Detection and Evaluation of Blast Resistance Genes in Backbone Indica Rice Varieties from South China
Abstract
1. Introduction
2. Results
2.1. Variability of Resistance to Rice Blast
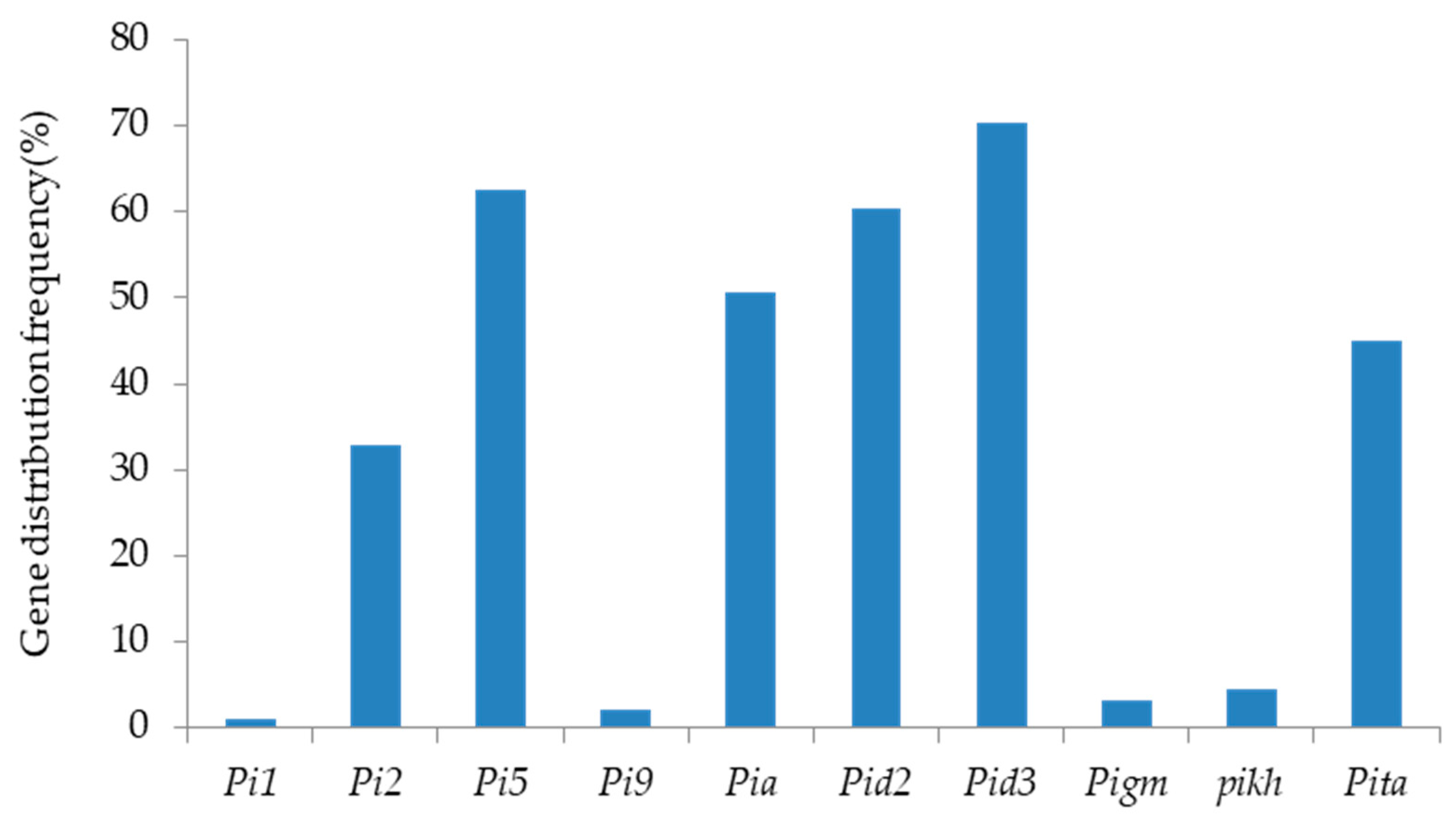
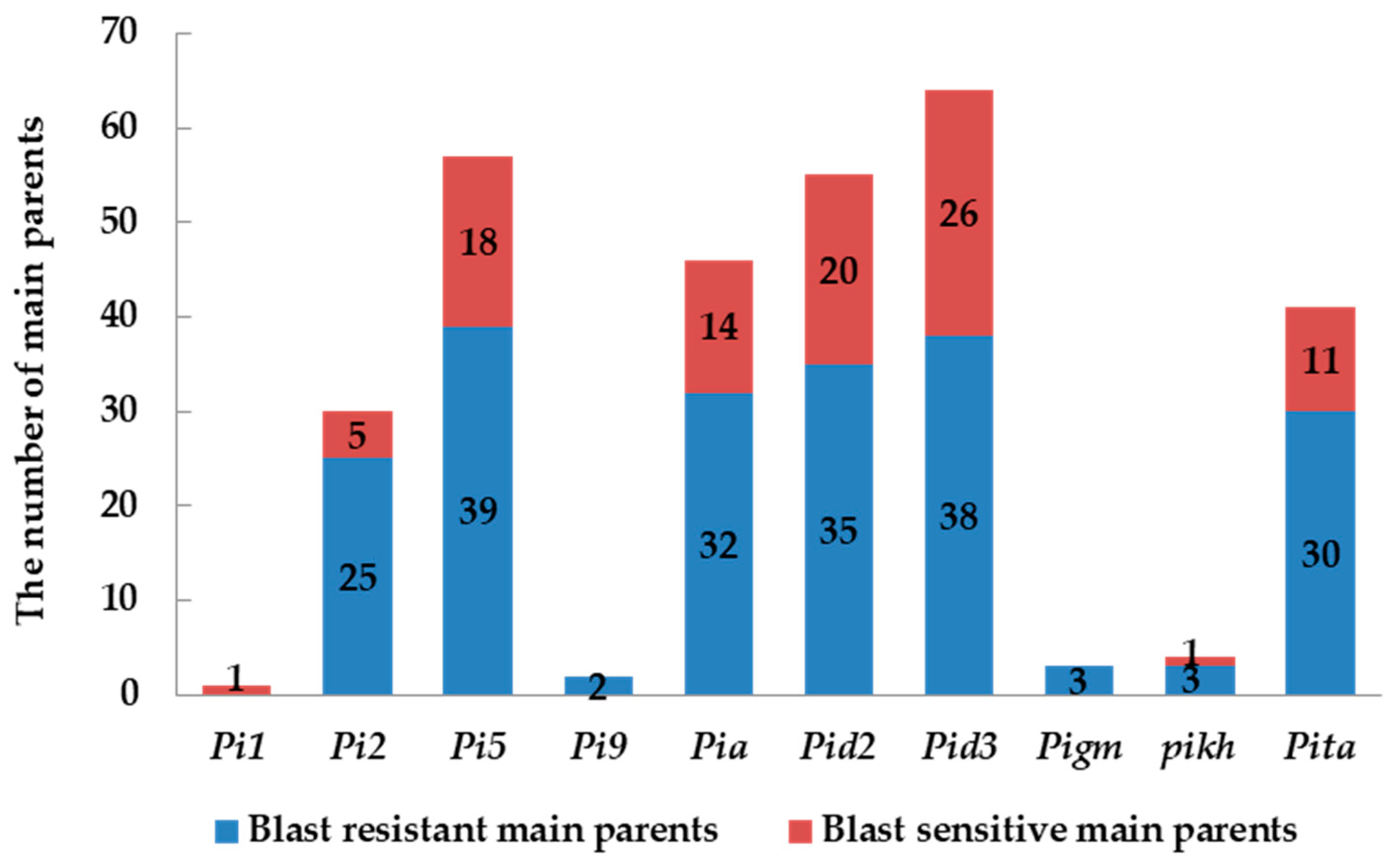
2.2. Identification and Analysis of Rice Blast Resistance Genes among the Backbone Rice Varieties in South China
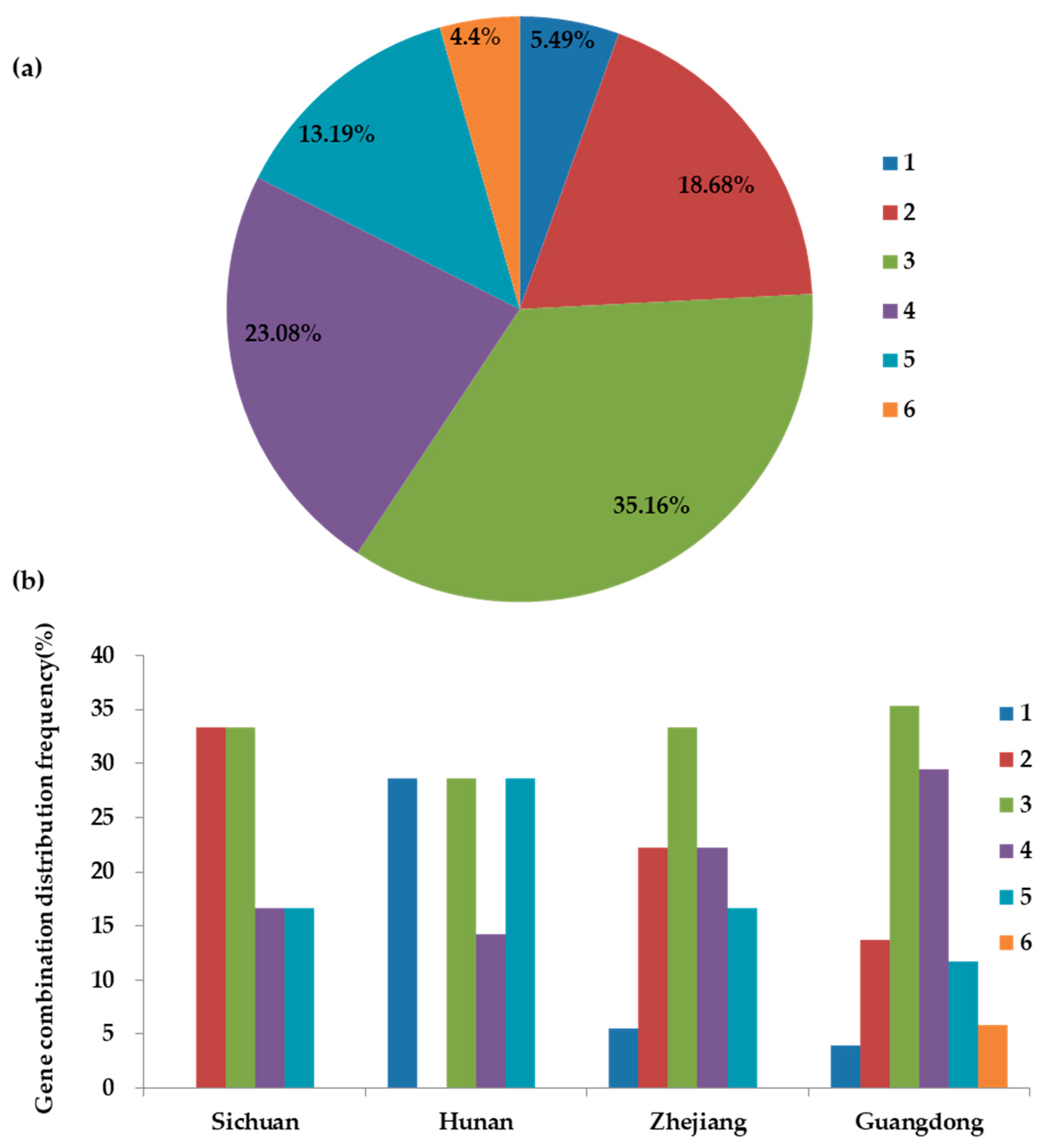
2.3. Correlation Analysis of Gene Combination Distributions and Resistance Phenotypes
3. Discussion
3.1. Distribution and Manifestation of Blast Resistance Genes in Rice Varieties
3.2. Gene Pyramiding for Broad-Spectrum Rice Blast Resistance
4. Materials and Methods
4.1. Materials and Strains
4.2. Resistance Identification
4.3. Rice Blast Resistance Gene Detection
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pennisi, E. Armed and Dangerous. Science 2010, 327, 804–805. [Google Scholar] [PubMed]
- Devanna, B.N.; Jain, P.; Solanke, A.U.; Das, A.; Thakur, S.; Singh, P.K.; Kumari, M.; Dubey, H.; Jaswal, R.; Pawar, D.; et al. Understanding the Dynamics of Blast Resistance in Rice-Magnaporthe oryzae Interactions. J. Fungi 2022, 8, 584. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.; Sundaram, K.T.; Singh, U.M.; Srinivas Prasad, M.; Laha, G.S.; Sinha, P.; Singh, V.K. Superior haplotypes towards the development of blast and bacterial blight-resistant rice. Front. Plant Sci. 2024, 15, 1272326. [Google Scholar] [CrossRef] [PubMed]
- Khush, G.S.; Jena, K.K. Current Status and Future Prospects for Research on Blast Resistance in Rice (Oryza sativa L.). In Advances in Genetics, Genomics and Control of Rice Blast Disease; Springer: Dordrecht, The Netherlands, 2009. [Google Scholar]
- Li, W.; Chern, M.; Yin, J.; Wang, J.; Chen, X. Recent advances in broad-spectrum resistance to the rice blast disease. Curr. Opin. Plant Biol. 2019, 50, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Skamnioti, P.; Gurr, S.J. Against the grain: Safeguarding rice from rice blast disease. Trends Biotechnol. 2009, 27, 141–150. [Google Scholar] [CrossRef]
- Kamoun, S.; Furzer, O.; Jones, J.D.; Judelson, H.S.; Ali, G.S.; Dalio, R.J.; Roy, S.G.; Schena, L.; Zambounis, A.; Panabières, F.; et al. The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 2015, 16, 413–434. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Kou, Y.; Shi, H.; Qiu, J.; Tao, Z.; Wang, W. Effectors and environment modulating rice blast disease: From understanding to effective control. Trends Microbiol. 2024. [Google Scholar] [CrossRef]
- Yin, J.; Zou, L.; Zhu, X.; Cao, Y.; He, M.; Chen, X. Fighting the enemy: How rice survives the blast pathogen’s attack. Crop. J. 2021, 9, 10. [Google Scholar] [CrossRef]
- Tanweer, F.A.; Rafii, M.Y.; Sijam, K.; Rahim, H.A.; Ahmed, F.; Latif, M.A. Current advance methods for the identification of blast resistance genes in rice. Comptes Rendus Biol. 2015, 338, 321–334. [Google Scholar] [CrossRef]
- Ashkani, S.; Rafii, M.Y.; Shabanimofrad, M.; Ghasemzadeh, A.; Ravanfar, S.A.; Latif, M.A. Molecular progress on the mapping and cloning of functional genes for blast disease in rice (Oryza sativa L.): Current status and future considerations. Crit. Rev. Biotechnol. 2016, 36, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhai, K.; Xie, Z.; Yang, D.; Zhu, X.; Liu, J.; Wang, X.; Qin, P.; Yang, Y.; Zhang, G.; et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 2017, 355, 962–965. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, J.R.; Lacorte-Apostol, V.; Kroj, T.; Padilla, J.; Telebanco-Yanoria, M.J.; Glaus, A.N.; Roulin, A.; Padilla, A.; Zhou, B.; Keller, B.; et al. Genome-wide association analysis uncovers rice blast resistance alleles of Ptr and Pia. Commun. Biol. 2024, 7, 607. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.K.; Sao, R.; Choudhary, D.K.; Thada, A.; Kumar, V.; Mondal, S.; Das, B.K.; Jankuloski, L.; Sharma, D. Advancement in the Breeding, Biotechnological and Genomic Tools towards Development of Durable Genetic Resistance against the Rice Blast Disease. Plants 2022, 11, 2386. [Google Scholar] [CrossRef] [PubMed]
- Hua, L.; Wu, J.; Chen, C.; Wu, W.; He, X.; Lin, F.; Wang, L.; Ashikawa, I.; Matsumoto, T.; Wang, L.; et al. The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor. Appl. Genet. 2012, 125, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Feng, Y.; Bao, L.; Li, X.; Gao, G.; Zhang, Q.; Xiao, J.; Xu, C.; He, Y. Improving blast resistance of Jin 23B and its hybrid rice by marker-assisted gene pyramiding. Mol. Breed. 2012, 30, 1679–1688. [Google Scholar] [CrossRef]
- Zhou, Y.; Lei, F.; Wang, Q.; He, W.; Yuan, B.; Yuan, W. Identification of Novel Alleles of the Rice Blast-Resistance Gene Pi9 through Sequence-Based Allele Mining. Rice 2020, 13, 80. [Google Scholar] [CrossRef]
- Wang, K.; Li, S.; Chen, L.; Tian, H.; Chen, C.; Fu, Y.; Du, H.; Hu, Z.; Li, R.; Du, Y.; et al. E3 ubiquitin ligase OsPIE3 destabilises the B-lectin receptor-like kinase PID2 to control blast disease resistance in rice. New Phytol. 2023, 237, 1826–1842. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhu, X.; Shen, Y.; He, Z. Genetic characterization and fine mapping of the blast resistance locus Pigm(t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety. Theor. Appl. Genet. 2006, 113, 705–713. [Google Scholar] [CrossRef]
- Tian, D.; Deng, Y.; Yang, X.; Li, G.; Li, Q.; Zhou, H.; Chen, Z.; Guo, X.; Su, Y.; Luo, Y.; et al. Association analysis of rice resistance genes and blast fungal avirulence genes for effective breeding resistance cultivars. Front. Microbiol. 2022, 13, 1007492. [Google Scholar] [CrossRef]
- Feng, Z.; Li, M.; Xu, Z.; Gao, P.; Wu, Y.; Wu, K.; Zhao, J.; Wang, X.; Wang, J.; Li, M.; et al. Development of Rice Variety with Durable and Broad-Spectrum Resistance to Blast Disease Through Marker-Assisted Introduction of Pigm Gene. Front. Plant Sci. 2022, 13, 937767. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, F.; Luo, X.; Kong, D.; Zhang, A.; Wang, F.; Pan, Z.; Wang, J.; Bi, J.; Luo, L.; et al. Molecular Breeding of a Novel PTGMS Line of WDR for Broad-Spectrum Resistance to Blast Using Pi9, Pi5, and Pi54 Genes. Rice 2021, 14, 96. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, N.; Mitsunaga, T.; Hayashi, K.; Koizumi, S.; Fujita, Y. Effects of Pyramiding Quantitative Resistance Genes pi21, Pi34, and Pi35 on Rice Leaf Blast Disease. Plant Dis. 2015, 99, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Lin, X.; Xiang, X.; Ren, H.; Fan, X.; Chen, K. Characterization and Evaluation of Transgenic Rice Pyramided with the Pi Genes Pib, Pi25 and Pi54. Rice 2021, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Jamaloddin, M.; Durga Rani, C.V.; Swathi, G.; Anuradha, C.; Vanisri, S.; Rajan, C.P.D.; Krishnam Raju, S.; Bhuvaneshwari, V.; Jagadeeswar, R.; Laha, G.S.; et al. Marker Assisted Gene Pyramiding (MAGP) for bacterial blight and blast resistance into mega rice variety “Tellahamsa”. PLoS ONE 2020, 15, e0234088. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Singh, A.; Singh, S.P.; Ellur, R.K.; Choudhary, V.; Sarkel, S.; Singh, D.; Krishnan, S.G.; Nagarajan, M.; Vinod, K.K.; et al. Incorporation of blast resistance into “PRR78”, an elite Basmati rice restorer line, through marker assisted backcross breeding. Field Crops Res. 2012, 128, 8–16. [Google Scholar] [CrossRef]
- Khanna, A.; Sharma, V.; Ellur, R.K.; Shikari, A.B.; Singh, A.K. Marker assisted pyramiding of major blast resistance genes Pi9 and Pita in the genetic background of an elite Basmati rice variety, Pusa Basmati 1. Indian J. Genet. Plant Breed. 2015, 75, 417. [Google Scholar] [CrossRef]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Oladosu, Y.; Kolapo, K.; Musa, I.; Halidu, J.; Muhammad, I.I.; Ahmed, M. Marker-Assisted Introgression of Multiple Resistance Genes Confers Broad Spectrum Resistance against Bacterial Leaf Blight and Blast Diseases in PUTRA-1 Rice Variety. Agronomy 2020, 10, 42. [Google Scholar] [CrossRef]
- Zhai, C.; Lin, F.; Dong, Z.; He, X.; Yuan, B.; Zeng, X.; Wang, L.; Pan, Q. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 2011, 189, 321–334. [Google Scholar] [CrossRef]
- Ma, J.; Lei, C.; Xu, X.; Hao, K.; Wang, J.; Cheng, Z.; Ma, X.; Ma, J.; Zhou, K.; Zhang, X.; et al. Pi64, Encoding a Novel CC-NBS-LRR Protein, Confers Resistance to Leaf and Neck Blast in Rice. Mol. Plant-Microbe Interact. MPMI 2015, 28, 558–568. [Google Scholar] [CrossRef]
- Manojkumar, H.B.; Deepak, C.A.; Harinikumar, K.M.; Rajanna, M.P.; Chethana, B. Molecular profiling of blast resistance genes and evaluation of leaf and neck blast disease reaction in rice. J. Genet. 2020, 99, 52. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Chen, Z.; Chen, Z.; Zhou, Y.; Wang, Z.; Wang, F.; Chen, S. Allele-specific marker-based assessment revealed that the rice blast resistance genes Pi2 and Pi9 have not been widely deployed in Chinese indica rice cultivars. Rice 2016, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xiao, N.; Chen, Y.; Yu, L.; Pan, C.; Li, Y.; Zhang, X.; Huang, N.; Ji, H.; Dai, Z.; et al. Comprehensive evaluation of resistance effects of pyramiding lines with different broad-spectrum resistance genes against Magnaporthe oryzae in rice (Oryza sativa L.). Rice 2019, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Shafi, A.; Khan, R.S.; Mir, S.; Khan, G.H.; Masoodi, K.Z.; Sofi, N.R.; Mohidin, F.A.; Lone, J.A.; Shikari, A.B. Gene expression of near-isogenic lines (NILs) carrying blast resistance genes Pi9 and Pi54 in the background of rice cultivar Mushk Budji. Mol. Biol. Rep. 2023, 50, 5901–5915. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.M.; Luo, L.X.; Wang, H.; Guo, T.; Liu, Y.Z.; Zhou, J.Y.; Zhu, X.Y.; Yang, Q.Y.; Chen, Z.Q. Pyramiding of Pi46 and Pita to improve blast resistance and to evaluate the resistance effect of the two R genes. J. Integr. Agric. 2016, 10, 9. [Google Scholar] [CrossRef]
- Jiang, J.; Mou, T.; Yu, H.; Zhou, F. Molecular breeding of thermo-sensitive genic male sterile (TGMS) lines of rice for blast resistance using Pi2 gene. Rice 2015, 8, 11. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peng, P.; Jiang, H.; Luo, L.; Ye, C.; Xiao, Y. Pyramiding of Multiple Genes to Improve Rice Blast Resistance of Photo-Thermo Sensitive Male Sterile Line, without Yield Penalty in Hybrid Rice Production. Plants 2023, 12, 1389. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.S.; Tian, Y.H.; YU, H.-Q.; Sun, Y.J.; Cao, G.C.; Chen, B.; Fang, Z.B.; Fan, B. The Methods of Isolation and Preservation of the Single Spore of Rice Blast Fungus. Hub Agric. Ences 2015, 54, 3. [Google Scholar]
- Yang, N.; Yu, J.; Wang, A.; Tang, J.; Kwabena, O.P. A rapid rice blast detection and identification method based on crop disease spores’ diffraction fingerprint texture. J. Sci. Food Agric. 2020, 100, 3608–3621. [Google Scholar] [CrossRef]
- Workshop, R.B. Proceedings of the Rice Blast Workshop; International Rice Research Institute: Laguna, Philippines, 1979. [Google Scholar]
- Gouda, P.K.; Saikumar, S.; Varma, C.M.K.; Nagesh, K.; Thippeswamy, S.; Shenoy, V.; Ramesha, M.S.; Shashidhar, H.E.; Ahn, S.N. Marker-assisted breeding of Pi-1 and Piz-5 genes imparting resistance to rice blast in PRR78, restorer line of Pusa RH-10 Basmati rice hybrid. Plant Breed. 2013, 132, 61–69. [Google Scholar] [CrossRef]

| Number | Germplasm Origin | Rice Blast Resistance Gene | Blast Resistance Level |
|---|---|---|---|
| 23P-1 | Sichuan | Pia+Pid2+Pid3 | MR |
| 23P-2 | Sichuan | Pia+Pid3 | MS |
| 23P-3 | Sichuan | Pid3+Pita | MR |
| 23P-4 | Sichuan | Pia+Pid2+Pid3+Pigm | R |
| 23P-5 | Sichuan | Pia+Pid2+Pid3+Pigm+pikh | R |
| 23P-6 | Sichuan | Pia+Pid3+pikh | MS |
| 23P-7 | Hubei | Pi1+Pi5 | MS |
| 23P-8 | Hubei | Pi2+Pid2+Pid3 | MR |
| 23P-9 | Hunan | Pi2+Pi5+Pita | MS |
| 23P-10 | Hunan | Pi5+Pia+Pid2+Pid3+Pita | MR |
| 23P-11 | Hunan | Pia | MS |
| 23P-12 | Hunan | Pi5+Pi9+Pid2+Pid3 | MR |
| 23P-13 | Hunan | Pi5+Pi9+Pia+Pid2+Pid3 | R |
| 23P-14 | Hunan | Pi5 | MR |
| 23P-15 | Hunan | Pia+Pid2+Pid3 | MS |
| 23P-16 | Jiangxi | Pi2+Pi5 | MS |
| 23P-17 | Jiangxi | Pid2+Pid3 | MS |
| 23P-18 | Jiangxi | Pi2+Pi5+Pia+Pid2+Pid3+Pita | MR |
| 23P-19 | Anhui | Pia+Pid2+Pid3 | MS |
| 23P-20 | Anhui | Pia+Pigm | MR |
| 23P-21 | Fujian | Pi2+Pid2+Pid3 | R |
| 23P-22 | Zhejiang | Pi5+Pia+Pid2+Pid3+Pita | R |
| 23P-23 | Zhejiang | Pi5+Pia | R |
| 23P-24 | Zhejiang | Pi2+Pi5+Pita | R |
| 23P-25 | Zhejiang | Pi2+Pi5 | MR |
| 23P-26 | Zhejiang | Pi5+Pid2+Pid3+Pita | MR |
| 23P-27 | Zhejiang | Pid2+Pid3+Pita | MR |
| 23P-28 | Zhejiang | Pia+Pid2+Pid3 | MS |
| 23P-29 | Zhejiang | Pi5+Pid2+Pid3 | MS |
| 23P-30 | Zhejiang | Pia+Pid2+Pid3 | R |
| 23P-31 | Zhejiang | Pi5+Pid3 | MS |
| 23P-32 | Zhejiang | Pi5+Pia+Pid2+Pid3+Pita | MR |
| 23P-33 | Zhejiang | Pi5+Pia+Pid2+Pid3 | MR |
| 23P-34 | Zhejiang | Pi2+Pi5 | MR |
| 23P-35 | Zhejiang | Pi5+Pia+Pid2+Pid3+Pita | MR |
| 23P-36 | Zhejiang | Pia+Pid2+Pid3 | MS |
| 23P-37 | Zhejiang | Pia | MR |
| 23P-38 | Zhejiang | Pi2+Pi5+Pia+Pita | MR |
| 23P-39 | Zhejiang | Pi5+Pia+Pid2+Pid3 | MR |
| 23P-40 | Guangdong | Pi2+Pi5+Pita | R |
| 23P-41 | Guangdong | Pia+Pid2+Pid3 | MR |
| 23P-42 | Guangdong | Pid3 | MS |
| 23P-43 | Guangdong | Pi5+Pia+Pid2+Pid3+Pita | MR |
| 23P-44 | Guangdong | Pi5+Pid2+Pid3 | MS |
| 23P-45 | Guangdong | Pi2+Pi5+Pia+Pid2+Pid3+Pita | MR |
| 23P-46 | Guangdong | Pi2+Pi5+Pia | R |
| 23P-47 | Guangdong | Pid2+Pid3+Pita | MS |
| 23P-48 | Guangdong | Pid2+Pid3+Pita | MS |
| 23P-49 | Guangdong | Pi2+Pi5+Pia | MR |
| 23P-50 | Guangdong | Pi5+Pid2+Pid3 | MR |
| 23P-51 | Guangdong | Pi5+Pia+Pid2+Pid3+Pita | MR |
| 23P-52 | Guangdong | Pia+Pid3+pikh | R |
| 23P-53 | Guangdong | Pia+Pid3+pikh | R |
| 23P-54 | Guangdong | Pi2+Pi5+Pid2+Pid3 | MR |
| 23P-55 | Guangdong | Pi2+Pi5+Pia+Pita | R |
| 23P-56 | Guangdong | Pia+Pid2+Pid3+Pita | MR |
| 23P-57 | Guangdong | Pi2+Pi5+Pita | R |
| 23P-58 | Guangdong | Pi5+Pia+Pid2+Pid3 | MR |
| 23P-59 | Guangdong | Pi5+Pid2+Pid3 | MR |
| 23P-60 | Guangdong | Pi5+Pid2+Pid3+Pita | MR |
| 23P-61 | Guangdong | Pi2+Pi5+Pid2+Pid3 | R |
| 23P-62 | Guangdong | Pi5+Pid2+Pid3+Pita | MR |
| 23P-63 | Guangdong | Pia+Pid2+Pid3+Pita | MR |
| 23P-64 | Guangdong | Pi2+Pita | MR |
| 23P-65 | Guangdong | Pi5+Pid2+Pid3 | MS |
| 23P-66 | Guangdong | Pi5+Pid2+Pid3+Pita | MS |
| 23P-67 | Guangdong | Pi5+Pia+Pid2+Pid3 | S |
| 23P-68 | Guangdong | Pid2+Pid3+Pita | MS |
| 23P-69 | Guangdong | Pi5+Pia+Pid2+Pid3+Pita | MR |
| 23P-70 | Guangdong | Pi2+Pi5+Pita | MR |
| 23P-71 | Guangdong | Pi2+Pi5+Pia+Pid2+Pid3+Pita | MR |
| 23P-72 | Guangdong | Pi2+Pi5+Pia+Pid2+Pid3+Pita | R |
| 23P-73 | Guangdong | Pi2+Pi5 | R |
| 23P-74 | Guangdong | Pid2+Pid3+Pita | R |
| 23P-75 | Guangdong | Pi2 | MS |
| 23P-76 | Guangdong | Pi5+Pid2+Pid3+Pita | MS |
| 23P-77 | Guangdong | Pi2+Pi5+Pid2+Pid3+Pita | MS |
| 23P-78 | Guangdong | Pi2+Pi5+Pita | R |
| 23P-79 | Guangdong | Pi5+Pid3 | MS |
| 23P-80 | Guangdong | Pi2+Pi5+Pia | MR |
| 23P-81 | Guangdong | Pi5+Pia+Pid2+Pid3+Pita | S |
| 23P-82 | Guangdong | Pia+Pid2+Pid3+Pita | S |
| 23P-83 | Guangdong | Pi2+Pi5+Pia+Pita | MS |
| 23P-84 | Guangdong | Pi5+Pia+Pid2+Pid3+Pita | MS |
| 23P-85 | Guangdong | Pi5+Pia+Pid2+Pid3 | S |
| 23P-86 | Guangdong | Pi5+Pia+Pid2+Pid3 | S |
| 23P-87 | Guangdong | Pi5+Pid3 | MS |
| 23P-88 | Guangdong | Pi2+Pita | MR |
| 23P-89 | Guangdong | Pi2+Pi5 | MR |
| 23P-90 | Guangdong | Pi2+Pita | MR |
| 23P-91 | Guangxi | Pid2+Pid3+Pita | MR |
| The Number of Resistance Genes | The Number of Varieties | Resistance Frequency (%) | Resistance Grade of Varieties | |||
|---|---|---|---|---|---|---|
| R | MR | MS | S | |||
| 1 | 5 | 40 | 0 | 2 | 3 | 0 |
| 2 | 17 | 58.82 | 2 | 8 | 7 | 0 |
| 3 | 32 | 62.5 | 10 | 10 | 12 | 0 |
| 4 | 21 | 66.68 | 3 | 11 | 3 | 4 |
| 5 | 12 | 75 | 3 | 6 | 1 | 1 |
| 6 | 4 | 100 | 1 | 3 | 0 | 0 |
| Gene Combination | Number of Varieties | Proportion (%) | Rice Blast Resistance Rate (%) |
|---|---|---|---|
| Pia+Pid2+Pid3 | 7 | 7.7 | 42.9 |
| Pia+Pid3 | 1 | 1.1 | 0 |
| Pid3+Pita | 1 | 1.1 | 100 |
| Pia+Pid2+Pid3+Pigm | 1 | 1.1 | 100 |
| Pia+Pid2+Pid3+Pigm+pikh | 1 | 1.1 | 100 |
| Pia+Pid3+pikh | 3 | 3.3 | 66.7 |
| Pi1+Pi5 | 1 | 1.1 | 0 |
| Pi2+Pid2+Pid3 | 2 | 2.2 | 100 |
| Pi2+Pi5+Pita | 6 | 6.6 | 83.3 |
| Pi5+Pia+Pid2+Pid3+Pita | 9 | 9.9 | 77.8 |
| Pia | 2 | 2.2 | 50 |
| Pi5+Pi9+Pid2+Pid3 | 1 | 1.1 | 100 |
| Pi5+Pi9+Pia+Pid2+Pid3 | 1 | 1.1 | 100 |
| Pi5 | 1 | 1.1 | 100 |
| Pi2+Pi5 | 5 | 5.5 | 80 |
| Pid2+Pid3 | 1 | 1.1 | 0 |
| Pi2+Pi5+Pia+Pid2+Pid3+Pita | 4 | 4.4 | 100 |
| Pia+Pigm | 1 | 1.1 | 100 |
| Pi5+Pia | 1 | 1.1 | 100 |
| Pi5+Pid2+Pid3+Pita | 5 | 5.5 | 60 |
| Pid2+Pid3+Pita | 6 | 6.6 | 50 |
| Pi5+Pid2+Pid3 | 5 | 5.5 | 40 |
| Pi5+Pid3 | 3 | 3.3 | 0 |
| Pi5+Pia+Pid2+Pid3 | 6 | 6.6 | 50 |
| Pi2+Pi5+Pia+Pita | 3 | 3.3 | 66.7 |
| Pid3 | 1 | 1.1 | 0 |
| Pi2+Pi5+Pia | 3 | 3.3 | 100 |
| Pi2+Pi5+Pid2+Pid3 | 2 | 2.2 | 100 |
| Pia+Pid2+Pid3+Pita | 3 | 3.3 | 33.3 |
| Pi2+Pita | 3 | 3.3 | 100 |
| Pi2 | 1 | 1.1 | 0 |
| Pi2+Pi5+Pid2+Pid3+Pita | 1 | 1.1 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, L.; Song, J.; Cui, Y.; Fan, H.; Wang, J. Detection and Evaluation of Blast Resistance Genes in Backbone Indica Rice Varieties from South China. Plants 2024, 13, 2134. https://doi.org/10.3390/plants13152134
Tang L, Song J, Cui Y, Fan H, Wang J. Detection and Evaluation of Blast Resistance Genes in Backbone Indica Rice Varieties from South China. Plants. 2024; 13(15):2134. https://doi.org/10.3390/plants13152134
Chicago/Turabian StyleTang, Liqun, Jian Song, Yongtao Cui, Honghuan Fan, and Jianjun Wang. 2024. "Detection and Evaluation of Blast Resistance Genes in Backbone Indica Rice Varieties from South China" Plants 13, no. 15: 2134. https://doi.org/10.3390/plants13152134
APA StyleTang, L., Song, J., Cui, Y., Fan, H., & Wang, J. (2024). Detection and Evaluation of Blast Resistance Genes in Backbone Indica Rice Varieties from South China. Plants, 13(15), 2134. https://doi.org/10.3390/plants13152134





