CIRCADIAN CLOCK-ASSOCIATED1 Delays Flowering by Directly Inhibiting the Transcription of BcSOC1 in Pak-choi
Abstract
1. Introduction
2. Results
2.1. Identification of BcCCA1
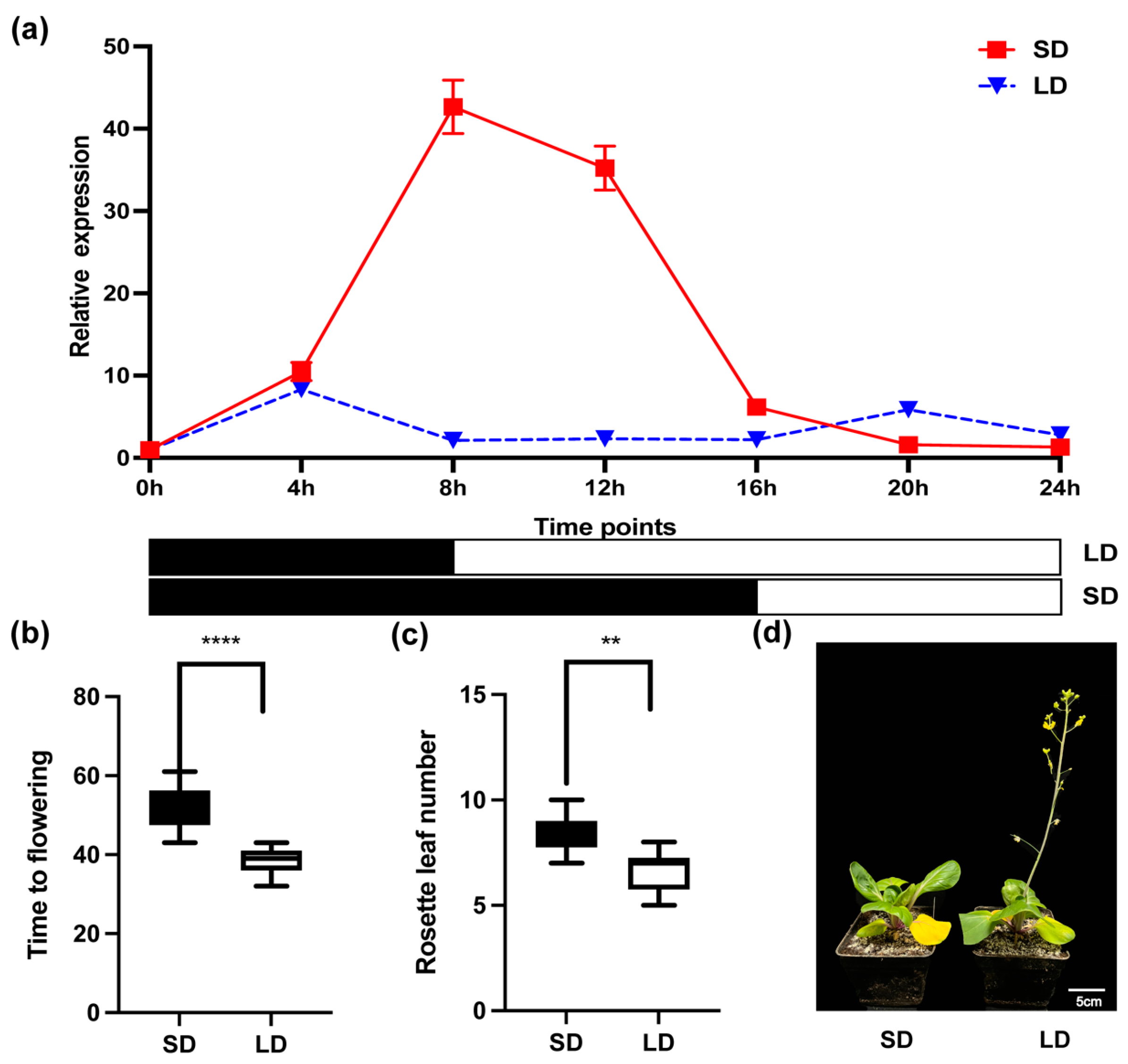
2.2. The Expression Patterns of BcCCA1
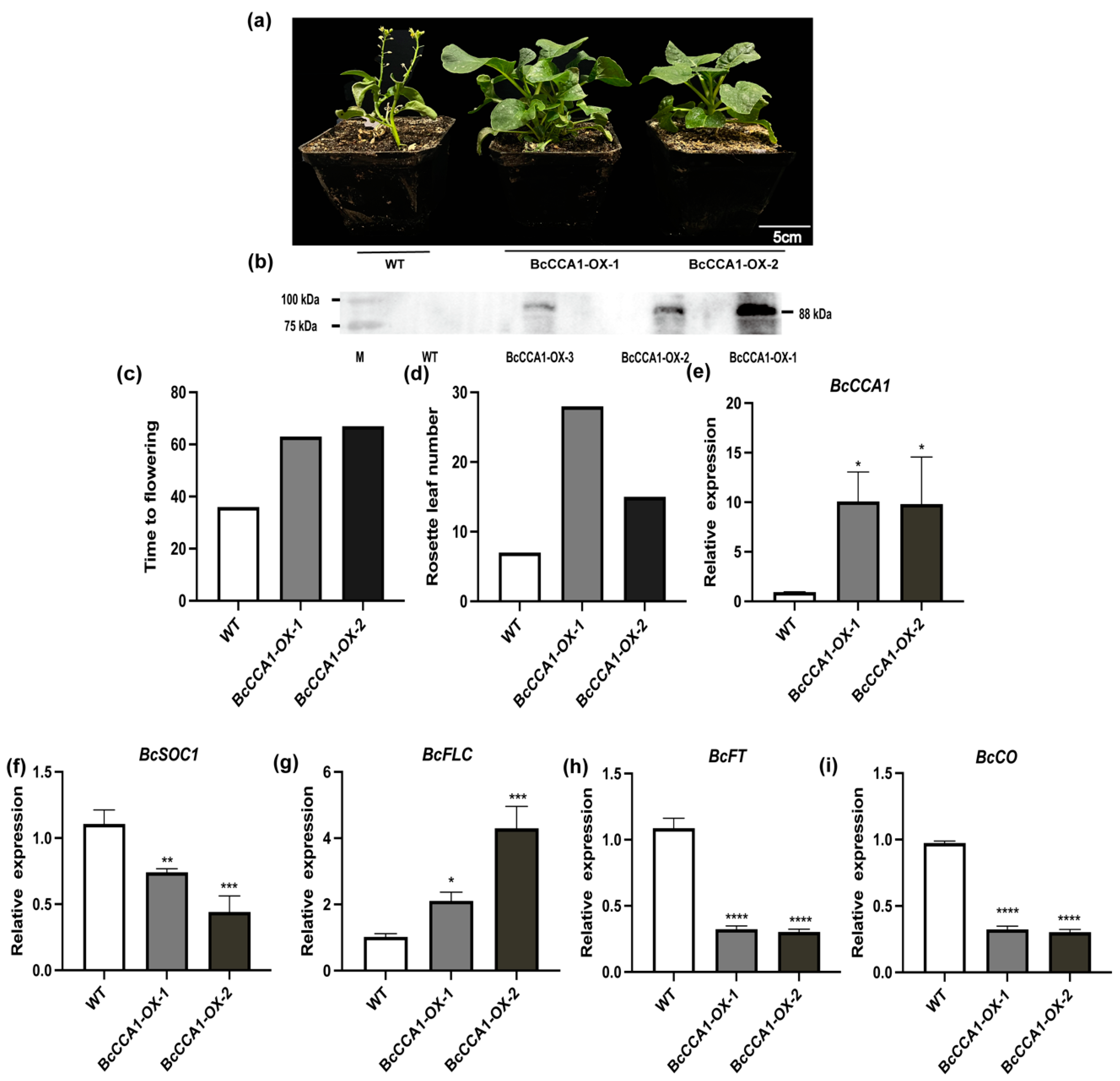
2.3. Analysis of BcCCA1-Overexpressed Plants
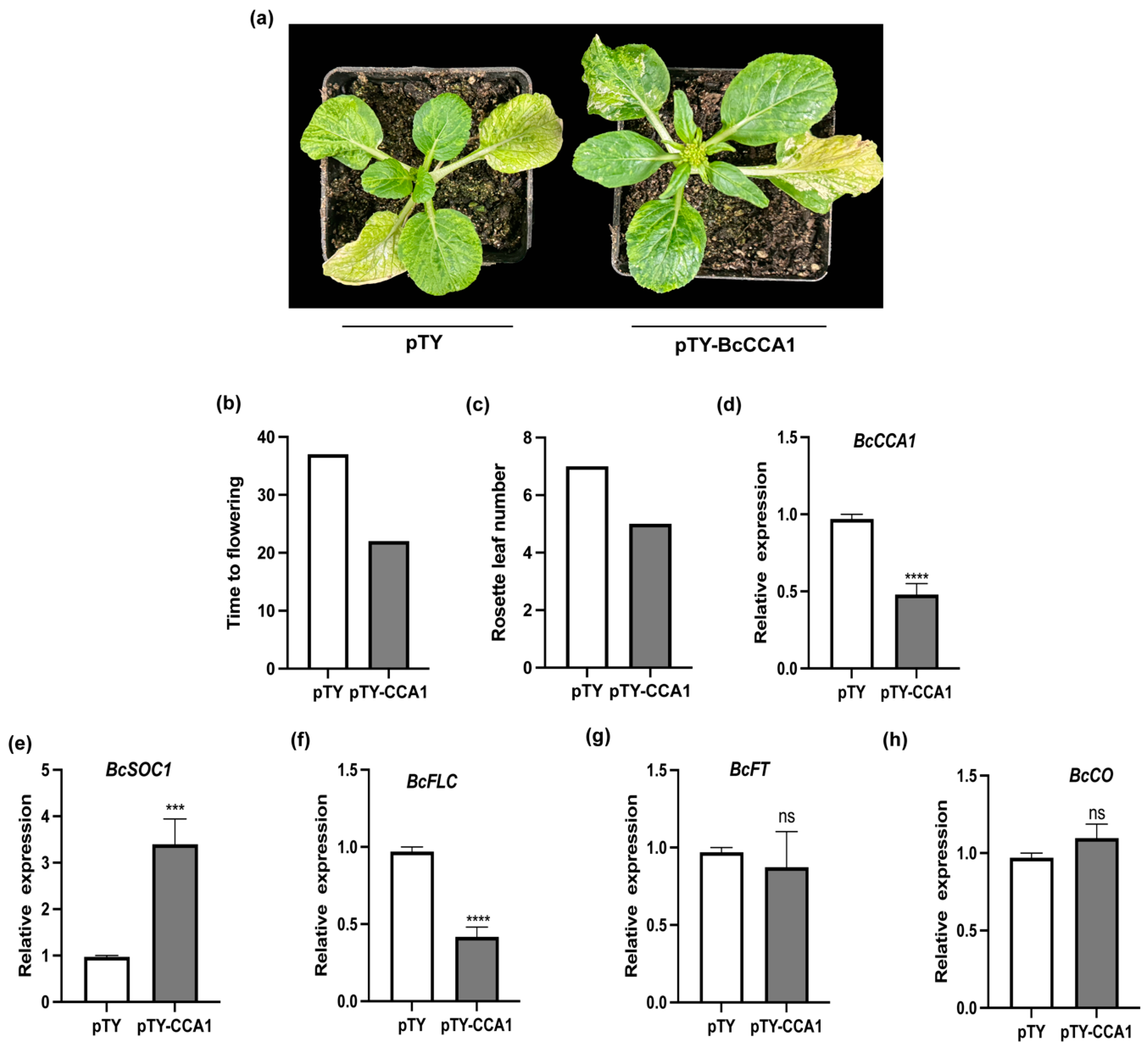
2.4. Analysis of BcCCA1-Silenced Plants
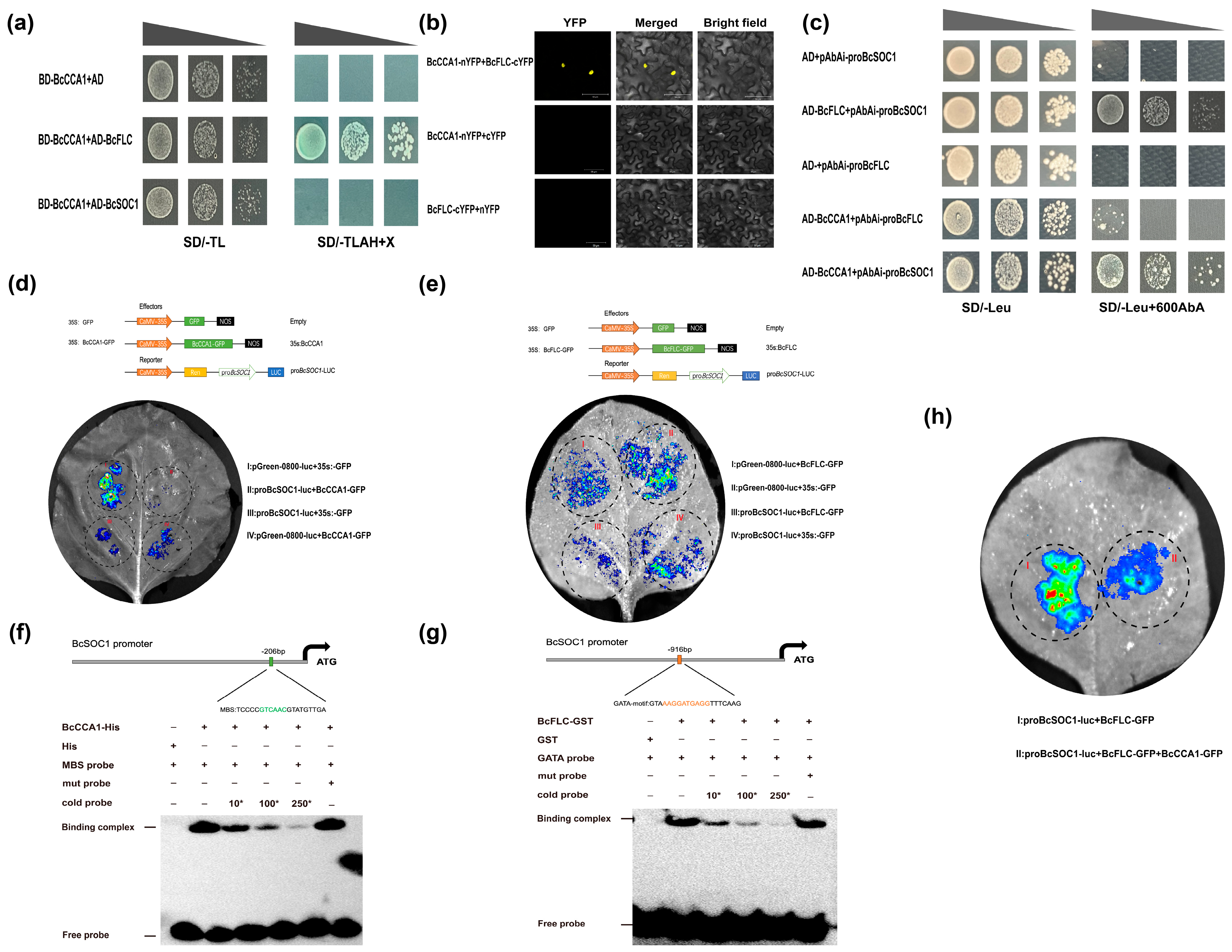
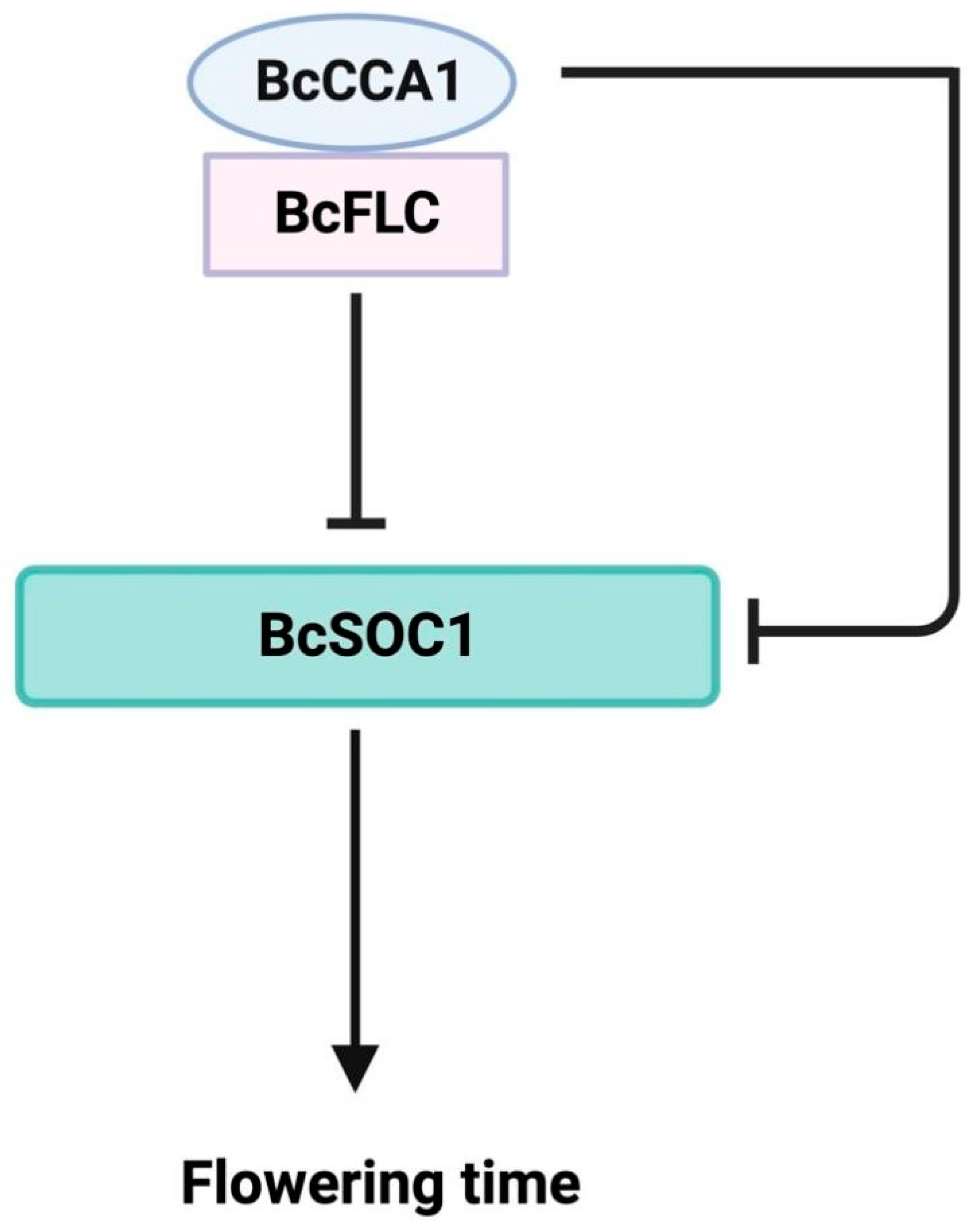
2.5. The BcCCA1-BcFLC Complex Can Repress the Expression of BcSOC1
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Cloning and Analysis of BcCCA1
4.3. Subcellular Localization of BcCCA1 in Tobacco
4.4. Vector Construction and Transgenic Plant Generation
4.5. Silencing BcCCA1 Expression by the VIGS System
4.6. mRNA Extraction and Quantitative Real-Time PCR
4.7. Yeast Two-Hybrid Assay
4.8. Bimolecular Fluorescence Complementation (BiFC) Assay
4.9. Yeast One-Hybrid Assay
4.10. Dual-Luciferase Assay
4.11. Electrophoretic Mobility Shift Assay
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Osnato, M.; Cota, I.; Nebhnani, P.; Cereijo, U.; Pelaz, S. Photoperiod Control of Plant Growth: Flowering Time Genes Beyond Flowering. Front. Plant Sci. 2021, 12, 805635. [Google Scholar] [CrossRef]
- Wijnen, H.; Young, M.W. Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 2006, 40, 409–448. [Google Scholar] [CrossRef]
- Hughes, C.L.; Harmer, S.L. Myb-like transcription factors have epistatic effects on circadian clock function but additive effects on plant growth. Plant Direct 2023, 7, e533. [Google Scholar] [CrossRef] [PubMed]
- Patke, A.; Young, M.W.; Axelrod, S. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2020, 21, 67–84. [Google Scholar] [CrossRef]
- Favero, D.S.; Lambolez, A.; Sugimoto, K. Molecular pathways regulating elongation of aerial plant organs: A focus on light, the circadian clock, and temperature. Plant J. 2021, 105, 392–420. [Google Scholar] [CrossRef]
- Nicotra, A.; Atkin, O.; Bonser, S.; Davidson, A.; Finnegan, E.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef]
- Zhao, L.; Richards, S.; Turck, F.; Kollmann, M. Information integration and decision making in flowering time control. PLoS ONE 2020, 15, e0239417. [Google Scholar] [CrossRef] [PubMed]
- Lee, Z.; Kim, S.; Choi, S.J.; Joung, E.; Kwon, M.; Park, H.J.; Shim, J.S. Regulation of Flowering Time by Environmental Factors in Plants. Plants 2023, 12, 3680. [Google Scholar] [CrossRef] [PubMed]
- Fernández, V.; Takahashi, Y.; Le Gourrierec, J.; Coupland, G. Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J. 2016, 86, 426–440. [Google Scholar] [CrossRef]
- Osnato, M.; Castillejo, C.; Matías-Hernández, L.; Pelaz, S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 2012, 3, 808. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 2001, 13, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Trevaskis, B.; Bagnall, D.J.; Ellis, M.H.; Peacock, W.J.; Dennis, E.S. MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. USA 2003, 100, 13099–13104. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Ye, M.; Sang, M.; Wu, R. A Regulatory Network for miR156-SPL Module in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 6166. [Google Scholar] [CrossRef] [PubMed]
- Riboni, M.; Robustelli Test, A.; Galbiati, M.; Tonelli, C.; Conti, L. Environmental stress and flowering time: The photoperiodic connection. Plant Signal. Behav. 2014, 9, e29036. [Google Scholar] [CrossRef]
- Hwang, K.; Susila, H.; Nasim, Z.; Jung, J.-Y.; Ahn, J.H. Arabidopsis ABF3 and ABF4 Transcription Factors Act with the NF-YC Complex to Regulate SOC1 Expression and Mediate Drought-Accelerated Flowering. Mol. Plant 2019, 12, 489–505. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, Y.; Han, C.; Zhang, W.; Jia, H.; Li, X. GA signaling and CO/FT regulatory module mediate salt-induced late flowering in Arabidopsis thaliana. Plant Growth Regul. 2007, 53, 195–206. [Google Scholar] [CrossRef]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef] [PubMed]
- Harmer, S.L.; Kay, S.A. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 2005, 17, 1926–1940. [Google Scholar] [CrossRef]
- Farinas, B.; Mas, P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011, 66, 318–329. [Google Scholar] [CrossRef]
- Hsu, P.Y.; Harmer, S.L. Wheels within wheels: The plant circadian system. Trends Plant Sci. 2014, 19, 240–249. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, Y.; Zhang, H.; Xu, J.; Gao, X.; Zhang, T.; Liu, X.; Guo, L.; Zhao, D. Environmental F actors coordinate circadian clock function and rhythm to regulate plant development. Plant Signal. Behav. 2023, 18, 2231202. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.X.; Webb, C.J.; Knowles, S.M.; Kim, S.H.; Wang, Z.; Tobin, E.M. CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012, 158, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.K.; Chung, K.S.; Kim, J.; Lee, J.H.; Hong, S.M.; Yoo, S.J.; Yoo, S.Y.; Lee, J.S.; Ahn, J.H. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 2005, 139, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Tajima, T.; Yoshida, R.; Oda, A.; Mizoguchi, T. Possible involvement of FLC in natural variation of activity to enhance the late flowering phenotype of the clock mutant lhy cca1 under continuous light. Plant Biotechnol. 2010, 27, 455–461. [Google Scholar] [CrossRef][Green Version]
- Li, Y.; Liu, G.-F.; Ma, L.-M.; Liu, T.-K.; Zhang, C.-W.; Xiao, D.; Zheng, H.-K.; Chen, F.; Hou, X.-L. A chromosome-level reference genome of non-heading Chinese cabbage [Brassica campestris (syn. Brassica rapa) ssp. chinensis]. Hortic. Res. 2020, 7, 212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, Q.; Liu, W.; Wu, X.; Li, Z.; Xu, Y.; Li, Y.; Imaizumi, T.; Hou, X.; Liu, T. BrABF3 promotes flowering through the direct activation of CONSTANS transcription in pak choi. Plant J. 2022, 111, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Chen, X.; Li, Y.; Shah, S.H.A.; Xiao, D.; Wang, J.; Hou, X.; Liu, T.; Li, Y. ETHYLENE RESPONSE FACTOR 070 inhibits flowering in Pak-choi by indirectly impairing BcLEAFY expression. Plant Physiol. 2024, 195, 986–1004. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Shen, H.-R.; Zhao, J.-J.; Wei, Y.-P.; Liu, D.-R.; Hou, X.-L.; Bonnema, G. Genetic dissection of flowering time in Brassica rapa responses to temperature and photoperiod. Plant Sci. 2019, 280, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Yin, M.; He, Y. Molecular Genetic Understanding of Photoperiodic Regulation of Flowering Time in Arabidopsis and Soybean. Int. J. Mol. Sci. 2021, 23, 466. [Google Scholar] [CrossRef]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef]
- Lei, J.; Jayaprakasha, G.K.; Singh, J.; Uckoo, R.; Borrego, E.J.; Finlayson, S.A.; Kolomiets, M.V.; Patil, B.S.; Braam, J.; Zhu-Salzman, K. CIRCADIAN CLOCK-ASSOCIATED1 Controls Resistance to Aphids by Altering Indole Glucosinolate Production. Plant Physiol. 2019, 181, 1344–1359. [Google Scholar] [CrossRef]
- Sun, Q.; Wang, S.; Xu, G.; Kang, X.; Zhang, M.; Ni, M. SHB1 and CCA1 interaction desensitizes light responses and enhances thermomorphogenesis. Nat. Commun. 2019, 10, 3110. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, T.; Wheatley, K.; Hanzawa, Y.; Wright, L.; Mizoguchi, M.; Song, H.-R.; Carré, I.A.; Coupland, G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2002, 2, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Tobin, E.M. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 1998, 93, 1207–1217. [Google Scholar] [CrossRef] [PubMed]
- Karami-Moalem, S.; Ahmadikhah, A.; Nemati, Z.; Haghi, R. Transcriptome differential display of a drought-tolerant early flowering spineless mutant of safflower (Carthamus tinctorius) and identification of candidate genes. Crop Sci. 2023, 63, 2329–2346. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Q.; Dong, J.; Cai, C.; Zhu, H.; Li, S. Genome-wide identification and characterization of mungbean CIRCADIAN CLOCK ASSOCIATED 1 like genes reveals an important role of VrCCA1L26 in flowering time regulation. BMC Genom. 2022, 23, 374. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Kubota, A.; Imaizumi, T. Circadian Clock and Photoperiodic Flowering in Arabidopsis: CONSTANS Is a Hub for Signal Integration. Plant Physiol. 2017, 173, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Niwa, Y.; Ito, S.; Nakamichi, N.; Mizoguchi, T.; Niinuma, K.; Yamashino, T.; Mizuno, T. Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol. 2007, 48, 925–937. [Google Scholar] [CrossRef]
- Kim, J.A.; Kim, H.S.; Choi, S.H.; Jang, J.Y.; Jeong, M.J.; Lee, S.I. The importance of the circadian clock in regulating plant metabolism. Int. J. Mol. Sci. 2017, 18, 2680. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef]
- Searle, I.; He, Y.; Turck, F.; Vincent, C.; Fornara, F.; Kröber, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Kyung, J.; Jeon, M.; Jeong, G.; Shin, Y.; Seo, E.; Yu, J.; Kim, H.; Park, C.-M.; Hwang, D.; Lee, I. The two clock proteins CCA1 and LHY activate VIN3 transcription during vernalization through the vernalization-responsive cis-element. Plant Cell 2022, 34, 1020–1037. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, R.; Fekih, R.; Fujiwara, S.; Oda, A.; Miyata, K.; Tomozoe, Y.; Nakagawa, M.; Niinuma, K.; Hayashi, K.; Ezura, H.; et al. Possible role of early flowering 3 (ELF3) in clock-dependent floral regulation by short vegetative phase (SVP) in Arabidopsis thaliana. New Phytol. 2009, 182, 838–850. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Long, Y.; Xu, L.; Zhang, W.; Liu, T.; Zhang, C.; Hou, X.; Li, Y. CELL CYCLE SEITCH 52 regulates tillering by interacting with LATERAL SUPPRESSOR in non-heading Chinese cabbage. Plant Sci. 2021, 309, 110934. [Google Scholar] [CrossRef]
- Huang, F.; Liu, T.; Hou, X. Isolation and Functional Characterization of a Floral Repressor, BcMAF1, from Pak-choi (Brassica rapa ssp. Chinensis). Front. Plant Sci. 2018, 9, 290. [Google Scholar] [CrossRef]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Xiao, D.; Jiang, C.; Li, Y.; Hou, X. CIRCADIAN CLOCK-ASSOCIATED1 Delays Flowering by Directly Inhibiting the Transcription of BcSOC1 in Pak-choi. Plants 2024, 13, 2190. https://doi.org/10.3390/plants13162190
He Y, Xiao D, Jiang C, Li Y, Hou X. CIRCADIAN CLOCK-ASSOCIATED1 Delays Flowering by Directly Inhibiting the Transcription of BcSOC1 in Pak-choi. Plants. 2024; 13(16):2190. https://doi.org/10.3390/plants13162190
Chicago/Turabian StyleHe, Ying, Dong Xiao, Cheng Jiang, Yiran Li, and Xilin Hou. 2024. "CIRCADIAN CLOCK-ASSOCIATED1 Delays Flowering by Directly Inhibiting the Transcription of BcSOC1 in Pak-choi" Plants 13, no. 16: 2190. https://doi.org/10.3390/plants13162190
APA StyleHe, Y., Xiao, D., Jiang, C., Li, Y., & Hou, X. (2024). CIRCADIAN CLOCK-ASSOCIATED1 Delays Flowering by Directly Inhibiting the Transcription of BcSOC1 in Pak-choi. Plants, 13(16), 2190. https://doi.org/10.3390/plants13162190






