A Comparative Genetic Analysis of Phoenix atlantica in Cape Verde
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Sampling
2.2. DNA Isolation and Microsatellite Analysis
2.3. Data Analysis
2.4. Demographic Analysis
3. Results
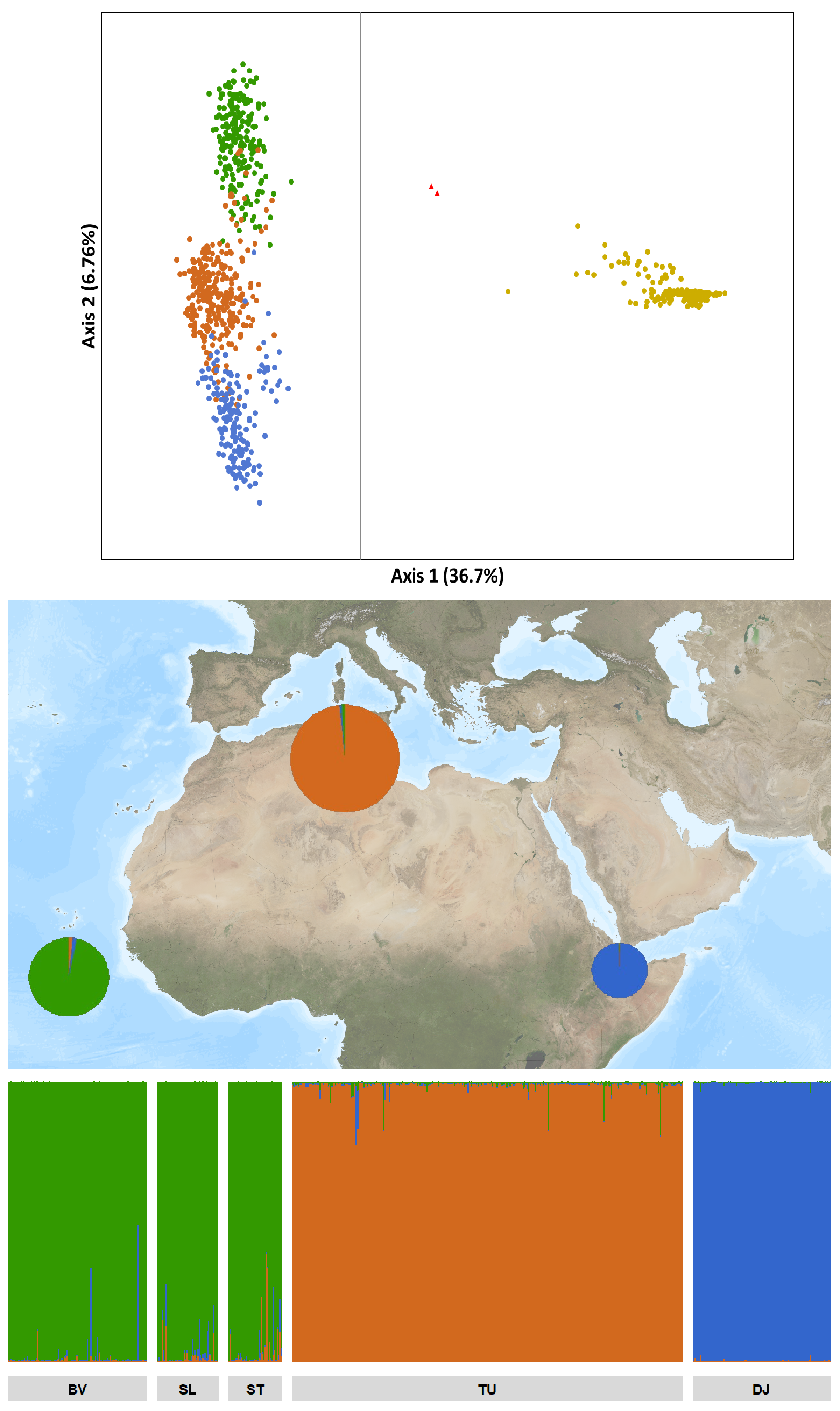
3.1. Genetic Relationship of P. atlantica with Other Phoenix Species
3.2. Demographic History
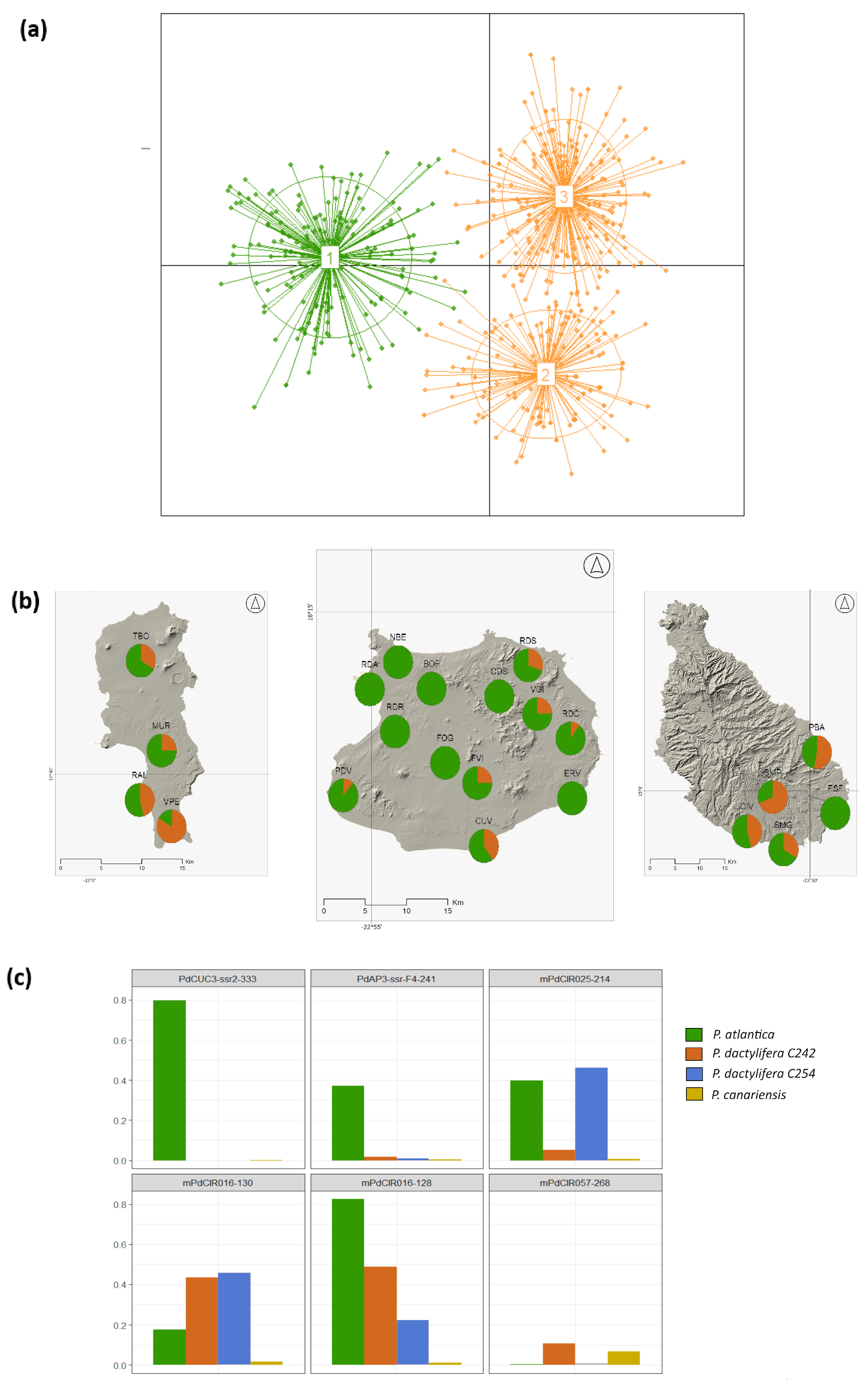
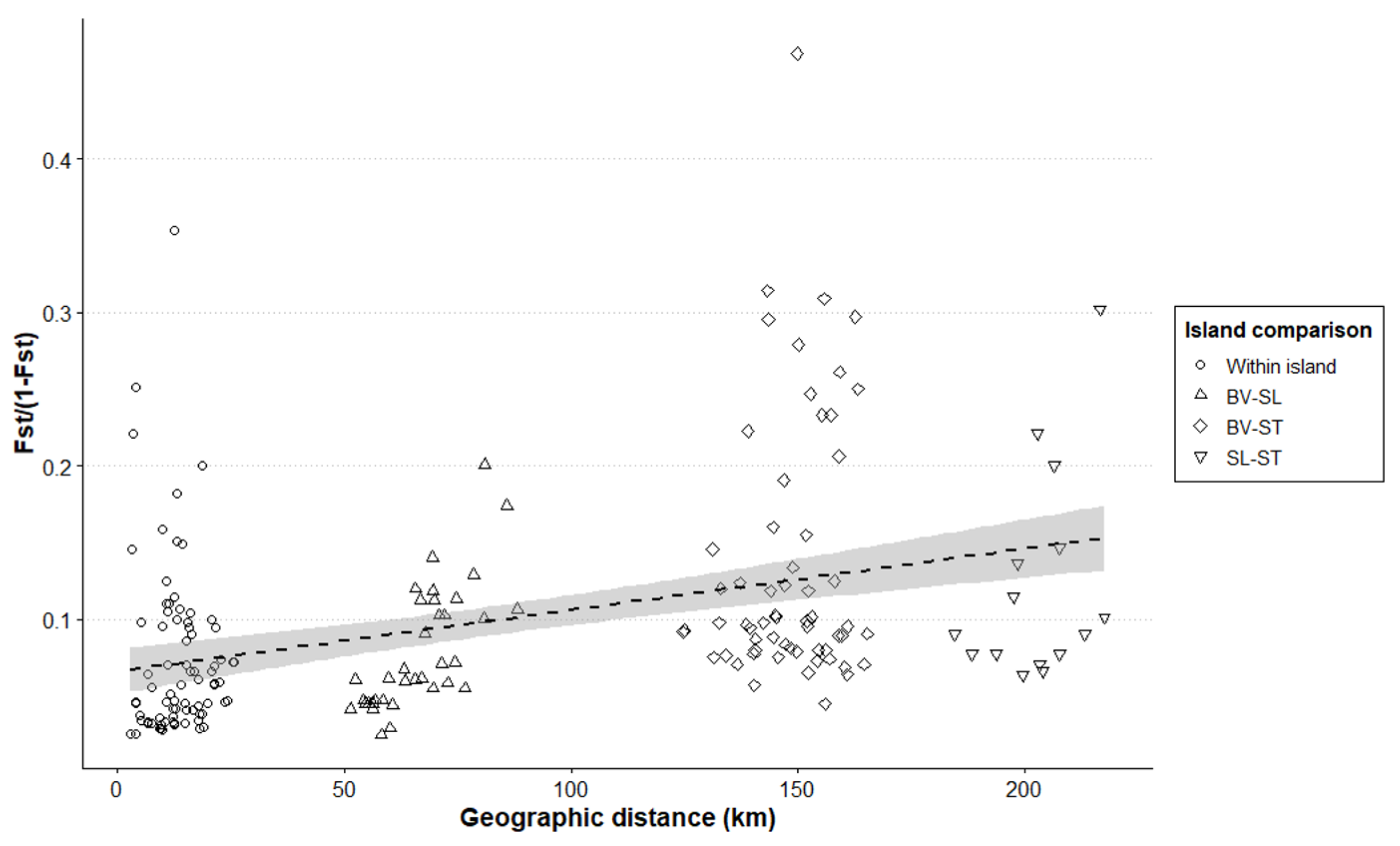
3.3. Characterization of P. atlantica in Cape Verde
4. Discussion
4.1. Genetic Characterization of P. atlantica
4.2. P. atlantica Is Closely Related to P. dactylifera from North Africa
4.3. Implications for P. atlantica Conservation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Gros-Balthazard, M.; Hazzouri, K.M.; Flowers, J.M. Genomic insights into Date palm origins. Genes 2018, 9, 502. [Google Scholar] [CrossRef] [PubMed]
- Gros-Balthazard, M. Hybridization in the genus Phoenix: A review. Emir. J. Food Agric. 2013, 25, 831–842. [Google Scholar] [CrossRef]
- Chao, C.T.; Krueger, R.R. The date palm (Phoenix dactylifera L.): Overview of biology, uses, and cultivation. HortScience 2007, 42, 1077–1082. [Google Scholar] [CrossRef]
- Gros-Balthazard, M.; Flowers, J.M. A Brief History of the Origin of Domesticated Date Palms; Springer: Berlin/Heidelberg, Germany, 2021; pp. 55–74. [Google Scholar]
- FAOSTAT. Agro-Statistics Database; Food and Agricultural Organization of the United Nations: Rome, Italy, 2022. [Google Scholar]
- Pintaud, J.; Zehdi, S.; Couvreur, T.; Barrow, S.; Henderson, S.; Aberlenc-Bertossi, F.; Tregear, J.; Billotte, N. Species delimitation in the genus Phoenix (Arecaceae) based on SSR markers, with emphasis on the identity of the date palm (Phoenix dactylifera L.). In Diversity, Phylogeny, and Evolution in the Monocotyledons; Seberg, O., Petersen, G., Barfod, A., Davis, J., Eds.; Aarhus University Press: Aarhus, Denmark, 2010. [Google Scholar]
- Rivera, D.; Obón, C.; García-Arteaga, J.; Egea, T.; Alcaraz, F.; Laguna, E.; Carreño, E.; Johnson, D.; Krueger, R.; Delgadillo, J.; et al. Carpological analysis of Phoenix (Arecaceae): Contributions to the taxonomy and evolutionary history of the genus. Bot. J. Linnean Soc. 2014, 175, 74–122. [Google Scholar] [CrossRef]
- Saro, I.; Rodríguez-Rodríguez, P.; Rivera, D.; Obón, C.; Aberlenc, F.; Díaz-Pérez, A.; Zehdi-Azouzi, S.; Curbelo, L.; Sosa, P.A. The genetic characterization of the Canarian endemic palm (Phoenix canariensis) by Simple Sequence Repeats and chloroplast markers: A tool for the molecular traceability of Phoenix hybridization. Diversity 2024, 16, 411. [Google Scholar] [CrossRef]
- Henderson, S.; Gomes, I.; Gomes, S.; Baker, W. Phoenix in the Cape Verde Islands. J. Int. Palm Soc. 2003, 47, 1–14. [Google Scholar]
- Barrow, S.C. A Monograph of Phoenix L. (Palmae: Coryphoideae). Kew Bull. 1998, 53, 513–575. [Google Scholar] [CrossRef]
- Henderson, S.A.; Billotte, N.; Pintaud, J.C. Genetic isolation of Cape Verde island Phoenix atlantica (Arecaceae) revealed by microsatellite markers. Conserv. Gen. 2006, 7, 213–223. [Google Scholar] [CrossRef]
- Chaluvadi, S.R.; Young, P.; Thompson, K.; Bahri, B.A.; Gajera, B.; Narayanan, S.; Krueger, R.; Bennetzen, J.L. Phoenix phylogeny, and analysis of genetic variation in a diverse collection of date palm (Phoenix dactylifera) and related species. Plant Divers. 2019, 41, 330–339. [Google Scholar] [CrossRef]
- Flowers, J.M.; Hazzouri, K.M.; Gros-Balthazard, M.; Mo, Z.; Koutroumpa, K.; Perrakis, A.; Ferrand, S.; Khierallah, H.S.M.; Fuller, D.Q.; Aberlenc, F.; et al. Cross-species hybridization and the origin of North African date palms. Proc. Natl. Acad. Sci. USA 2019, 116, 1651–1658. [Google Scholar] [CrossRef]
- Gros-Balthazard, M.; Galimberti, M.; Kousathanas, A.; Newton, C.; Ivorra, S.; Paradis, L.; Vigouroux, Y.; Carter, R.; Tengberg, M.; Battesti, V.; et al. The discovery of wild Date palms in Oman reveals a complex domestication history involving centers in the Middle East and Africa. Curr. Biol. 2017, 27, 2211–2218. [Google Scholar] [CrossRef] [PubMed]
- Duarte, M.C.; Rego, F.; Romeiras, M.M.; Moreira, I. Plant species richness in the Cape Verde Islands—eco-geographical determinants. Biodivers. Conserv. 2008, 17, 453–466. [Google Scholar] [CrossRef]
- Romeiras, M.M.; Monteiro, F.; Duarte, M.C.; Schaefer, H.; Carine, M. Patterns of genetic diversity in three plant lineages endemic to the Cape Verde Islands. AoB Plants 2015, 7, plv051. [Google Scholar] [CrossRef] [PubMed]
- Varela, D.; Romeiras, M.M.; Silva, L. Implications of climate change on the distribution and conservation of Cabo Verde endemic trees. Glob. Ecol. Conserv. 2022, 34, e02025. [Google Scholar] [CrossRef]
- Gomes, I.; Gomes, S.; Vera-Cruz, M.; Kilian, N.; Leyens, T.; Lobin, W. Plantas Endémicas e Árvores Indígenas de Cabo Verde; Instituto Nacional de Investigação e Desenvolvimento Agrário Praia: São Jorge dos Órgãos, Cape Verde, 1995.
- Cañizares, S.M.S.; Canalejo, A.M.C.; Tabales, J.M.N. Stakeholders’ perceptions of tourism development in Cape Verde, Africa. Curr. Issues Tour. 2016, 19, 966–980. [Google Scholar] [CrossRef]
- Zehdi-Azouzi, S.; Cherif, E.; Moussouni, S.; Gros-Balthazard, M.; Naqvi, S.A.; Ludeña, B.; Castillo, K.; Chabrillange, N.; Bouguedoura, N.; Bennaceur, M.; et al. Genetic structure of the date palm (Phoenix dactylifera) in the Old World reveals a strong differentiation between eastern and western populations. Ann. Bot. 2015, 116, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Saro, I.; González-Pérez, M.A.; García-Verdugo, C.; Sosa, P.A. Patterns of genetic diversity in Phoenix canariensis, a widespread oceanic palm (species) endemic from the Canarian archipelago. Tree Genet. Genomes 2015, 11, 1–13. [Google Scholar] [CrossRef]
- Billotte, N.; Marseillac, N.; Brottier, P.; Noyer, J.L.; Jacquemoud-Collet, J.P.; Moreau, C.; Couvreur, T.; Chevallier, M.H.; Pintaud, J.C.; Risterucci, A.M. Nuclear microsatellite markers for the date palm (Phoenix dactylifera L.): Characterization and utility across the genus Phoenix and in other palm genera. Mol. Ecol. Notes 2004, 4, 256–258. [Google Scholar] [CrossRef]
- Aberlenc-Bertossi, F.; Castillo, K.; Tranchant-Dubreuil, C.; Chérif, E.; Ballardini, M.; Abdoulkader, S.; Gros-Balthazard, M.; Chabrillange, N.; Santoni, S.; Mercuri, A.; et al. In silico mining of microsatellites in coding sequences of the date palm (Arecaceae) genome, characterization, and transferability. Appl. Plant Sci. 2014, 2, 1300058. [Google Scholar] [CrossRef]
- Sosa, P.A.; Saro, I.; Johnson, D.; Obón, C.; Alcaraz, F.; Rivera, D. Biodiversity and conservation of Phoenix canariensis: A review. Biodivers. Conserv. 2021, 30, 275–293. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.X. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Jombart, T.; Kamvar, Z.N.; Collins, C.; Lustrik, R.; Beugin, M.P.; Knaus, B.J.; Jombart, M.T. Package ‘adegenet’. Github Repository. 2018. Available online: https://github.com/thibautjombart/adegenet (accessed on 25 February 2024).
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Cornuet, J.M.; Pudlo, P.; Veyssier, J.; Dehne-Garcia, A.; Gautier, M.; Leblois, R.; Marin, J.M.; Estoup, A. DIYABC v2.0: A software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 2014, 30, 1187–1189. [Google Scholar] [CrossRef] [PubMed]
- Guillemaud, T.; Beaumont, M.A.; Ciosi, M.; Cornuet, J.M.; Estoup, A. Inferring introduction routes of invasive species using approximate Bayesian computation on microsatellite data. Heredity 2010, 104, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Cornuet, J.M.; Santos, F.; Beaumont, M.A.; Robert, C.P.; Marin, J.M.; Balding, D.J.; Guillemaud, T.; Estoup, A. Inferring population history with DIY ABC: A user-friendly approach to approximate Bayesian computation. Bioinformatics 2008, 24, 2713–2719. [Google Scholar] [CrossRef]
- Khanum, P.; Khan, A.A.; Khan, I.A.; Ghaffar, A.; Khan, Z. TPD1-like gene as a suitable marker for early sex determination in Date palm (Phoenix dactylifera L.). Genes 2023, 14, 907. [Google Scholar] [CrossRef]
- Janes, J.K.; Miller, J.M.; Dupuis, J.R.; Malenfant, R.M.; Gorrell, J.C.; Cullingham, C.I.; Andrew, R.L. The K = 2 conundrum. Mol. Ecol. 2017, 26, 3594–3602. [Google Scholar] [CrossRef]
- Fernández-Palacios, J.M.; Kreft, H.; Irl, S.D.H.; Norder, S.; Ah-Peng, C.; Borges, P.A.V.; Burns, K.C.; de Nascimento, L.; Meyer, J.Y.; Montes, E.; et al. Scientists’ warning—The outstanding biodiversity of islands is in peril. Glob. Ecol. Conserv. 2021, 31, e01847. [Google Scholar] [CrossRef]
- Kvist, L.; Broggi, J.; Illera, J.C.; Koivula, K. Colonisation and diversification of the blue tits (Parus caeruleus teneriffae-group) in the Canary Islands. Mol. Phylogenet. Evol. 2005, 34, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Barrado, M.R.; Villaverde, T.; Perez, M.F.; Sanmartín, I.; Riina, R. The sweet tabaiba or there and back again: Phylogeographic history of the Macaronesian Euphorbia balsamifera. Ann. Bot. 2024, 133, 883–904. [Google Scholar] [CrossRef]
- Madeira, J.P. Cape Verde: Dimensions in nation-building. Humania del Sur 2016, 11, 93–105. [Google Scholar]
- Pintaud, J.C.; Ludeña, B.; Aberlenc-Bertossi, F.; Zehdi, S.; Gros-Balthazard, M.; Ivorra, S.; Terral, J.F.; Newton, C.; Tengberg, M.; Abdoulkader, S.; et al. Biogeography of the date palm (Phoenix dactylifera L., Arecaceae): Insights on the origin and on the structure of modern diversity. Proc. Int. Symp. Date Palm 2011, 994, 19–38. [Google Scholar] [CrossRef]
- de Nascimento, L.; Nogué, S.; Criado, C.; Ravazzi, C.; Whittaker, R.J.; Willis, K.J.; Fernández-Palacios, J.M. Reconstructing Holocene vegetation on the island of Gran Canaria before and after human colonization. Holocene 2016, 26, 113–125. [Google Scholar] [CrossRef]
- van Zeista, W. Fruits in foundation deposits of two temples. J. Archaeol. Sci. 1983, 10, 351–354. [Google Scholar] [CrossRef]
- Mercuri, A.M.; Bosi, G.; Buldrini, F. Seeds, Fruits and Charcoal from the Fewet Compound. In Life and Death of a Rural Village in Garamantian Times: Archaeological Investigations in the Oasis of Fewet (Libyan Sahara); Arid Zone Archaeology Monographs; All’Insegna del Giglio: Sesto Fiorentino, Italy, 2013; Volume 6, pp. 177–190. ISSN 2035-5459. [Google Scholar]
- Fuller, D.Q. Long and attenuated: Comparative trends in the domestication of tree fruits. Veg. Hist. Archaeobot. 2018, 27, 165–176. [Google Scholar] [CrossRef]
- der Veen, M.V. Ancient agriculture in Libya: A review of the evidence. Acta Palaeobot. 1995, 35, 85–98. [Google Scholar]
- Munier, P. The Date palm. Le Palmier-Dattier; Number 24 in Techniques Agricoles et Productions Tropicales; Maisonneuve et Larose: Paris, France, 1973; p. 221. [Google Scholar]
- Manel, S.; Holderegger, R. Ten years of landscape genetics. Trends Ecol. Evol. 2013, 28, 614–621. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarmiento Cabello, S.; Rodríguez-Rodríguez, P.; Arbelo Ramírez, G.; Naranjo-Cigala, A.; Curbelo, L.; da Graca Gomes, M.d.M.; Brito, J.; Aberlenc, F.; Zehdi-Azouzi, S.; Sosa, P.A. A Comparative Genetic Analysis of Phoenix atlantica in Cape Verde. Plants 2024, 13, 2209. https://doi.org/10.3390/plants13162209
Sarmiento Cabello S, Rodríguez-Rodríguez P, Arbelo Ramírez G, Naranjo-Cigala A, Curbelo L, da Graca Gomes MdM, Brito J, Aberlenc F, Zehdi-Azouzi S, Sosa PA. A Comparative Genetic Analysis of Phoenix atlantica in Cape Verde. Plants. 2024; 13(16):2209. https://doi.org/10.3390/plants13162209
Chicago/Turabian StyleSarmiento Cabello, Sonia, Priscila Rodríguez-Rodríguez, Guacimara Arbelo Ramírez, Agustín Naranjo-Cigala, Leticia Curbelo, Maria de Monte da Graca Gomes, Juliana Brito, Frédérique Aberlenc, Salwa Zehdi-Azouzi, and Pedro A. Sosa. 2024. "A Comparative Genetic Analysis of Phoenix atlantica in Cape Verde" Plants 13, no. 16: 2209. https://doi.org/10.3390/plants13162209
APA StyleSarmiento Cabello, S., Rodríguez-Rodríguez, P., Arbelo Ramírez, G., Naranjo-Cigala, A., Curbelo, L., da Graca Gomes, M. d. M., Brito, J., Aberlenc, F., Zehdi-Azouzi, S., & Sosa, P. A. (2024). A Comparative Genetic Analysis of Phoenix atlantica in Cape Verde. Plants, 13(16), 2209. https://doi.org/10.3390/plants13162209




