Precision Detection of Salt Stress in Soybean Seedlings Based on Deep Learning and Chlorophyll Fluorescence Imaging
Abstract
:1. Introduction
2. Materials and Methods
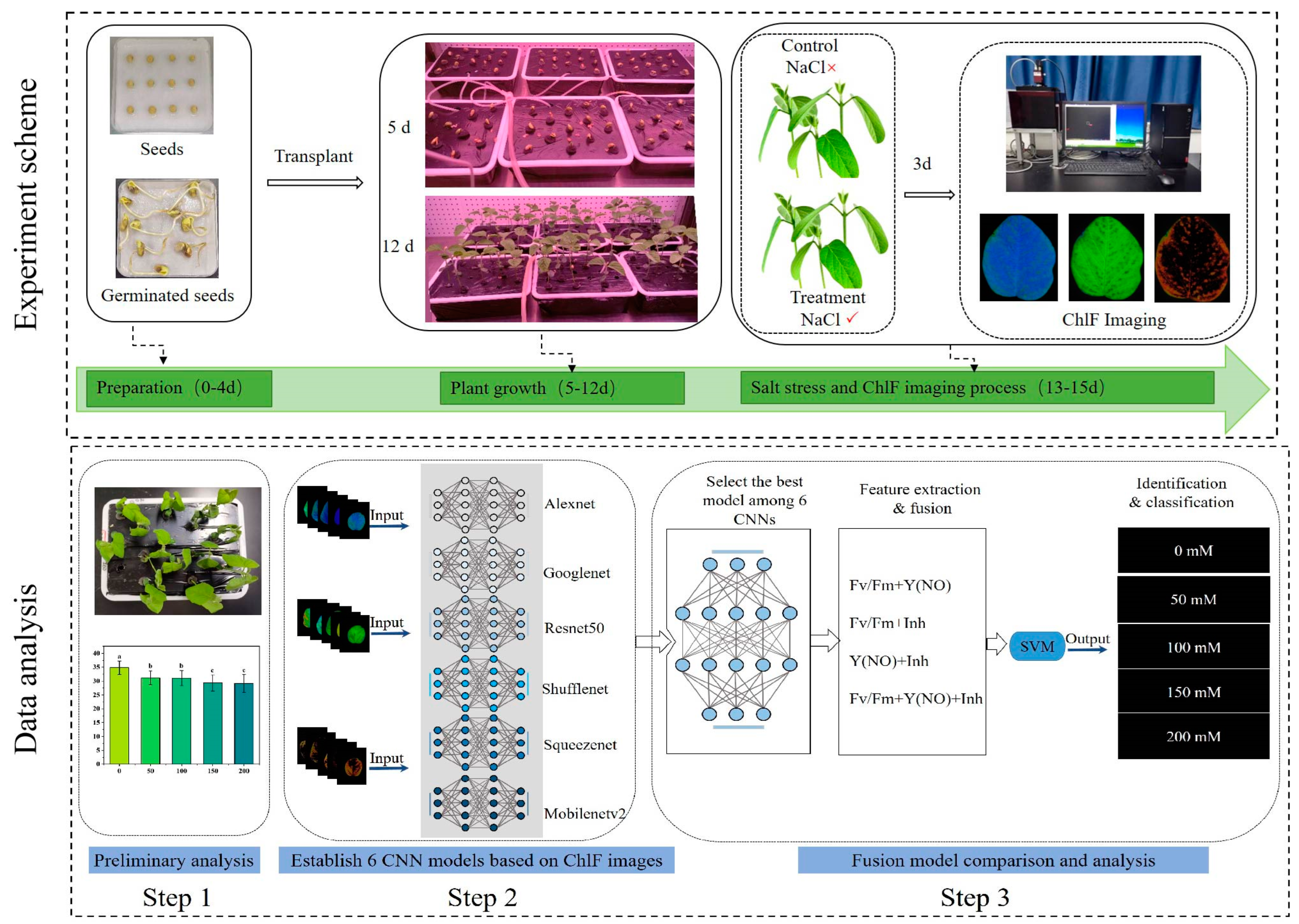
2.1. Plant Material and Workflow of Experiment
2.2. Chlorophyll Fluorescence Image Acquisition and Augmentation
2.3. SPAD Value Determination
2.4. Software
3. Results
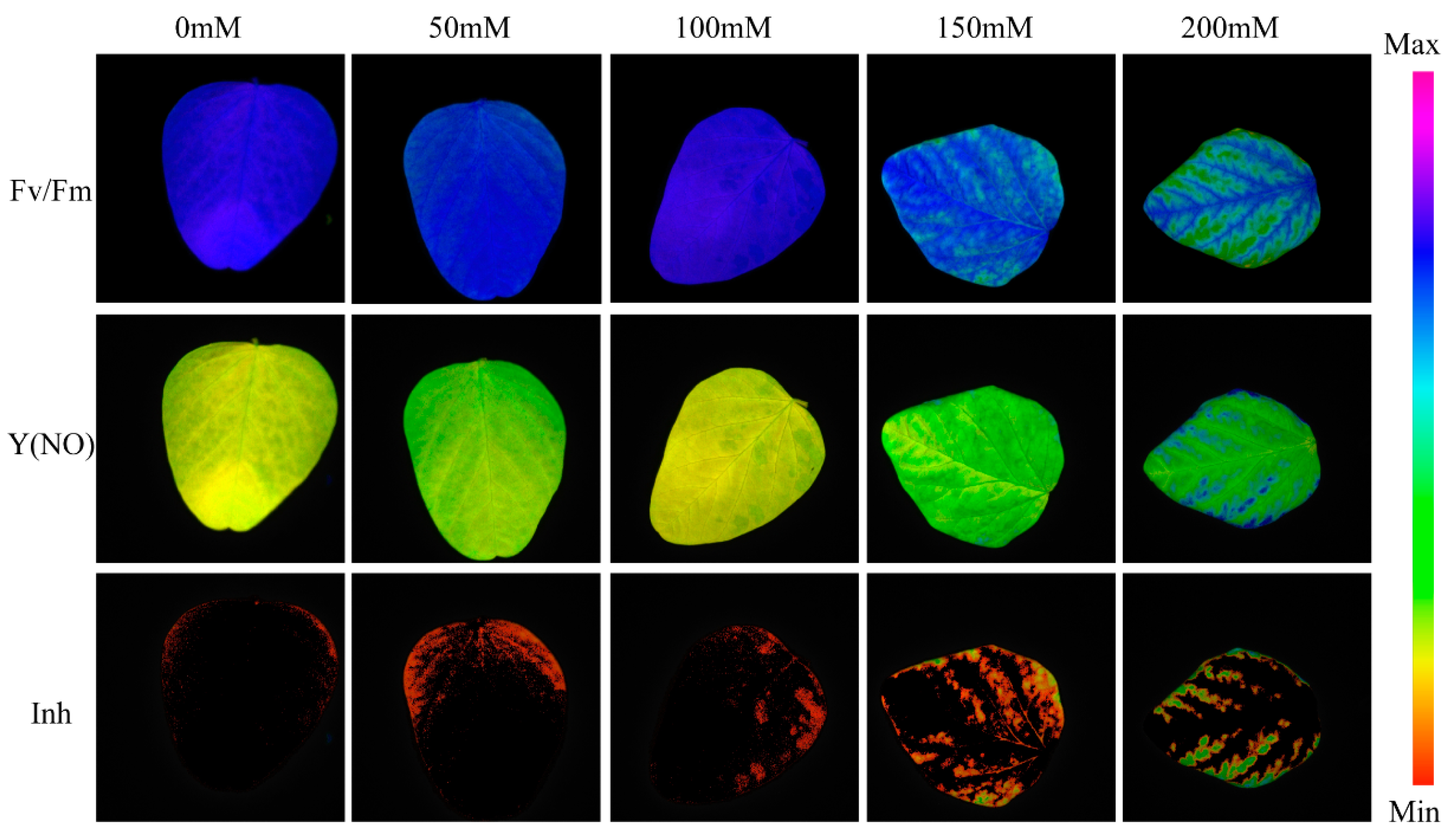
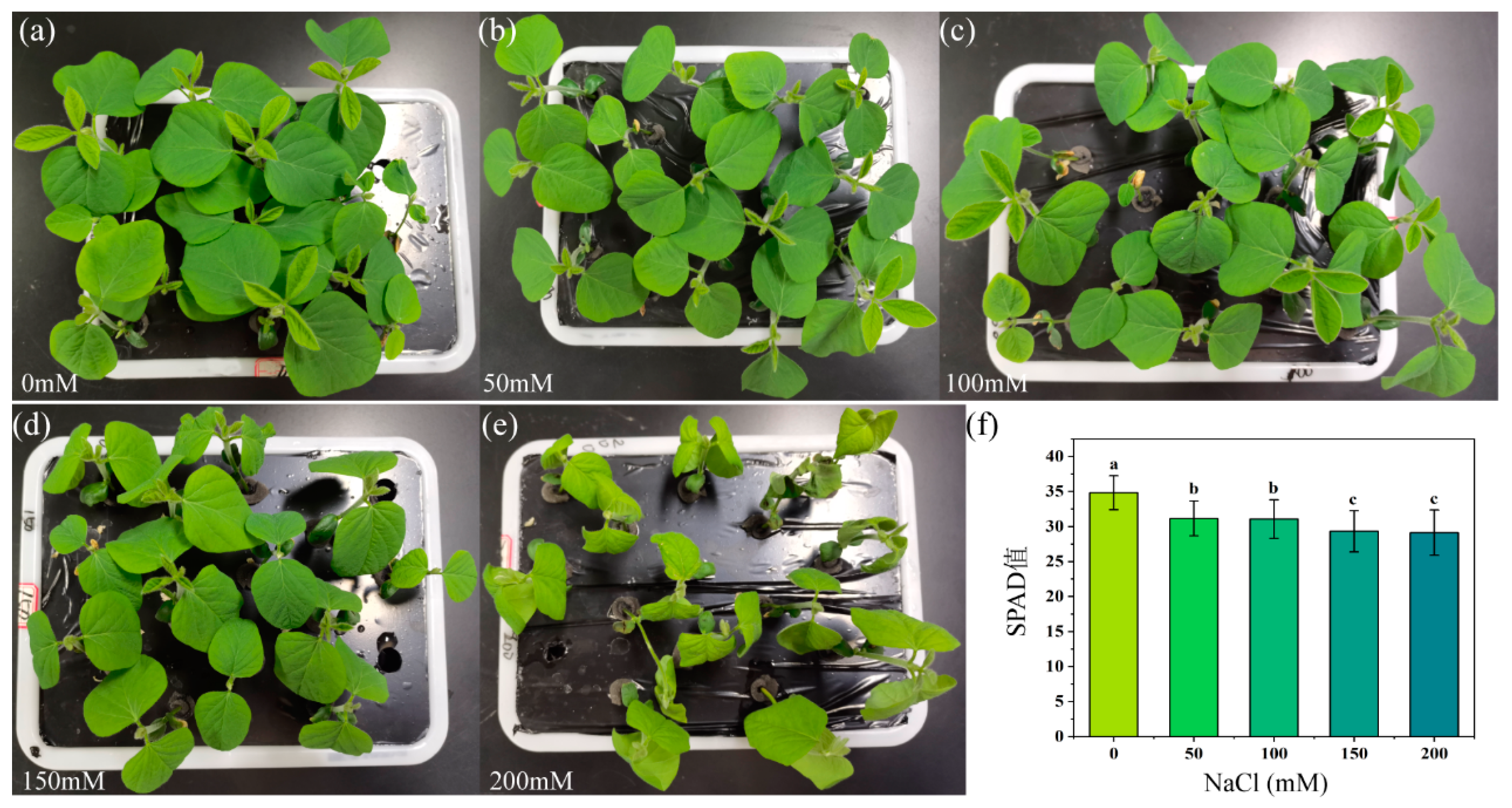
3.1. Changes in Leaf Phenotype and SPAD Values
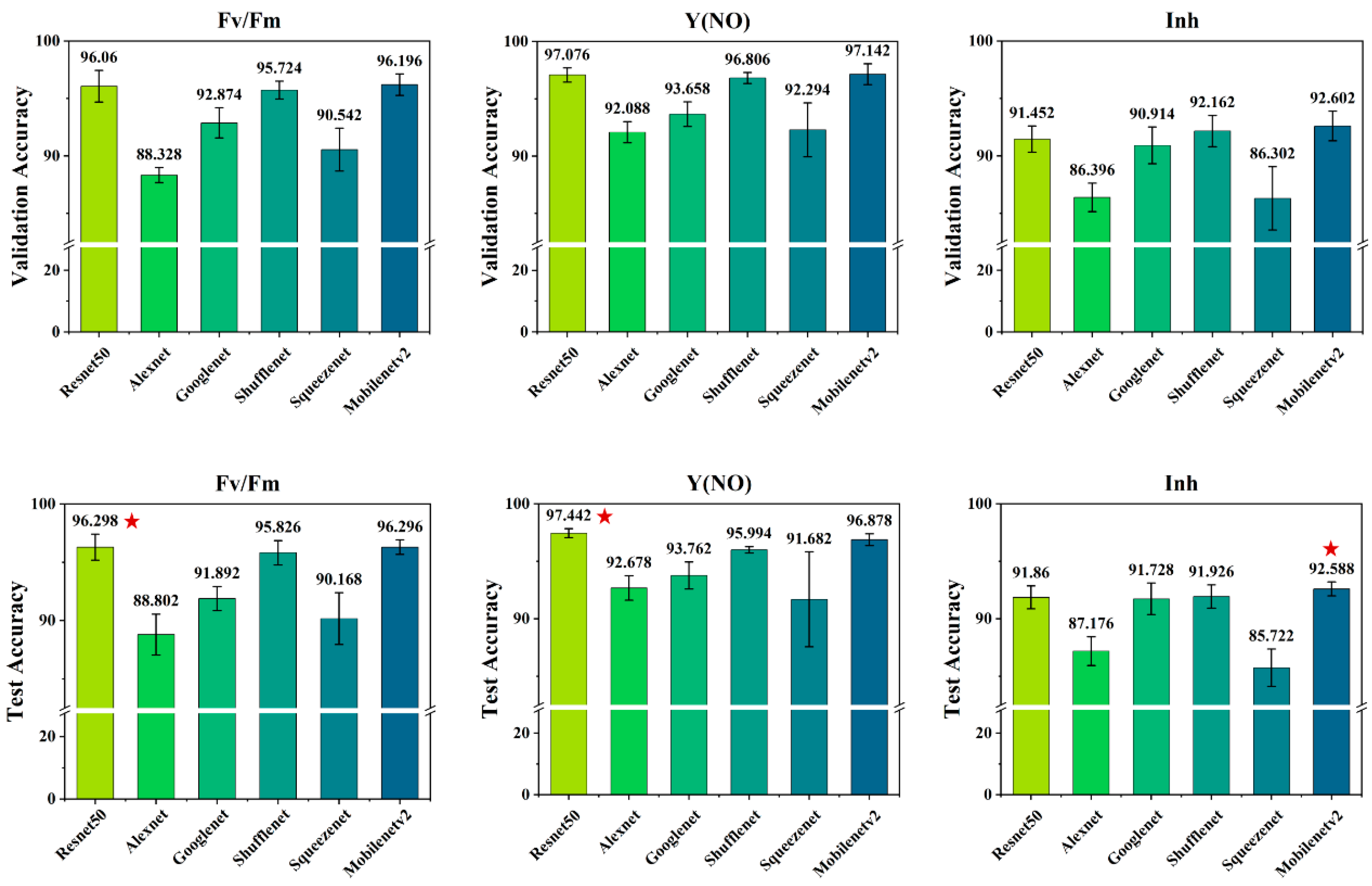
3.2. Salt Stress Recognition Results Using Various CNN Models
3.3. Various Salt Stress Recognition Results Based on Feature Fusion Models
3.4. Evaluating Indicator
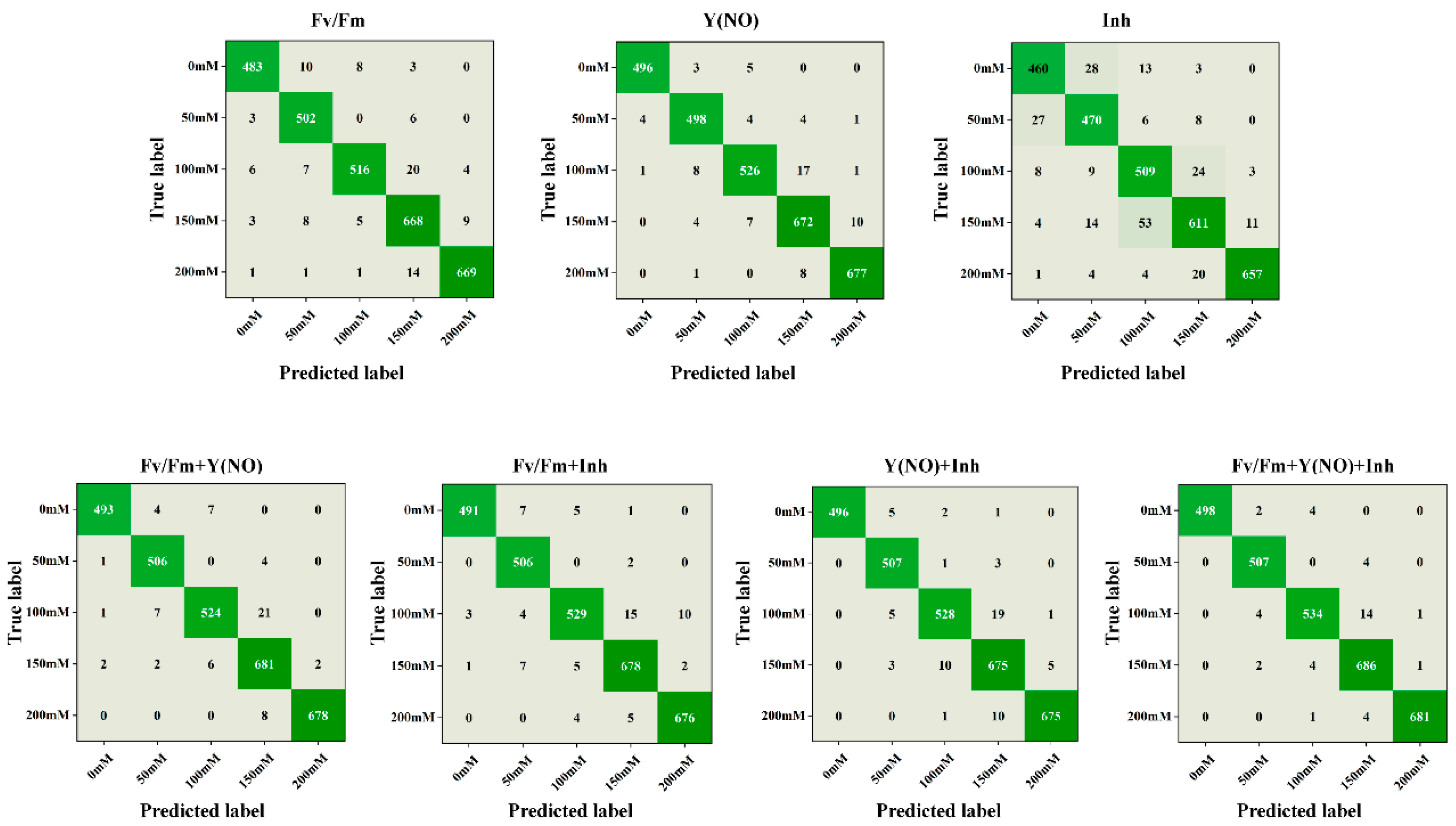
3.5. Confusion Matrix Analysis
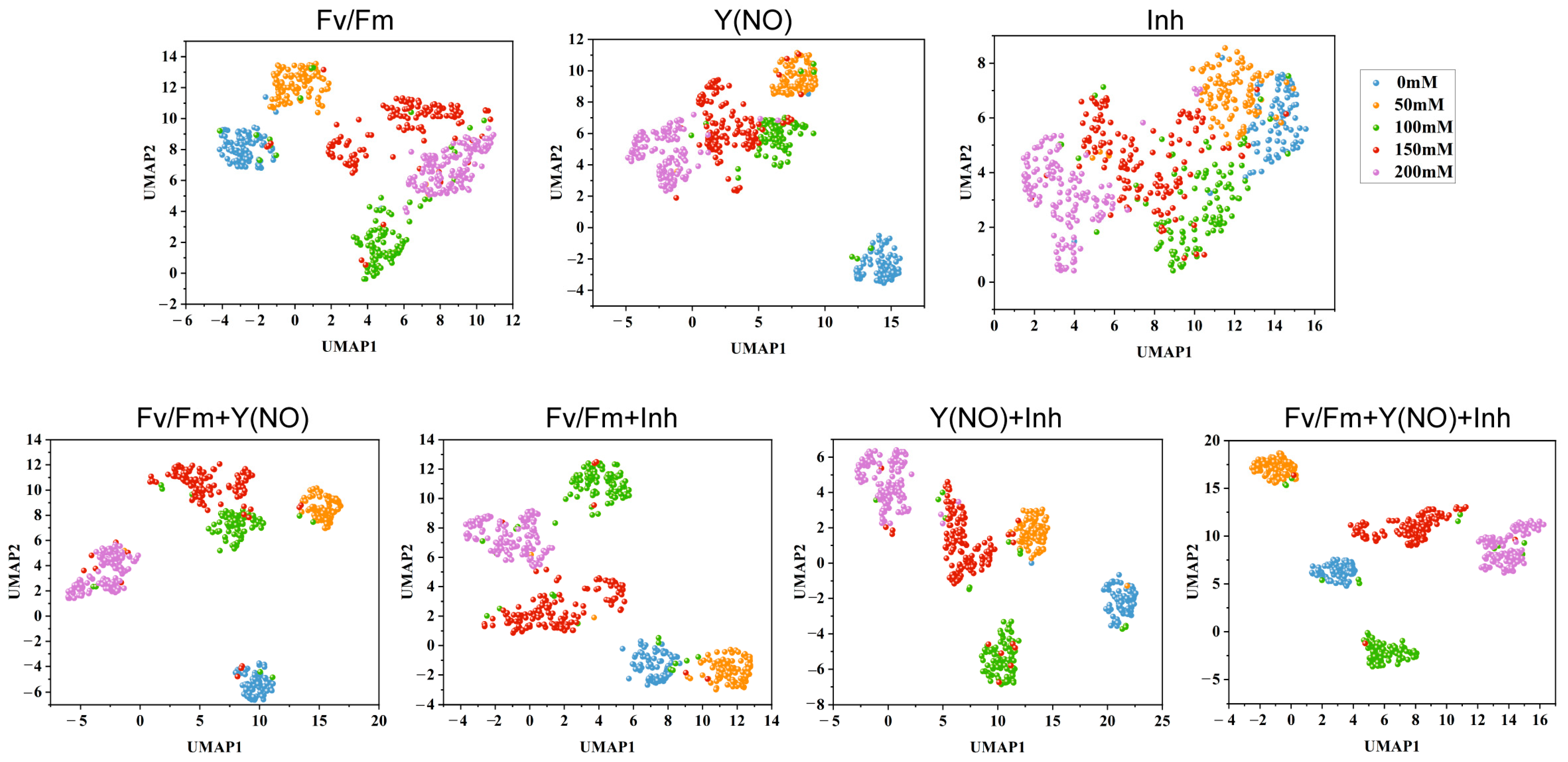
3.6. UMAP Dimensionality Reduction Analysis
4. Discussion
4.1. Insights into Plant Responses to Salt Stress through ChlF Imaging
4.2. Enhancing Salt Stress Recognition with Integrated ChlF Image Feature Fusion
4.3. Future Directions
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Majeed, A.; Muhammad, Z. Salinity: A Major Agricultural Problem—Causes, Impacts on Crop Productivity and Management Strategies. In Plant Abiotic Stress Tolerance: Agronomic, Molecular and Biotechnological Approaches; Hasanuzzaman, M., Hakeem, K.R., Nahar, K., Alharby, H.F., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 83–99. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Sarkar, B.; Jat, H.S.; Sharma, P.C.; Bolan, N.S. Soil Salinity under Climate Change: Challenges for Sustainable Agriculture and Food Security. J. Environ. Manag. 2021, 280, 111736. [Google Scholar] [CrossRef] [PubMed]
- Ivushkin, K.; Bartholomeus, H.; Bregt, A.K.; Pulatov, A.; Kempen, B.; de Sousa, L. Global Mapping of Soil Salinity Change. Remote Sens. Environ. 2019, 231, 111260. [Google Scholar] [CrossRef]
- FAO; ITPS. Status of the World’s Soil Resources (SWSR)—Main Report; Food and Agriculture Organization of the United Nations and Intergovernmental Technical Panel on Soils: Rome, Italy, 2015. [Google Scholar]
- Li, S.; Chen, J.; Hao, X.; Ji, X.; Zhu, Y.; Chen, X.; Yao, Y. A Systematic Review of Black Soybean (Glycine max (L.) Merr.): Nutritional Composition, Bioactive Compounds, Health Benefits, and Processing to Application. Food Front. 2024, 5, 1188–1211. [Google Scholar] [CrossRef]
- Shawquat, M.; Chowdhury, J.A.; Razzaque, M.A.; Ali, M.Z.; Paul, S.K.; Aziz, M. Dry Matter Production and Seed Yield of Soybean as Affected by Post-Flowering Salinity and Water Stress. Bangladesh Agron. J. 2017, 19, 21. [Google Scholar] [CrossRef]
- Negrao, S.; Schmockel, S.M.; Tester, M. Evaluating Physiological Responses of Plants to Salinity Stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine Learning for High-Throughput Stress Phenotyping in Plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Naik, H.S.; Zhang, J.; Lofquist, A.; Assefa, T.; Sarkar, S.; Ackerman, D.; Singh, A.; Singh, A.K.; Ganapathysubramanian, B. A Real-Time Phenotyping Framework Using Machine Learning for Plant Stress Severity Rating in Soybean. Plant Methods 2017, 13, 23. [Google Scholar] [CrossRef]
- Alom, M.Z.; Taha, T.M.; Yakopcic, C.; Westberg, S.; Sidike, P.; Nasrin, M.S.; Hasan, M.; Van Essen, B.C.; Awwal, A.A.S.; Asari, V.K. A State-of-the-Art Survey on Deep Learning Theory and Architectures. Electronics 2019, 8, 292. [Google Scholar] [CrossRef]
- Singh, A.K.; Ganapathysubramanian, B.; Sarkar, S.; Singh, A. Deep Learning for Plant Stress Phenotyping: Trends and Future Perspectives. Trends Plant Sci. 2018, 23, 883–898. [Google Scholar] [CrossRef]
- Ghosal, S.; Blystone, D.; Singh, A.K.; Ganapathysubramanian, B.; Singh, A.; Sarkar, S. An Explainable Deep Machine Vision Framework for Plant Stress Phenotyping. Proc. Natl. Acad. Sci. USA 2018, 115, 4613–4618. [Google Scholar] [CrossRef]
- Long, Y.; Ma, M. Recognition of Drought Stress State of Tomato Seedling Based on Chlorophyll Fluorescence Imaging. IEEE Access 2022, 10, 48633–48642. [Google Scholar] [CrossRef]
- Yao, J.; Sun, D.; Cen, H.; Xu, H.; Weng, H.; Yuan, F.; He, Y. Phenotyping of Arabidopsis Drought Stress Response Using Kinetic Chlorophyll Fluorescence and Multicolor Fluorescence Imaging. Front. Plant Sci. 2018, 9, 603. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Lu, R. Detection of Chilling Injury in Pickling Cucumbers Using Dual-Band Chlorophyll Fluorescence Imaging. Foods 2021, 10, 1094. [Google Scholar] [CrossRef] [PubMed]
- Rolfe, S.A.; Scholes, J.D. Chlorophyll Fluorescence Imaging of Plant-Pathogen Interactions. Protoplasma 2010, 247, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Herritt, M.T.; Pauli, D.; Mockler, T.C.; Thompson, A.L. Chlorophyll Fluorescence Imaging Captures Photochemical Efficiency of Grain Sorghum (Sorghum bicolor) in a Field Setting. Plant Methods 2020, 16, 109. [Google Scholar] [CrossRef]
- Küpper, H.; Benedikty, Z.; Morina, F.; Andresen, E.; Mishra, A.; Trtílek, M. Analysis of OJIP Chlorophyll Fluorescence Kinetics and QA Reoxidation Kinetics by Direct Fast Imaging. Plant Physiol. 2019, 179, 369–381. [Google Scholar] [CrossRef]
- Liu, L.; Martín-Barragán, B.; Prieto, F.J. A projection Multi-Objective SVM Method for Multi-Class Classification. Comput. Ind. Eng. 2021, 158, 107425. [Google Scholar] [CrossRef]
- Osório, J.; Osório, M.L.; Correia, P.J.; de Varennes, A.; Pestana, M. Chlorophyll Fluorescence Imaging as a Tool to Understand the Impact of Iron Deficiency and Resupply on Photosynthetic Performance of Strawberry Plants. Sci. Hortic. 2014, 165, 148–155. [Google Scholar] [CrossRef]
- Cen, H.; Weng, H.; Yao, J.; He, M.; Lv, J.; Hua, S.; Li, H.; He, Y. Chlorophyll Fluorescence Imaging Uncovers Photosynthetic Fingerprint of Citrus Huanglongbing. Front. Plant Sci. 2017, 8, 1509. [Google Scholar] [CrossRef]
- Hou, P.; Yang, W.; Du, W.; Aiqin, Y.; Zhao, J.; Zhang, Y.; Gao, C.; Wang, M. Effects of Different Degree Salt Stress on Biomass and Physiological Indexes of Soybean Seedling. Soybean Sci. 2020, 39, 422–430. (In Chinese) [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2012, 60, 84–90. [Google Scholar] [CrossRef]
- Szegedy, C.; Wei, L.; Yangqing, J.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. ShuffleNet: An Extremely Efficient Convolutional Neural Network for Mobile Devices. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6848–6856. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. Squeezenet: Alexnet-Level Accuracy with 50x Fewer Parameters and <0.5 MB Model Size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Mishra, A.; Matous, K.; Mishra, K.B.; Nedbal, L. Towards Discrimination of Plant Species by Machine Vision: Advanced Statistical Analysis of Chlorophyll Fluorescence Transients. J. Fluoresc. 2009, 19, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Shu, S.; Li, S.; He, L.; Li, H.; Du, N.; Sun, J.; Guo, S. Effects of Exogenous Putrescine on Chlorophyll Fluorescence Imaging and Heat Dissipation Capacity in Cucumber (Cucumis sativus L.) Under Salt Stress. J. Plant Growth Regul. 2014, 33, 798–808. [Google Scholar] [CrossRef]
- Awlia, M.; Nigro, A.; Fajkus, J.; Schmoeckel, S.M.; Negrao, S.; Santelia, D.; Trtilek, M.; Tester, M.; Julkowska, M.M.; Panzarova, K. High-Throughput Non-destructive Phenotyping of Traits that Contribute to Salinity Tolerance in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1414. [Google Scholar] [CrossRef] [PubMed]
- Rigó, G.; Valkai, I.; Faragó, D.; Kiss, E.; Van Houdt, S.; Van de Steene, N.; Hannah, M.A.; Szabados, L. Gene Mining in Halophytes: Functional Identification of Stress Tolerance Genes in Lepidium Crassifolium. Plant Cell Environ. 2016, 39, 2074–2084. [Google Scholar] [CrossRef]
- Tian, Y.; Xie, L.; Wu, M.; Yang, B.; Ishimwe, C.; Ye, D.; Weng, H. Multicolor Fluorescence Imaging for the Early Detection of Salt Stress in Arabidopsis. Agronomy 2021, 11, 2577. [Google Scholar] [CrossRef]
- Mishra, P.; Mohd Asaari, M.S.; Herrero-Langreo, A.; Lohumi, S.; Diezma, B.; Scheunders, P. Close Range Hyperspectral Imaging of Plants: A Review. Biosyst. Eng. 2017, 164, 49–67. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Murchie, E.H.; Lawson, T. Chlorophyll Fluorescence Analysis: A Guide to Good Practice and Understanding Some New Applications. J. Exp. Bot. 2013, 64, 3983–3998. [Google Scholar] [CrossRef]
- Hernández, J.A.; Almansa, M.S. Short-Term Effects of Salt Stress on Antioxidant Systems and Leaf Water Relations of Pea Leaves. Physiol. Plant. 2002, 115, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhu, Y.; Xu, H.; He, Y.; Cen, H. Time-Series Chlorophyll Fluorescence Imaging Reveals Dynamic Photosynthetic Fingerprints of sos Mutants to Drought Stress. Sensors 2019, 19, 2649. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-W.; Lee, T.-Y.; Nah, G.; Kim, D.-S. Potential of Thermal Image Analysis for Screening Salt Stress-Tolerant Soybean (Glycine max). Plant Genet. Resour. 2014, 12, S134–S136. [Google Scholar] [CrossRef]
- Wang, W.; Wang, C.; Pan, D.; Zhang, Y.; Luo, B.; Ji, J. Effects of Drought Stress on Photosynthesis and Chlorophyll Fluorescence Images of Soybean (Glycine max) Seedlings. Int. J. Agric. Biol. Eng. 2018, 11, 196–201. [Google Scholar] [CrossRef]
| Model | Layer | Mini Batch Size | Initial Learn Rate | Validation Frequency | Image Input Size |
|---|---|---|---|---|---|
| AlexNet | 25 | 32 | 1 × 10−4 | 64 | 227 × 227 × 3 |
| GoogLeNet | 144 | 32 | 1 × 10−4 | 64 | 224 × 224 × 3 |
| ResNet50 | 177 | 32 | 1 × 10−4 | 64 | 224 × 224 × 3 |
| ShuffleNet | 172 | 32 | 1 × 10−4 | 64 | 224 × 224 × 3 |
| SqueezeNet | 68 | 32 | 1 × 10−4 | 64 | 227 × 227 × 3 |
| MobileNetv2 | 154 | 32 | 1 × 10−4 | 64 | 224 × 224 × 3 |
| Class | Original Images | Augmented Images | Total |
|---|---|---|---|
| 0 mM | 72 | 432 | 504 |
| 50 mM | 73 | 438 | 511 |
| 100 mM | 79 | 474 | 553 |
| 150 mM | 99 | 594 | 693 |
| 200 mM | 98 | 588 | 686 |
| Feature Fused Model | Validation Accuracy (%) | Test Accuracy (%) |
|---|---|---|
| Fv/Fm + Y(NO) | 97.72 ± 1.01 | 97.79 ± 0.54 |
| Fv/Fm + Inh | 97.52 ± 0.85 | 97.73 ± 0.51 |
| Y(NO) + Inh | 97.76 ± 0.38 | 97.76 ± 0.35 |
| Fv/Fm + Y(NO) + Inh | 98.37 ± 0.63 | 98.61 ± 0.37 |
| Measures | Single Type ChIF Images | Fusion of Two Type ChIF Image Features | Fusion of Three ChIF Images Features | ||||
|---|---|---|---|---|---|---|---|
| Fv/Fm | Y(NO) | Inh | Fv/Fm + Y(NO) | Fv/Fm + Inh | Y(NO) + Inh | Fv/Fm + Y(NO) + Inh | |
| Precision (%) | 96.41 ± 2.57 | 97.43 ± 1.69 | 91.76 ± 4.71 | 97.89 ± 2.06 | 97.80 ± 2.21 | 97.90 ± 2.13 | 98.70 ± 1.67 |
| Recall (%) | 96.25 ± 2.63 | 97.33 ± 1.68 | 91.86 ± 3.72 | 97.74 ± 2.24 | 97.70 ± 2.19 | 97.78 ± 2.16 | 98.57 ± 1.85 |
| F1 Score (%) | 96.29 ± 1.83 | 97.37 ± 1.27 | 91.72 ± 3.17 | 97.79 ± 1.47 | 97.72 ± 1.36 | 97.82 ± 1.49 | 98.62 ± 1.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, Y.; Xin, N.; Zhao, L.; Shi, H.; Deng, L.; Han, Z.; Wu, G. Precision Detection of Salt Stress in Soybean Seedlings Based on Deep Learning and Chlorophyll Fluorescence Imaging. Plants 2024, 13, 2089. https://doi.org/10.3390/plants13152089
Deng Y, Xin N, Zhao L, Shi H, Deng L, Han Z, Wu G. Precision Detection of Salt Stress in Soybean Seedlings Based on Deep Learning and Chlorophyll Fluorescence Imaging. Plants. 2024; 13(15):2089. https://doi.org/10.3390/plants13152089
Chicago/Turabian StyleDeng, Yixin, Nan Xin, Longgang Zhao, Hongtao Shi, Limiao Deng, Zhongzhi Han, and Guangxia Wu. 2024. "Precision Detection of Salt Stress in Soybean Seedlings Based on Deep Learning and Chlorophyll Fluorescence Imaging" Plants 13, no. 15: 2089. https://doi.org/10.3390/plants13152089





