Plants Restoration Drives the Gobi Soil Microbial Diversity for Improving Soil Quality
Abstract
1. Introduction
2. Results
2.1. Physicochemical Properties
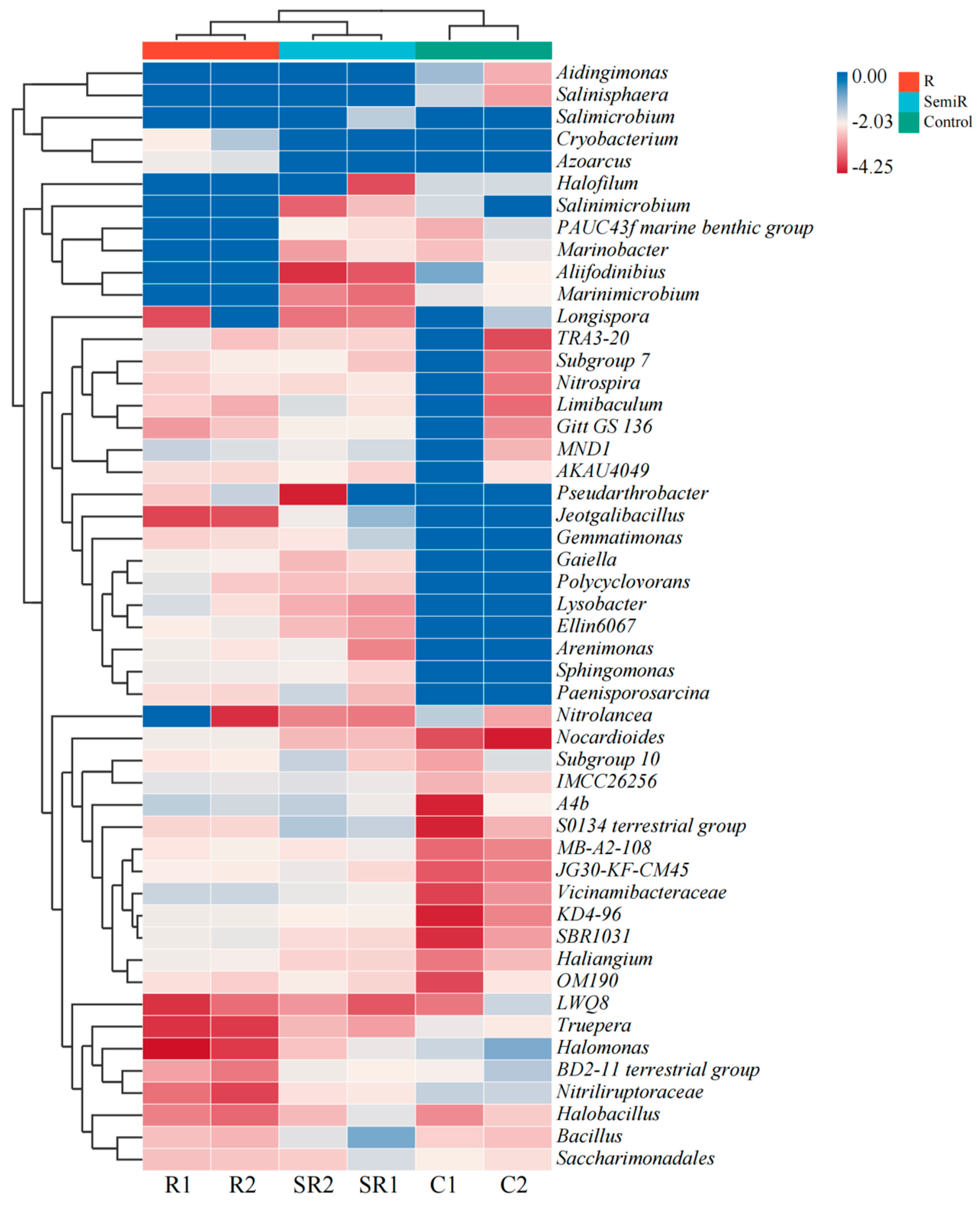
2.2. Bacterial Diversity
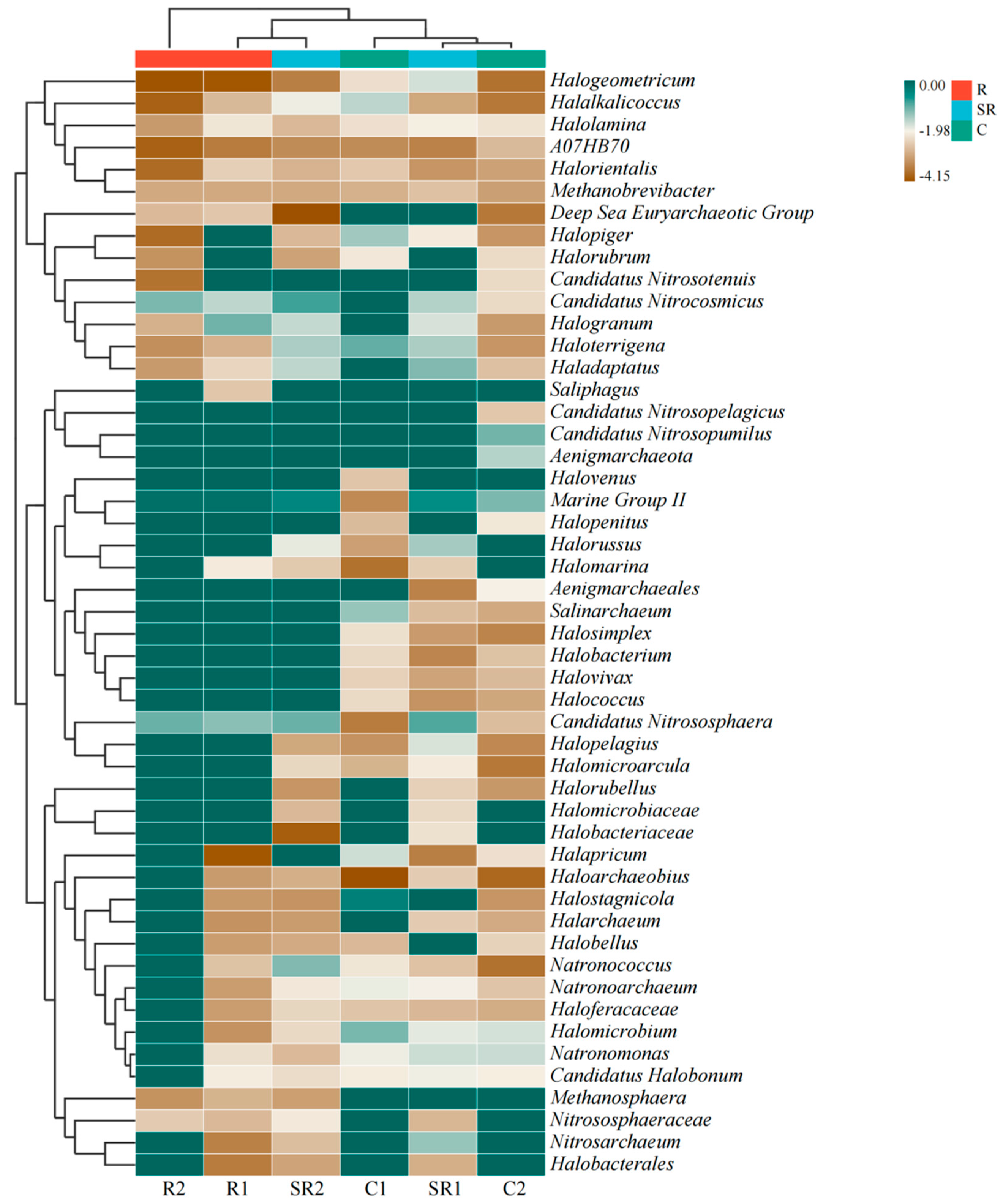
2.3. Archaeal Diversity
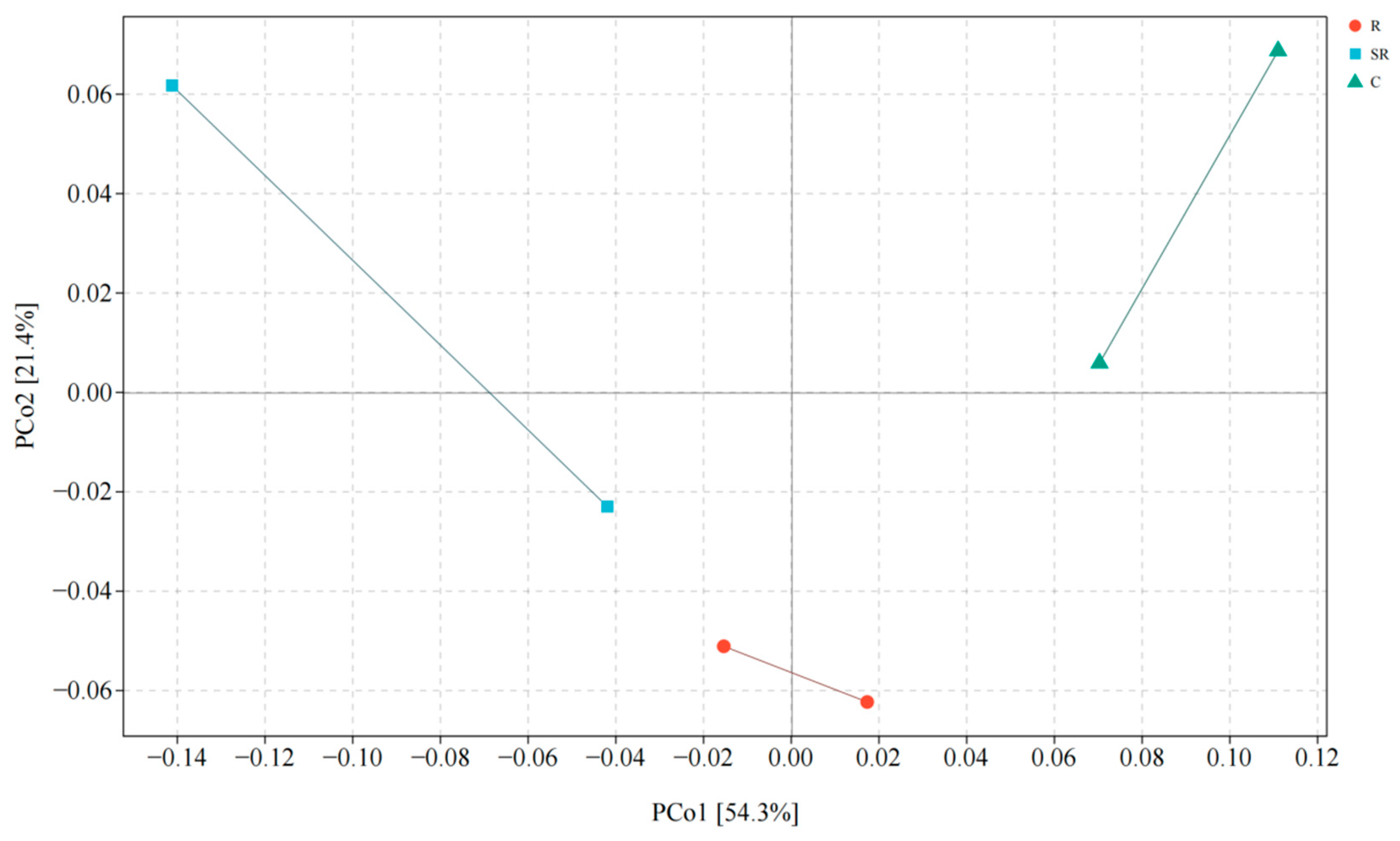
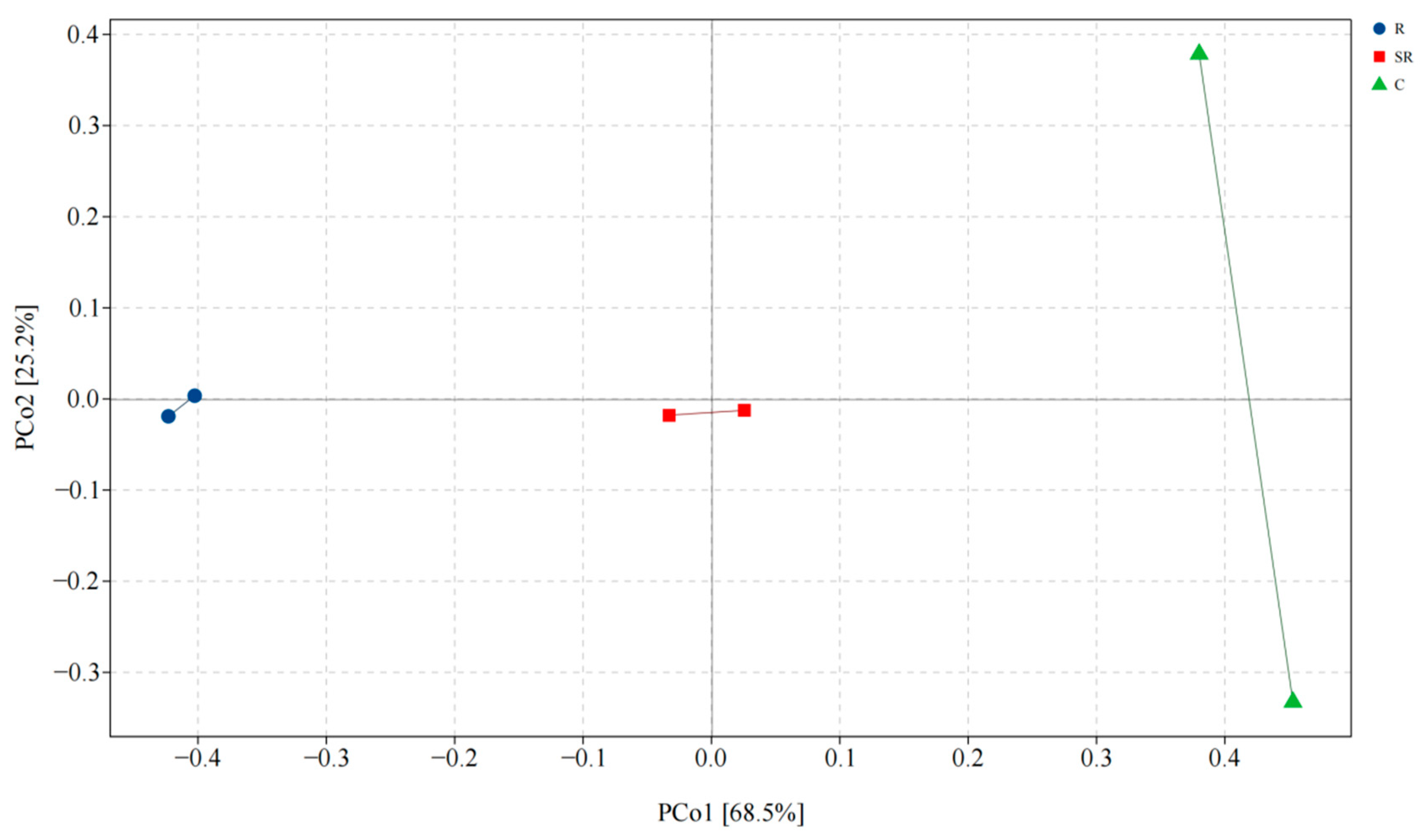
2.4. Principal Coordinate Analysis for Bacteria and Archaea
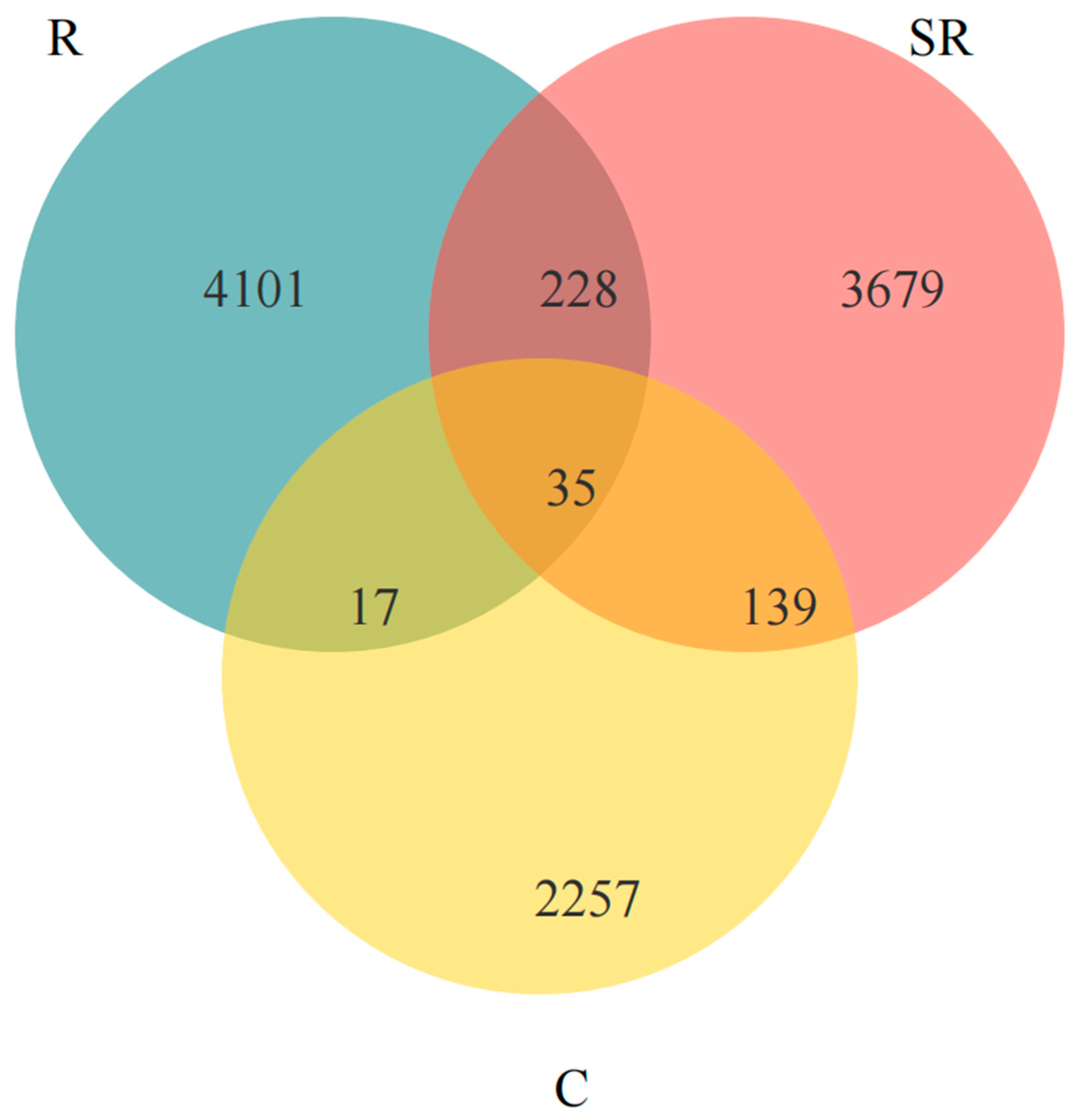
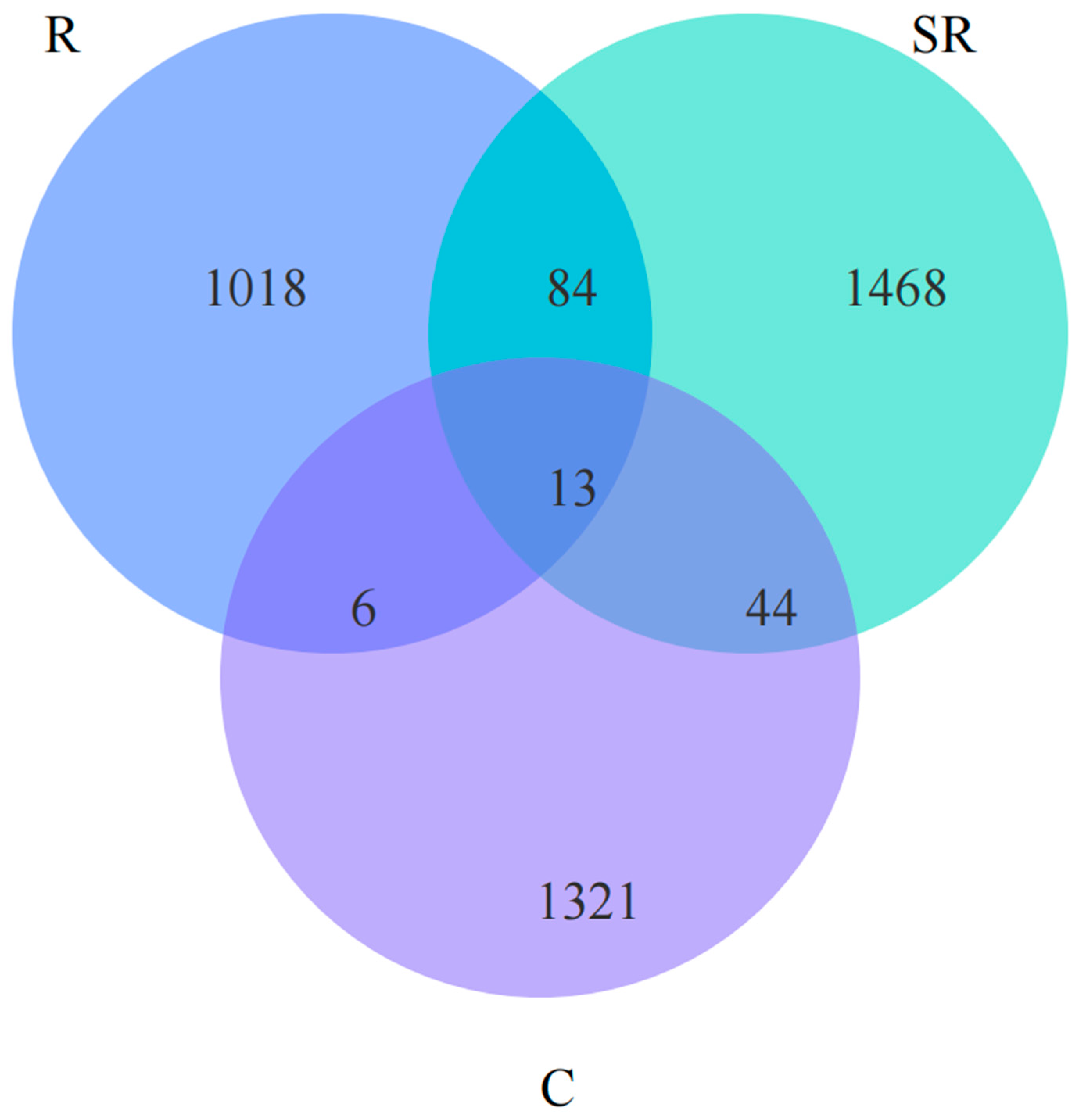
2.5. Venn Analysis of Seasonal and Rooting Conditions
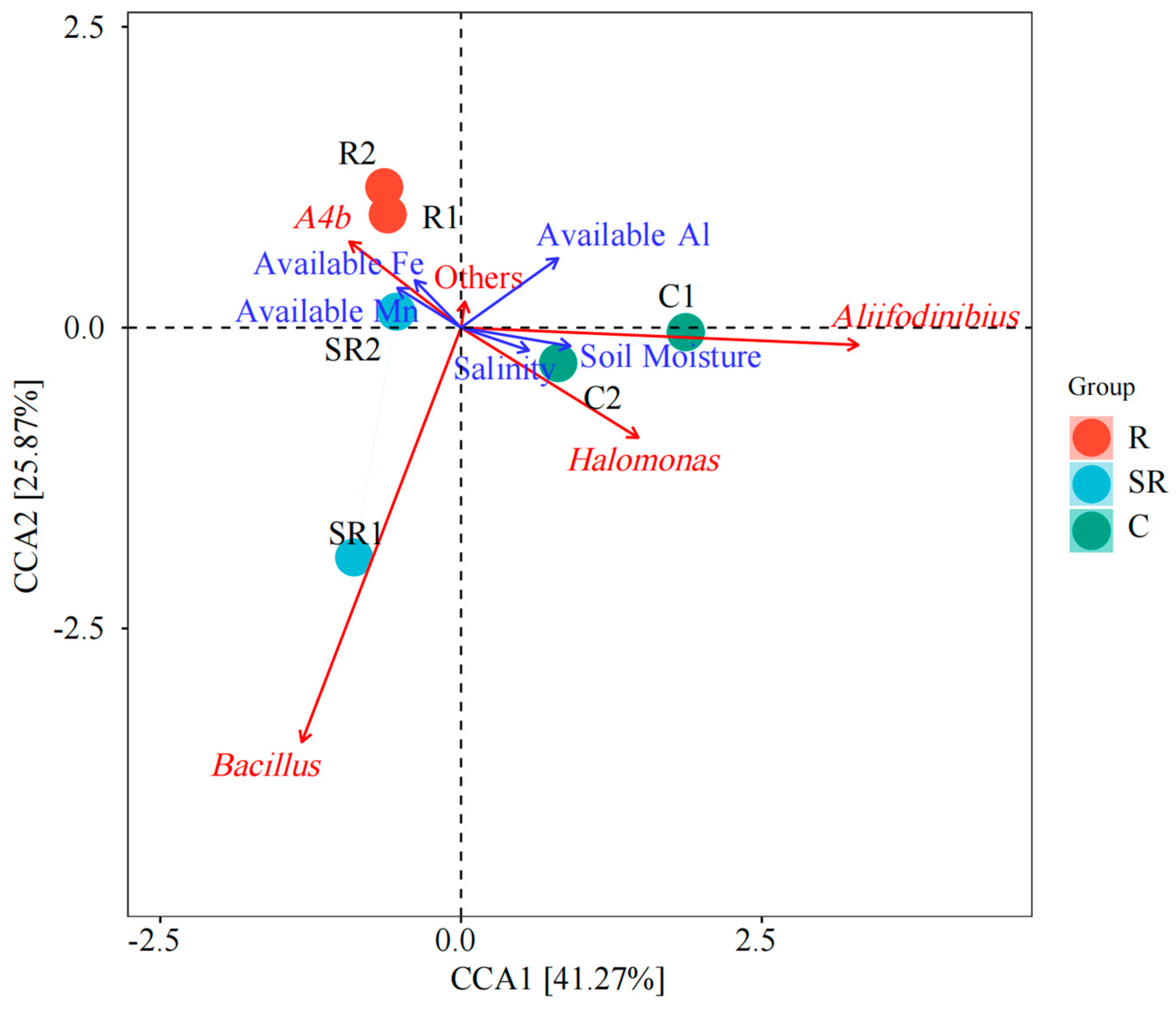
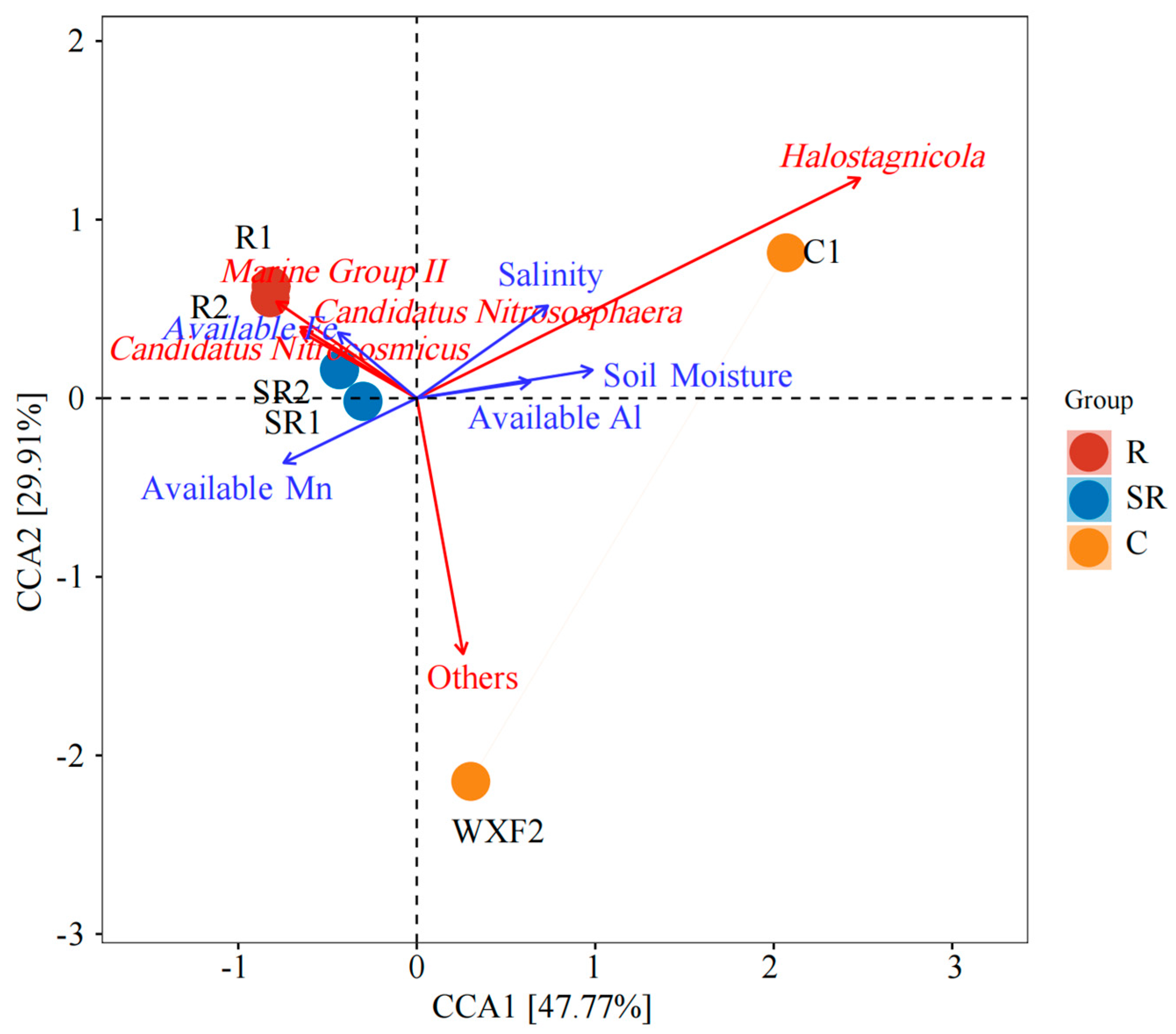
2.6. Canonical Correlation Analysis of Bacteria and Archaea
3. Discussion
4. Materials and Methods
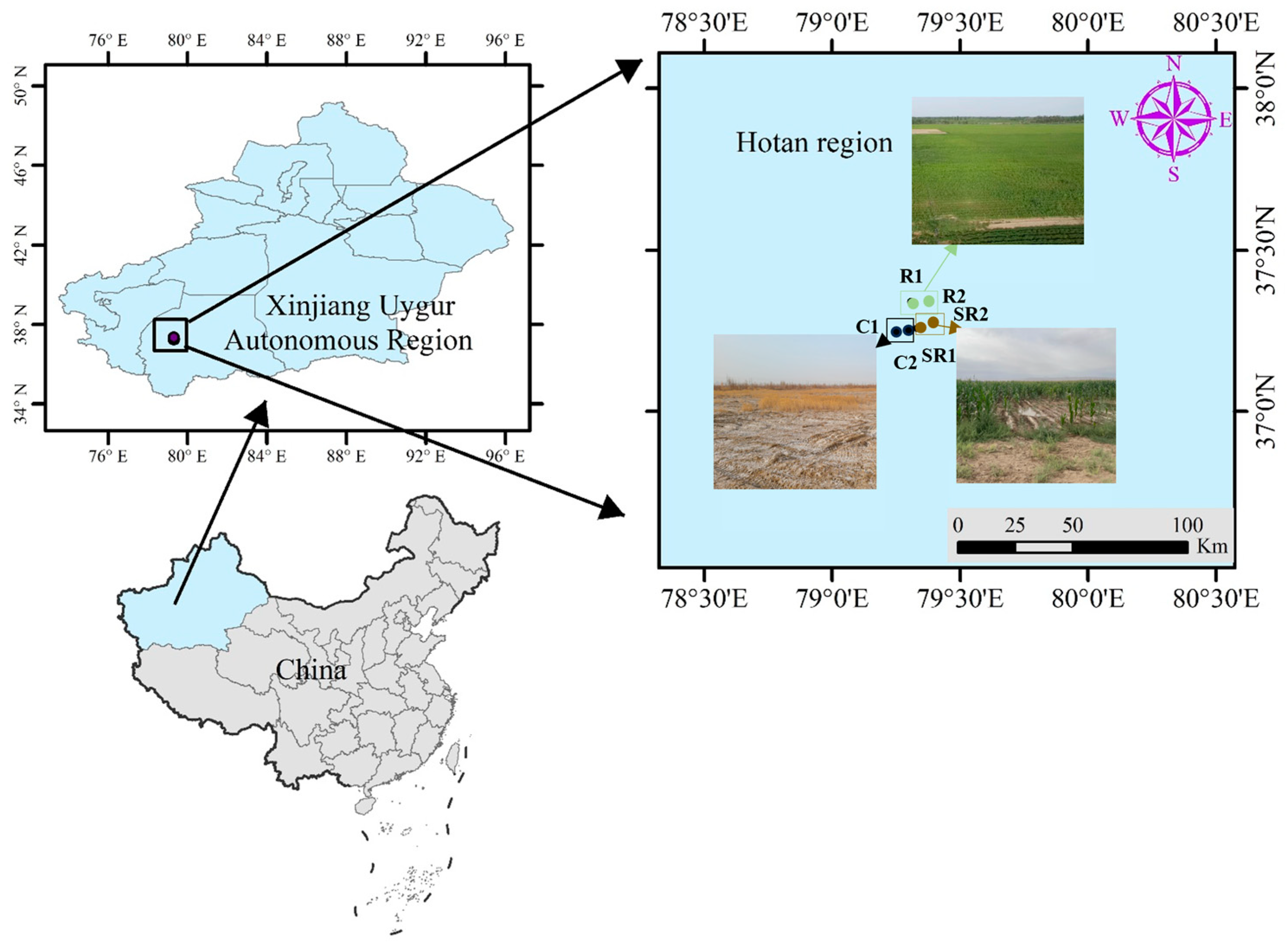
4.1. Study Area
4.2. Samples
4.3. Determination of Properties of the Samples
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, B.; Xia, H.; Chaoqiang, J.; Cuncang, J.; Muhammad, R.; Li, Y.; Yunfeng, C.; Xianpeng, F.; Zhiyi, Z.; Xiaoli, D. Straw Addition Enhances Crop Yield, Soil Aggregation, and Soil Microorganisms in a 14-Year Wheat-Rice Rotation System in Central China. Plants 2024, 13, 985. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Fu, B.; Wang, S.; Stringer, L.C.; Wang, Y.; Li, Z.; Yanxu, L.; Wenxin, Z. Drivers and Impacts of Changes in China Drylands. Nat. Rev. Earth Environ. 2021, 2, 858–873. [Google Scholar] [CrossRef]
- Abdelrahman, M.A.E. An Overview of Land Degradation, Desertification and Sustainable Land Management Using Gis and Remote Sensing Applications. Rend. Lincei-Sci. Fis. 2023, 34, 767–808. [Google Scholar] [CrossRef]
- Wu, S.; Liu, L.; Li, D.; Zhang, W.; Liu, K.; Shen, J.; Zhang, L. Global Desert Expansion During the 21st Century: Patterns, Predictors and Signals. Land. Degrad. Dev. 2023, 34, 377–388. [Google Scholar] [CrossRef]
- Jonathan, M.C.; Shane, A.B.; Tiffany, M.K.; Katharina, G.; Felix, M. Ecosystem Decay Exacerbates Biodiversity Loss with Habitat Loss. Nature 2020, 584, 238–243. [Google Scholar]
- Cristina, B.L.; Robert, M.E.; Hollie, F.T.; Adam, F. Countering the Effects of Habitat Loss, Fragmentation, and Degradation Through Habitat Restoration. One Earth 2020, 3, 672–676. [Google Scholar]
- Xu, K.; He, L.; Hu, H.; Liu, S.; Du, Y.; Wang, Z.; Li, Y.; Li, L.; Khan, A.; Wang, G. Positive Ecological Effects of Wind Farms on Vegetation in China Gobi Desert. Sci. Rep. 2019, 9, 6341. [Google Scholar]
- Aihong, F.; Weihong, L.; Yaning, C.; Yi, W.; Haichao, H.; Yupeng, L.; Fan, S.; Honghua, Z.; Chenggang, Z.; Xingming, H. The Effects of Ecological restoration Projects on the Resilience of an Extremely Drought-Prone Desert Riparian Forest Ecosystem in the Tarim River Basin, Xinjiang, China. Sci. Rep. 2021, 11, 18485. [Google Scholar]
- Xue, P.P.; Carrillo, Y.; Pino, V.; Minasny, B.; McBratney, A.B. Soil Properties Drive Microbial Community Structure in a Large Scale Transect in South Eastern Australia. Sci. Rep. 2018, 8, 11725. [Google Scholar] [CrossRef]
- Liao, J.; Dou, Y.; Yang, X.; An, S. Soil Microbial Community and Their Functional Genes During Grassland Restoration. J. Environ. Manag. 2022, 325 Pt A, 116488. [Google Scholar] [CrossRef]
- Cao, S. Impact of China Large-Scale Ecological Restoration Program on the Environment and Society in Arid and Semiarid Areas of China: Achievements, Problems, Synthesis, and Applications. Crit. Rev. Environ. Sci. Technol. 2011, 41, 317–335. [Google Scholar] [CrossRef]
- Han, C.; Liang, D.; Zhou, W.; Xu, Q.; Xiang, M.; Gu, Y.; Siddique, K.H. Soil, Plant, and Microorganism Interactions Drive Secondary Succession in Alpine Grassland Restoration. Plants 2024, 13, 780. [Google Scholar] [CrossRef]
- Xia, H.; Shen, J.; Riaz, M.; Jiang, C.; Zu, C.; Jiang, C.; Liu, B. Effects of Biochar and Straw Amendment on Soil Fertility and Microbial Communities in Paddy Soils. Plants 2024, 13, 1478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Dong, J.; Xiao, X.; Hu, Z.; Sheldon, S. Effectiveness of Ecological Restoration Projects in Horqin Sandy Land, China Based on Spot-Vgt Ndvi Data. Ecol. Eng. 2012, 38, 20–29. [Google Scholar] [CrossRef]
- Masciandaro, G.; Macci, C.; Peruzzi, E.; Ceccanti, B.; Doni, S. Organic Matter–Microorganism–Plant in Soil Biorestoration: A Synergic Approach. Rev. Environ. Sci. Bio 2013, 12, 399–419. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, K. Effect of Intertidal Vegetation (Suaeda salsa) Restoration on Microbial Diversity in the Offshore Areas of the Yellow River Delta. Plants 2024, 13, 213. [Google Scholar] [CrossRef] [PubMed]
- Hoffland, E.; Kuyper, T.W.; Comans, R.N.; Creamer, R.E. Eco-Functionality of Organic Matter in Soils. Plant Soil 2020, 455, 1–22. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiao, Y.; Fu, Z. Shifts of Bacterial Community and Predictive Functional Profiling of Denitrifying Phosphorus Removal–Partial Nitrification–Anammox Three-Stage Nitrogen and Phosphorus Removal Before and After Coupling for Treating Simulated Wastewater with Low C/N. Chem. Eng. J. 2023, 451, 138601. [Google Scholar] [CrossRef]
- Yu, M.; Su, W.Q.; Huang, L.; Parikh, S.J.; Tang, C.; Dahlgren, R.A.; Xu, J. Bacterial Community Structure and Putative Nitrogen-Cycling Functional Traits Along a Charosphere Gradient under Waterlogged Conditions. Soil Boil. Biochem. 2021, 162, 108420. [Google Scholar] [CrossRef]
- Huber, K.J.; Overmann, J. Vicinamibacteraceae Fam. Nov., the First Described Family within the Subdivision 6 Acidobacteria. Int. J. Syst. Evol. Microbiol. 2018, 68, 2331–2334. [Google Scholar] [CrossRef]
- Qian, G.L.; Hu, B.S.; Jiang, Y.H.; Liu, F.Q. Identification and Characterization of Lysobacter Enzymogenes as a Biological Control Agent Against Some Fungal Pathogens. Agric. Sci. China 2009, 8, 68–75. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, L.; Zeng, Y.; Xing, T.; Xu, B.; Wang, N. Genomic Insights of Cryobacterium Isolated from Ice Core Reveal Genome Dynamics for Adaptation in Glacier. Front. Microbiol. 2020, 11, 1530. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Peng, W.; Yin, X.; Wang, X.; Liu, Z.; Liu, Q.; Deng, Z.; Lin, S.; Liang, R. Identification of an Efficient Phenanthrene-Degrading Pseudarthrobacter sp. l1sw and Characterization of Its Metabolites and Catabolic Pathway. J. Hazard. Mater. 2024, 465, 133138. [Google Scholar] [CrossRef] [PubMed]
- Celandroni, F.; Vecchione, A.; Cara, A.; Mazzantini, D.; Lupetti, A.; Ghelardi, E. Identification of Bacillus Species: Implication on the Quality of Probiotic Formulations. PLoS ONE 2019, 14, e0217021. [Google Scholar] [CrossRef] [PubMed]
- Yaakop, A.S.; Chan, K.G.; Ee, R.; Lim, Y.L.; Lee, S.K.; Manan, F.A.; Goh, K.M. Characterization of the Mechanism of Prolonged Adaptation to Osmotic Stress of Jeotgalibacillus Malaysiensis Via Genome and Transcriptome Sequencing Analyses. Sci. Rep. 2016, 6, 33660. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Qiu, L.; Ishii, S.; Jia, X.; Su, F.; Song, Y.; Hao, M.; Shao, M.; Wei, X. Contrasting Response of Soil Microbiomes to Long-Term Fertilization in Various Highland Cropping Systems. ISME Commun. 2023, 3, 81. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Luo, Y.; Tang, C.; Li, Y.; Guggenberger, G.; Xu, J. Succession of the Soil Bacterial Community as Resource Utilization Shifts from Plant Residues to Rhizodeposits. Soil Boil. Biochem. 2022, 173, 108785. [Google Scholar] [CrossRef]
- Zheng, X.; Lin, C.; Guo, B.; Yu, J.; Ding, H.; Peng, S.; Sveen, T.R.; Zhang, Y. Effects of Re-Vegetation Restoration on Soil Bacterial Community Structure in Degraded Land in Subtropical China. Eur. J. Soil Biol. 2020, 98, 103184. [Google Scholar] [CrossRef]
- Kim, K.K.; Lee, J.S.; Stevens, D.A. Microbiology and Epidemiology of Halomonas Species. Future Microbiol. 2013, 8, 1559–1573. [Google Scholar] [CrossRef]
- Shiratori-Takano, H.; Yamada, K.; Beppu, T.; Ueda, K. Longispora Fulva sp. nov., Isolated from a Forest Soil, and Emended Description of the Genus Longispora. Int. J. Syst. Evol. Microbiol. 2011, 61, 804–809. [Google Scholar] [CrossRef]
- Rinke, C.; Rubino, F.; Messer, L.F.; Youssef, N.; Parks, D.H.; Chuvochina, M.; Brown, M.; Jeffries, T.; Tyson, G.W.; Seymour, J.R.; et al. A Phylogenomic and Ecological Analysis of the Globally Abundant Marine Group II Archaea (Ca. Poseidoniales Ord. Nov.). ISME J. 2019, 13, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.L.; Xie, W.; Martin-Cuadrado, A.B.; Rodriguez-Valera, F. Marine group II archaea, potentially important players in the global ocean carbon cycle. Front. Microbiol. 2015, 6, 1108. [Google Scholar] [CrossRef] [PubMed]
- Sauder, L.A.; Albertsen, M.; Engel, K.; Schwarz, J.; Nielsen, P.H.; Wagner, M.; Neufeld, J.D. Cultivation and Characterization of Candidatus Nitrosocosmicus Exaquare, an Ammonia-Oxidizing Archaeon from a Municipal Wastewater Treatment System. ISME J. 2017, 11, 1142–1157. [Google Scholar] [CrossRef] [PubMed]
- Zhalnina, K.V.; Dias, R.; Leonard, M.T.; Dorr de Quadros, P.; Camargo, F.A.; Drew, J.C.; Farmerie, W.G.; Daroub, S.H.; Triplett, E.W. Genome sequence of candidatus nitrososphaera evergladensis from group i. 1b enriched from everglades soil reveals novel genomic features of the ammonia-oxidizing archaea. PLoS ONE 2014, 9, e101648. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.J.; Tao, C.Q.; Zeng, C.L.; Zhu, L.; Cui, H.L. Salinigranum Halophilum sp. nov., Isolated From Marine Solar Salterns. Int. J. Syst. Evol. Microbiol. 2020, 70, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Kanekar, P.; Kulkarni, S.; Jagtap, C.; Kadam, V.; Pathan, H. A Novel Approach for the Development of Bio-Sensitized Solar Cell Using Cell Lysate of a Haloarchaeon Halostagnicola Larsenii Rg2. 14 (Mcc 2809) Containing Bacteriorhodopsin. Sol. Energy 2020, 212, 326–331. [Google Scholar] [CrossRef]
- Flores, N.; Hoyos, S.; Venegas, M.; Galetović, A.; Zúñiga, L.M.; Fábrega, F.; Paredes, B.; Salazar-Ardiles, C.; Vilo, C.; Ascaso, C.; et al. Haloterrigena sp. Strain SGH1, a Bacterioruberin-Rich, Perchlorate-tolerant Halophilic Archaeon Isolated from Halite Microbial Communities, Atacama Desert, Chile. Front. Microbiol. 2020, 11, 324. [Google Scholar]
- Hu, X.; Zeng, C.; Hou, Z.; Wang, Y.; Xu, Q.; Isobe, K.; Senoo, K.; Zhu, L. The Complete Genome Sequence of the Archaeal Isolate Halomicrobium Sp. Zps1 Reveals the Nitrogen Metabolism Characteristics under Hypersaline Conditions. Ann. Microbiol. 2020, 70, 29. [Google Scholar] [CrossRef]
- Qin, W.; Martens-Habbena, W.; Kobelt, J.N.; Stahl, D.A. Candidatus Nitrosopumilus. In Bergey Manual of Systematics of Archaea and Bacteria; 2016; Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/9781118960608.gbm01290 (accessed on 1 June 2024).
- Baker, B.J.; De Anda, V.; Seitz, K.W.; Dombrowski, N.; Santoro, A.E.; Lloyd, K.G. Diversity, Ecology and Evolution of Archaea. Nat. Microbiol. 2020, 5, 887–900. [Google Scholar] [CrossRef]
- Qiao, S.; Shi, X.; Wang, G.; Yang, G.; Hu, N.; Liu, S.; Liu, Y.; Zhu, A.; Li, C. Discussion on Grain-Size Characteristics of Seafloor Sediment and Transport Pattern in the Bohai Sea. Acta Oceanol. Sin. 2010, 32, 139–147. [Google Scholar]
- Hu, M.; Sardans, J.; Sun, D.; Yan, R.; Wu, H.; Ni, R.; Peñuelas, J. Microbial Diversity and Keystone Species Drive Soil Nutrient Cycling and Multifunctionality Following Mangrove Restoration. Environ. Res. 2024, 251, 118715. [Google Scholar] [CrossRef]
- Tziouvalekas, M.; Noulas, C.; Thalassinos, G.; Shaheen, S.M.; Rinklebe, J.; Antoniadis, V. Temperature-induced changes in dtpa-extractable trace elements: Predicting the potential impact of climate change on the availability of soil elements. Chemosphere 2024, 350, 141064. [Google Scholar] [CrossRef]
- Yang, B.; Gao, X.; Xing, Q. Geochemistry of Organic Carbon in Surface Sediments of a Summer Hypoxic Region in the Coastal Waters of Northern Shandong Peninsula. Cont. Shelf. Res. 2018, 171, 113–125. [Google Scholar] [CrossRef]
- Zheng, S.; Wang, B.; Xu, G.; Liu, F. Effects of Organic Phosphorus on Methylotrophic Methanogenesis in Coastal Lagoon Sediments with Seagrass (Zostera marina) Colonization. Front. Microbiol. 2020, 11, 1770. [Google Scholar] [CrossRef]
- Caporaso, J.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.; Costello, E.; Fierer, N.; Peña, A.; Goodrich, J.; Gordon, J.; et al. Qiime Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of Rrna Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
| pH | Salinity | Water Content, % | Effective Fe, mg/kg | Effective Mn, mg/kg | Effective Al, mg/kg | Effective Cu, mg/kg | Effective Zn, mg/kg | |
|---|---|---|---|---|---|---|---|---|
| R1 | 8.14 | 0.24 | 22.32 | 15.18 | 3.50 | 1.10 | 0.54 | 0.83 |
| R2 | 8.04 | 0.25 | 12.27 | 10.12 | 3.28 | 0.92 | 0.51 | 0.43 |
| SR1 | 8.18 | 1.31 | 12.66 | 8.63 | 1.97 | 0.32 | 0.45 | 0.37 |
| SR2 | 7.96 | 0.49 | 12.60 | 9.60 | 3.15 | 0.40 | 0.44 | 0.33 |
| C1 | 8.43 | 9.23 | 11.60 | 3.76 | 4.46 | 0.56 | 0.20 | 0.45 |
| C2 | 7.98 | 2.26 | 10.98 | 12.74 | 3.60 | 0.08 | 0.35 | 0.33 |
| Soil Particle Size | ||||||||
| 0–4 μm | 4–63 μm | >63 μm | TOM% | TON% | TOC% | |||
| R1 | 3.71 | 35.23 | 61.06 | 1.47 | 0.01 | 2.04 | ||
| R2 | 2.77 | 30.21 | 67.02 | 1.31 | 0.02 | 1.99 | ||
| SR1 | 4.11 | 37.94 | 57.95 | 3.17 | 0.03 | 2.12 | ||
| SR2 | 2.93 | 33.77 | 63.30 | 2.49 | 0.04 | 2.32 | ||
| C1 | 0.68 | 26.45 | 72.87 | 3.86 | 0.06 | 2.27 | ||
| C2 | 2.06 | 33.49 | 64.45 | 5.20 | 0.03 | 2.45 | ||
| R a | P b | |
|---|---|---|
| Salinity | 0.81 | 0.15 |
| Soil Moisture | 0.99 | 0.01 |
| Available Fe | 0.33 | 0.35 |
| Available Mn | 0.69 | 0.20 |
| Available Al | 0.41 | 0.52 |
| Available Cu | 0.66 | 0.14 |
| Available Zn | 0.99 | 0.01 |
| Grain Size 0–4 μm | 0.27 | 0.43 |
| Grain Size 4–63 μm | 0.74 | 0.16 |
| Grain Size > 63 μm | 0.54 | 0.35 |
| TOM | 0.58 | 0.33 |
| TON | 0.96 | 0.00 |
| TOC | 0.84 | 0.10 |
| R a | P b | |
|---|---|---|
| Salinity | 0.35 | 0.53 |
| Soil Moisture | 0.85 | 0.12 |
| Available Fe | 0.30 | 0.51 |
| Available Mn | 0.39 | 0.48 |
| Available Al | 0.98 | 0.00 |
| Available Cu | 0.61 | 0.25 |
| Available Zn | 0.96 | 0.01 |
| Grain Size 0–4 μm | 0.27 | 0.57 |
| Grain Size 4–63 μm | 0.92 | 0.02 |
| Grain Size > 63 μm | 0.77 | 0.12 |
| TOM | 0.81 | 0.08 |
| TON | 0.76 | 0.12 |
| TOC | 0.73 | 0.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Ma, J.; Wu, Q.; Hu, Y.; Feng, J. Plants Restoration Drives the Gobi Soil Microbial Diversity for Improving Soil Quality. Plants 2024, 13, 2159. https://doi.org/10.3390/plants13152159
Wang L, Ma J, Wu Q, Hu Y, Feng J. Plants Restoration Drives the Gobi Soil Microbial Diversity for Improving Soil Quality. Plants. 2024; 13(15):2159. https://doi.org/10.3390/plants13152159
Chicago/Turabian StyleWang, Lizhi, Junyong Ma, Qifeng Wu, Yongchao Hu, and Jinxiao Feng. 2024. "Plants Restoration Drives the Gobi Soil Microbial Diversity for Improving Soil Quality" Plants 13, no. 15: 2159. https://doi.org/10.3390/plants13152159
APA StyleWang, L., Ma, J., Wu, Q., Hu, Y., & Feng, J. (2024). Plants Restoration Drives the Gobi Soil Microbial Diversity for Improving Soil Quality. Plants, 13(15), 2159. https://doi.org/10.3390/plants13152159






