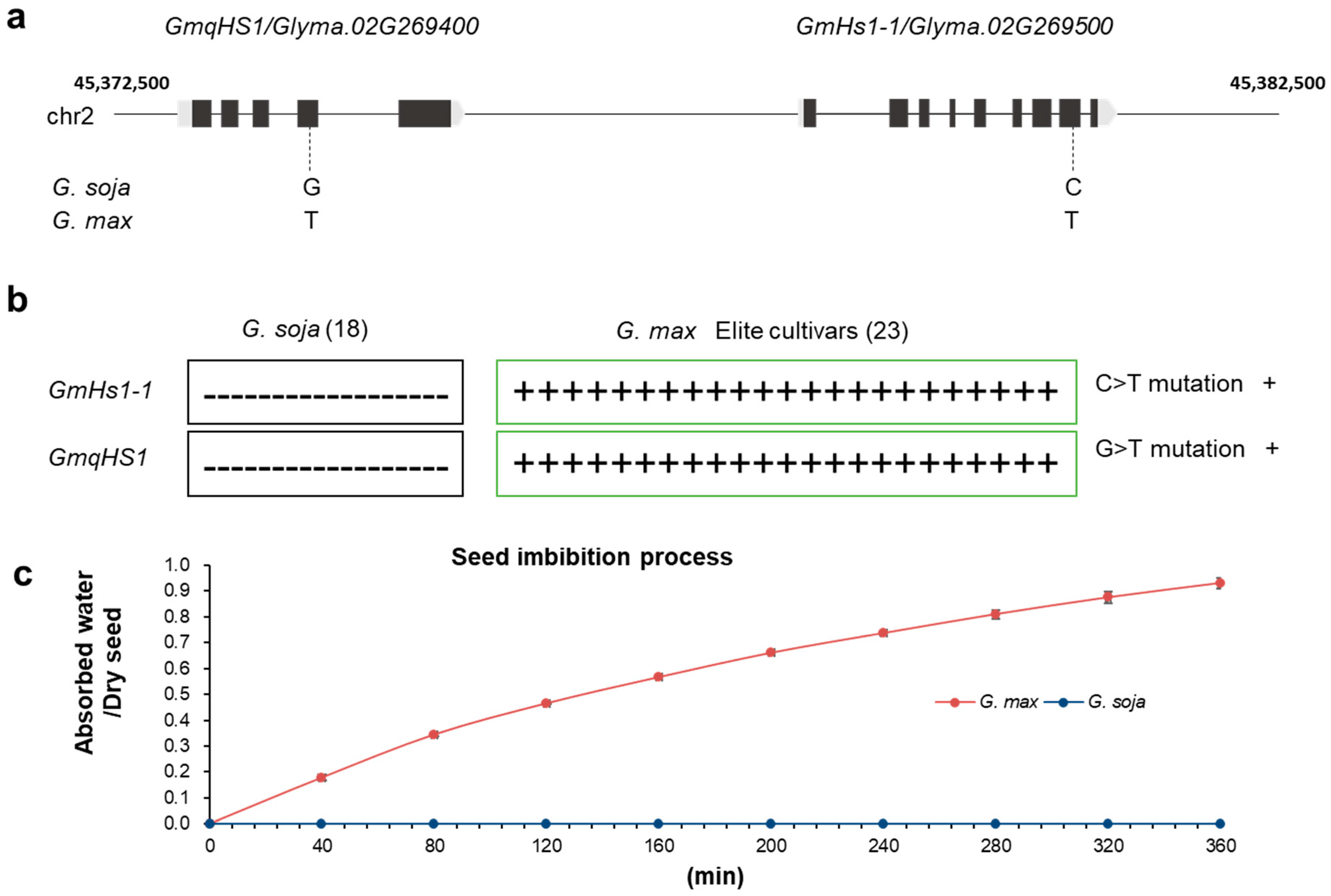
Hard-seededness is one of the crucial target traits during domestication of many legume crops [5,7,11]. Modern cultivated soybean, the most economically important legume crop, is commonly believed to have been domesticated from wild soybean in East Asia about 5000 years ago [13,14]. Previous studies indicated that several genes/QTLs control the hard-seededness [11]. Keim et al. [15] first used five RFLP markers to detect the QTLs of soybean hard-seededness, which were located on chromosomes 2, 3, 8 and 19, and explained 71.0% of the genetic variation. Watanabe et al. [16] identified three markers (RAS1-3) related to soybean hard-seededness and found that the dominant QTL (RAS2) was located on chromosome 2. In soybean PI594619, a single gene located between the markers Sat_202 and Satt459 on chromosome 2 was also identified to control seed coat impermeability [17]. Subsequently, two genes, qHS1 and GmHs1-1, related to soybean hard-seededness on chromosome 2 were cloned independently [9,10]. It is possible that the QTLs on chromosome 2 in soybean have the greatest effect on seed coat impermeability [15,18]. Interestingly, according to current research information, GmHs1-1 and GmqHS1 were mapped close to each other on chromosome 2 in the soybean genome (Figure 1a) by two different independent research groups [9,10].
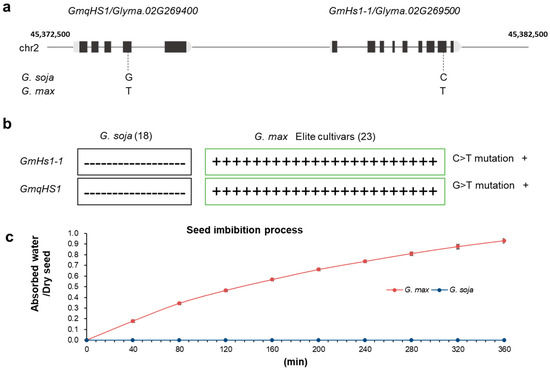
Figure 1.
Single-nucleotide polymorphism locations in G. soja and G. max. (a) Physical map of GmqHS1 and GmHs1-1 in the soybean genome from phytozome and the key SNP variations controlling seed coat impermeability. The physical distance between GmqHS1 and GmHs1-1 is 4175 bp. (b) Distribution of SNP mutations in GmHs1-1 and GmqHS1 among 18 different G. soja and 23 G. max populations. (c) Typical imbibition process of G. soja and G. max seeds.
Investigating the relationship between these two genes in cultivated soybean varieties will help us clearly understand the trait during soybean domestication. Thus, to test this speculation, we ordered 18 wild soybean (G. soja) and 23 cultivated soybean (G. max) accessions from the USDA which were used in the research of Sun et al. [9]. No mutations were found in either gene in the 18 wild soybean accessions. However, we observed SNPs in GmHs1-1 simultaneously with GmqHS1 in all 23 cultivated soybean accessions (Figure 1b). These results indicated that GmHs1-1 and GmqHS1 were selected together during soybean domestication, which might be the reason why functional GmHs1-1 or GmqHS1 only partially recovered the seed coat permeability phenotype in previous reported studies. Currently, the single-origin theory of the domestication of soybean appears to be widely accepted and also supported by marker analysis [19] and recent genome resequencing data [20,21]. The current understanding of the GmHs1-1 and GmqHS1 genes, together with advances for future research and utilization of the hard-seededness trait of soybean, was summarized recently [22].
During the process of soybean’s transition from wild to domesticated, the removal of hard-seededness characteristics affected the seed’s ability to absorb water and germinate. The classic imbibition process of wild soybean (G. soja) and cultivated soybean seeds (G. max) was described in this study (Figure 1c). Additionally, compared with wild soybean, the vigor and lifespan of cultivated soybean seeds after the breakdown of the impermeable seed coat are also affected. Based on previous reports of both hard and non-hard soybean seeds stored at room temperature (20 °C), it was found that non-hard seeds lost their vigor after approximately 2 years, while hard seeds still maintained a higher germination percentage of over 90% after 4 years [23]. As the main channel for germinating seeds to absorb water and exchange gases from the external environment, the seed coat and hilum are closely related to the maintenance of vigor and longer lifespan in hard soybean seeds. They hinder or restrict the influence of external water and gases on seeds, causing seeds to remain in a dormant or semi-dormant state for a long time, reducing the consumption of nutrients to the minimum level, thereby maintaining and extending seed vigor and lifespan. Overall, although the removal of hard-seededness characteristics in soybean is beneficial for seed germination, cultivation and production, it to some extent damages seed vigor and longevity under normal storage conditions. All these data indicate that both Hs1-1 and qHS1 are critical in controlling seed coat impermeability/hard-seededness. The simultaneous mutations of these two genes may have enabled artificial selection to break down hard-seededness during soybean domestication.
3. Materials and Methods
3.1. Plant Materials, Plant Growth and Growth Conditions
Wild soybean (G. soja) and cultivated soybean (G. max) accessions were requested from the USDA Soybean Germplasm Collection (Table 1). These populations were used for GmHs1-1 and GmqHS1 mutation variation analyses and association tests. Plants were grown at 22 °C day/20 °C night temperatures under a 16 h day/8 h night photoperiod in greenhouse conditions.

Table 1.
G. soja and G. max population samples from the USDA Soybean Germplasm Collection used for genotyping and phenotyping.
3.2. Seed Imbibition Phenotyping
Seeds of 18 wild soybean (G. soja) and 23 cultivated soybean (G. max) accessions were simultaneously harvested and used for seed coat impermeability tests. After the seeds were incubated in sterile water at room temperature for 2 h, the imbibed seeds were counted and the proportion of seeds with seed coat permeability was calculated. Cultivated soybean seeds showed imbibition, while wild soybean seeds did not. Additionally, the imbibition process of wild-type and mutant seeds was recorded by measuring the increased weight of imbibed seeds compared with their original dry weight at an interval of 40 min for 360 min.
3.3. DNA Isolation, PCR, Sequencing and Alignments
Genomic DNA was isolated from leaves of 4-week-old seedlings using a classical CTAB method (https://www.nature.com/articles/nprot.2006.384). PCR primers were designed to amplify the SNPs of the Hs1-1 and qHS1 genes (Table 2). The amplified fragment length was 190 bp for qHS1 and 388 bp for Hs1-1. The PCR cycling conditions included an initial denaturation step at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 15 s and annealing at 55 °C for 30 s. PCR products were directly sequenced, and alignment of the PCR product nucleotide sequences was carried out with DNASTAR Lasergene 12 (https://www.dnastar.com/software/).

Table 2.
Primers used in this study.
4. Conclusions
All the above data indicate that the Hs1-1 and qHS1 genes are critical in controlling seed coat impermeability/hard-seededness. The simultaneous mutations of these two genes may have enabled artificial selection to break down hard-seededness during soybean domestication.
Author Contributions
Data curation, D.T. and Q.Z.; Writing—original draft, H.Y. and M.C.; Writing—review & editing, J.W. and Z.-Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Hohhot Key R&D Project (2023-JBGS-S-1), the National Natural Science Foundation of China (grant no. 31972958), the China Scholarship Council (CSC), and the First-Class Grassland Science Discipline Program of Shandong Province, China.
Data Availability Statement
Data is contained within the article.
Acknowledgments
We would like to thank Qingzhen Jiang for assistance in paper revision.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Mohamed, Y.Y.; Barringer, S.A.; Splittstoesser, W.E. The role of seed coats in seed viability. Bot. Rev. 1994, 60, 426–439. [Google Scholar] [CrossRef]
- Potts, H.C.; Duangpatra, J.; Hairston, W.G. Some influences of hard seededness on soybean seed quality. Crop Sci. 1978, 18, 221–224. [Google Scholar] [CrossRef]
- Heatherly, L.G.; Kenty, M.M.; Kilen, T.C. Effects of storage environment and duration on impermeable seed coat in soybean. Field Crops Res. 1995, 40, 57–62. [Google Scholar] [CrossRef]
- Sedivy, E.J.; Wu, F.; Hanzawa, Y. Soybean domestication: The origin, genetic architecture and molecular bases. New Phytol. 2017, 214, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Abbo, S.; Shtienberg, D.; Lichtenzveig, J.; Lev-Yadun, S.; Gopher, A. The chickpea, summer cropping, and a new model for pulse domestication in the ancient near east. Q. Rev. Biol. 2003, 78, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Weeden, N.F. Genetic changes accompanying the domestication of Pisum sativum: Is there a common genetic basis to the ‘domestication syndrome’ for legumes? Ann. Bot. 2007, 100, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Andargie, M.; Pasquet, R.S.; Gowda, B.S.; Muluvi, G.M.; Timko, M.P. Molecular mapping of QTLs for domestication-related traits in cowpea (V. unguiculata (L.) Walp.). Euphytica 2014, 200, 401–412. [Google Scholar] [CrossRef]
- Mullin, W.J.; Xu, W. Study of soybean seed coat components and their relationship to water absorption. J. Agric. Food Chem. 2001, 49, 5331–5335. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Miao, Z.; Cai, C.; Zhang, D.; Zhao, M.; Wu, Y.; Zhang, X.; Swarm, S.A.; Zhou, L.; Zhang, Z.J.; et al. GmHs1-1, encoding a calcineurin-like protein, controls hard-seededness in soybean. Nat. Genet. 2015, 47, 939–943. [Google Scholar] [CrossRef]
- Jang, S.J.; Sato, M.; Sato, K.; Jitsuyama, Y.; Fujino, K.; Mori, H.; Takahashi, R.; Benitez, E.R.; Liu, B.; Yamada, T.; et al. A Single-Nucleotide Polymorphism in an endo-1,4-beta-glucanase gene controls seed coat permeability in soybean. PLoS ONE 2015, 10, e0128527. [Google Scholar] [CrossRef]
- Liu, B.; Fujita, T.; Yan, Z.H.; Sakamoto, S.; Xu, D.; Abe, J. QTL mapping of domestication-related traits in soybean (Glycine max). Ann. Bot. 2007, 100, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Lu, X.; Wen, J.; Wang, Z.; Chai, M. Physical seed dormancy in legumes: Molecular advances and perspectives. Plants 2024, 13, 1473. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guan, R.; Liu, Z.; Ma, Y.; Wang, L.; Li, L.; Lin, F.; Luan, W.; Chen, P.; Yan, Z.; et al. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor. Appl. Genet. 2008, 117, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Boerma, H.R.; Specht, J.E. Soybeans: Improvement, Production and Uses; American Society of Agronomy: Madison, WI, USA, 2004. [Google Scholar]
- Keim, P.; Diers, B.W.; Shoemaker, R.C. Genetic analysis of soybean hard seededness with molecular markers. Theor. Appl. Genet. 1990, 79, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Tajuddin, T.; Yamanaka, N.; Hayashi, M.; Harada, K. Analysis of QTLs for reproductive development and seed quality traits in soybean using recombinant inbred lines. Breed. Sci. 2004, 54, 399–407. [Google Scholar] [CrossRef]
- Kebede, H.; Smith, J.R.; Ray, J.D. Identification of a single gene for seed coat impermeability in soybean PI 594619. Theor. Appl. Genet. 2014, 127, 1991–2003. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, Y.; Kaga, A.; Tomooka, N.; Yano, H.; Takada, Y.; Kato, S.; Vaughan, D. QTL affecting fitness of hybrids between wild and cultivated soybeans in experimental fields. Ecol. Evol. 2013, 3, 2150–2168. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, Y.; Song, C.; Zhou, J.; Qiu, L.; Huang, H.; Wang, Y. A single origin and moderate bottleneck during domestication of soybean (Glycine max): Implications from microsatellites and nucleotide sequences. Ann. Bot. 2010, 106, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhao, X.; Liu, D.; Li, Y.; Lightfoot, D.A.; Yang, Z.; Zhao, L.; Zhou, G.; Wang, Z.; Huang, L.; et al. Domestication footprints anchor genomic regions of agronomic importance in soybeans. New Phytol. 2016, 209, 871–884. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef]
- Sun, Y.; Gong, Y. Research advances on the hard seededness trait of soybean and the underlying regulatory mechanisms. Front. Plant Sci. 2024, 15, 1419962. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Study on preservation of soybean germplasm using soybean hard seed. Soybean Sci. 1999, 18, 351–354. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).