Identification and Analysis of WRKY Transcription Factors in Response to Cowpea Fusarium Wilt in Cowpea
Abstract
:1. Introduction
2. Results
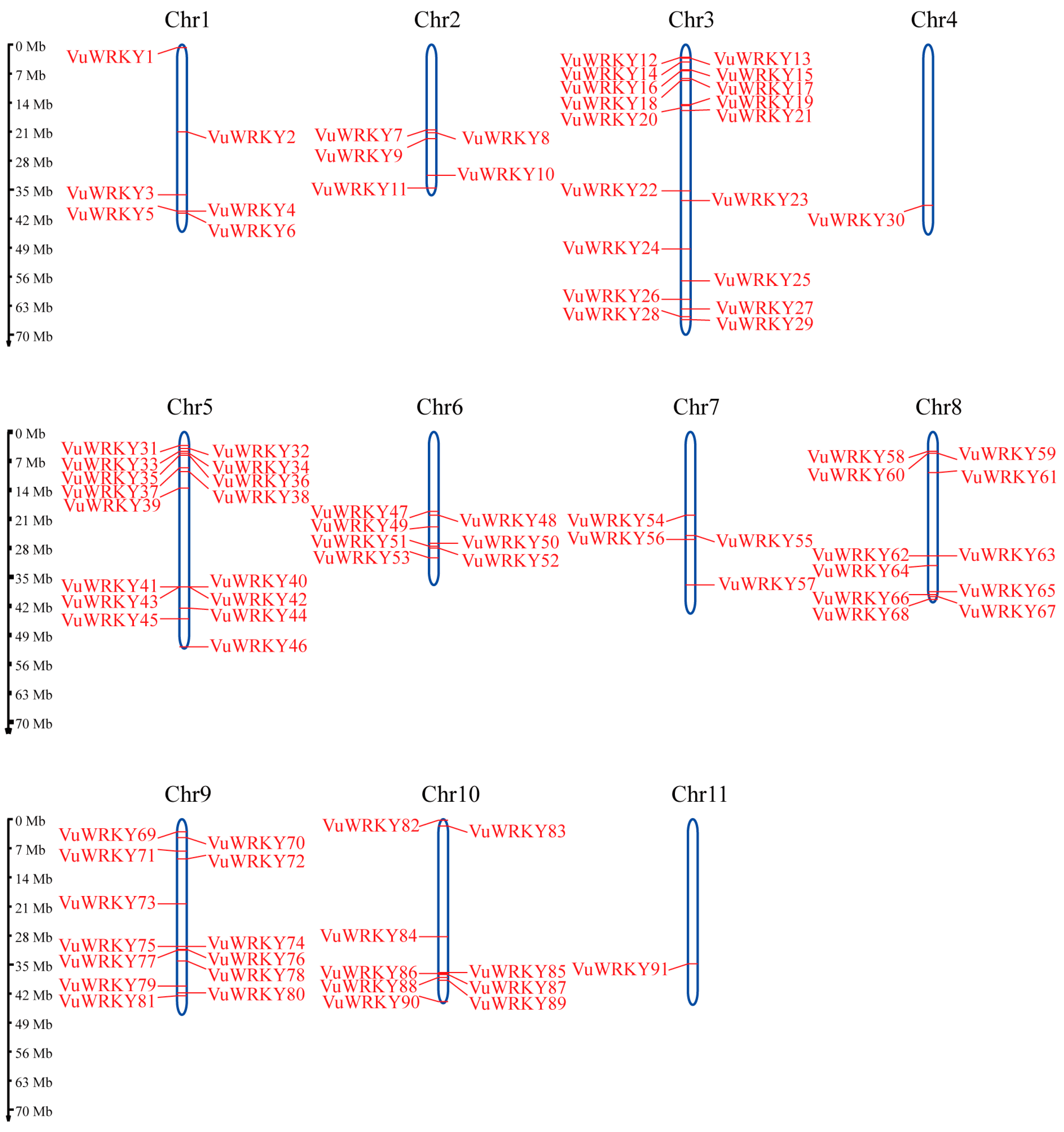
2.1. Genome-Wide Identification of WRKY Genes in Cowpea
2.2. Phylogenetic Tree Construction of VuWRKY Family Members and Analysis of Conserved Protein Domains in VuWRKY Proteins
2.3. Analysis of the Gene Structure and Conserved Motifs of Cowpea WRKY Genes
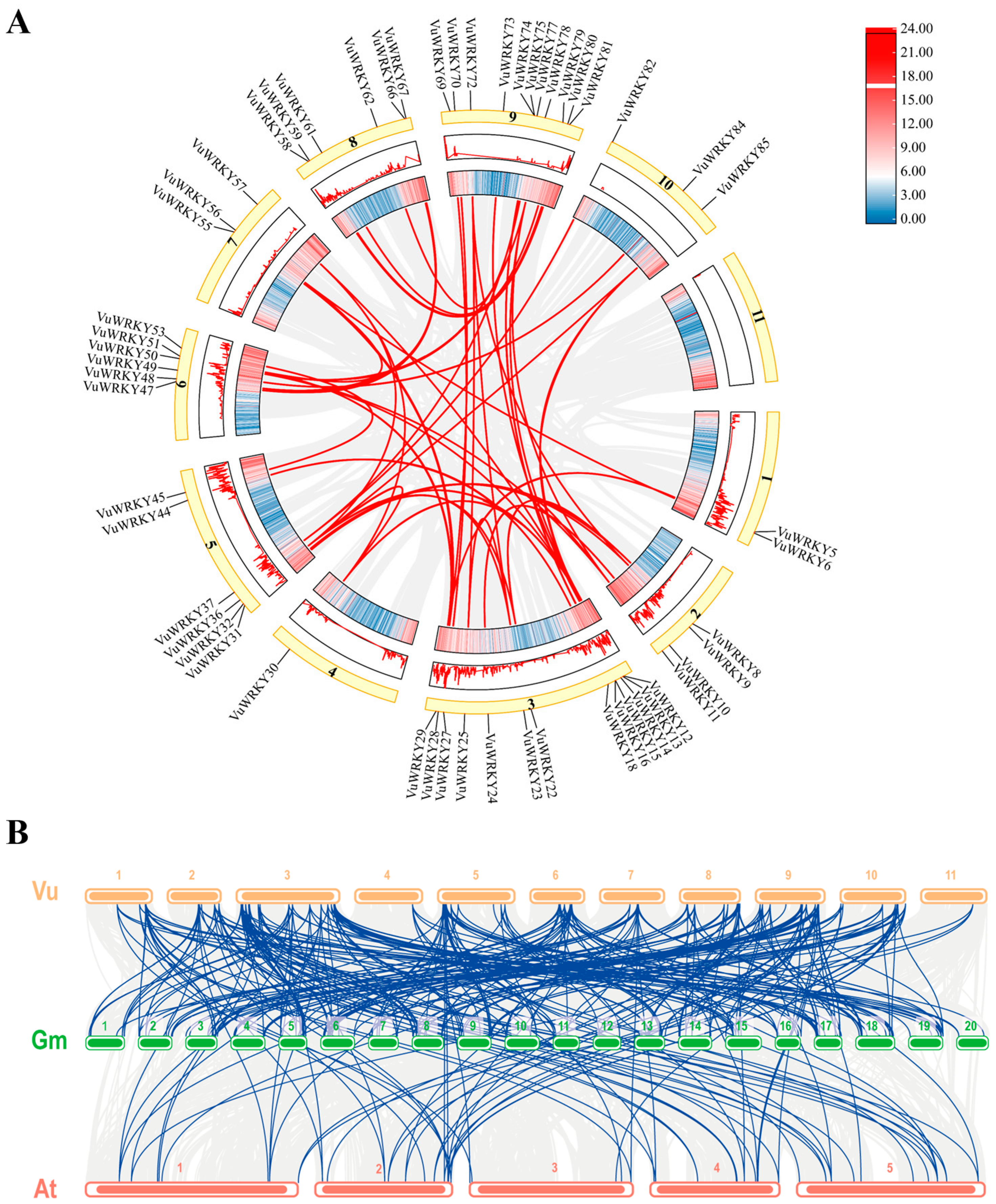
2.4. Homology Analysis of VuWRKY Genes
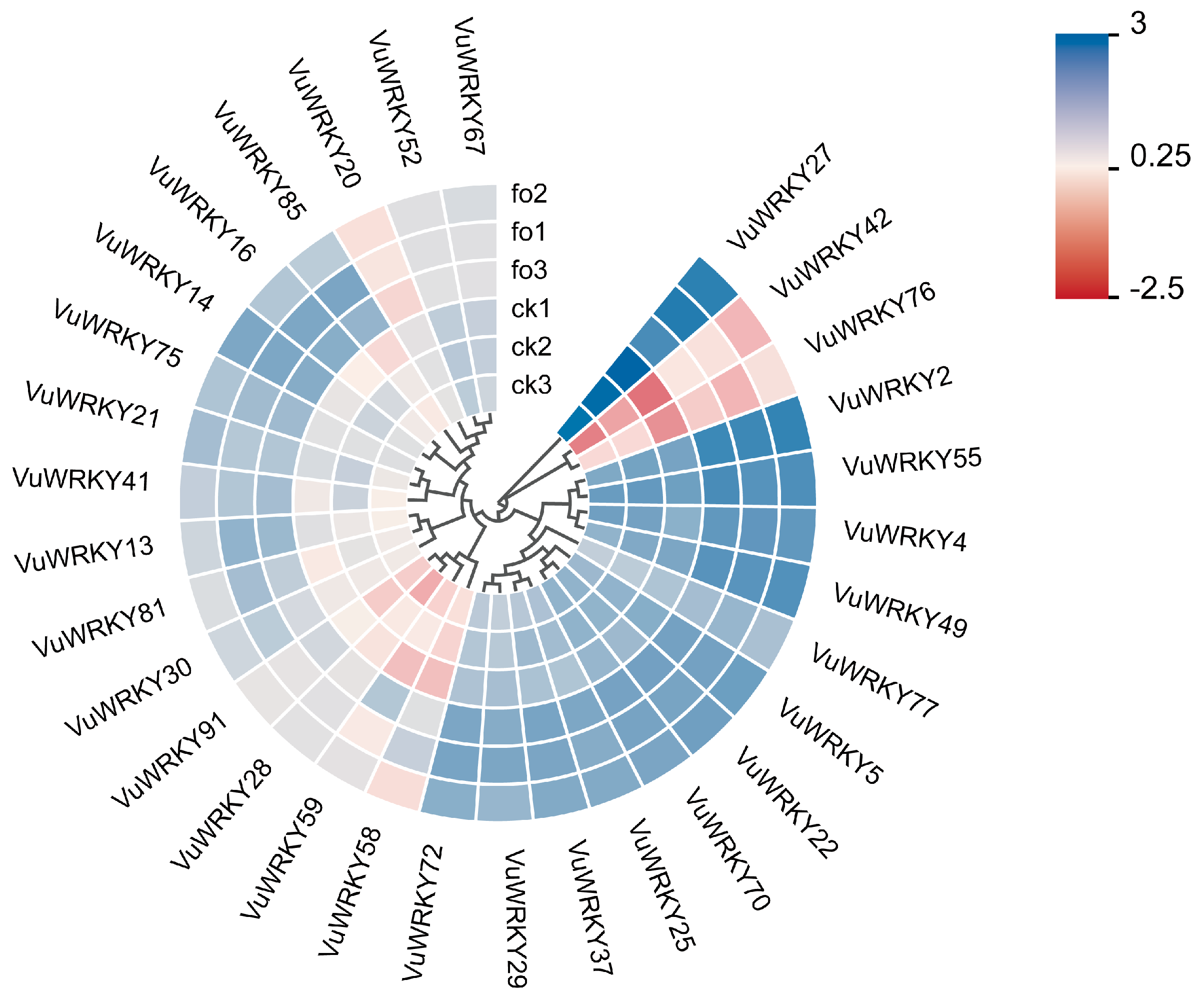
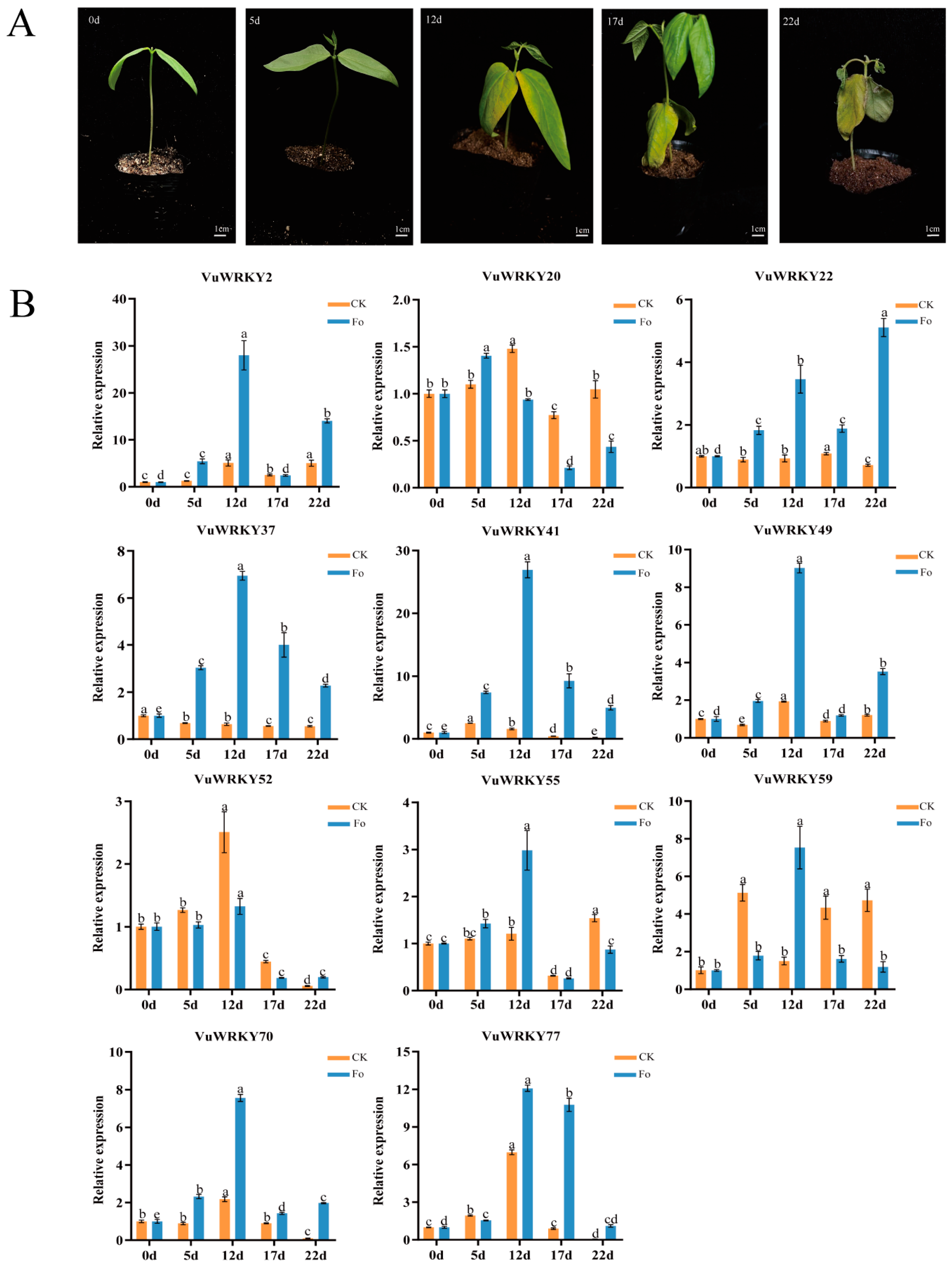
2.5. Expression Patterns of VuWRKY Genes in Response to Cowpea Fusarium Wilt Infection
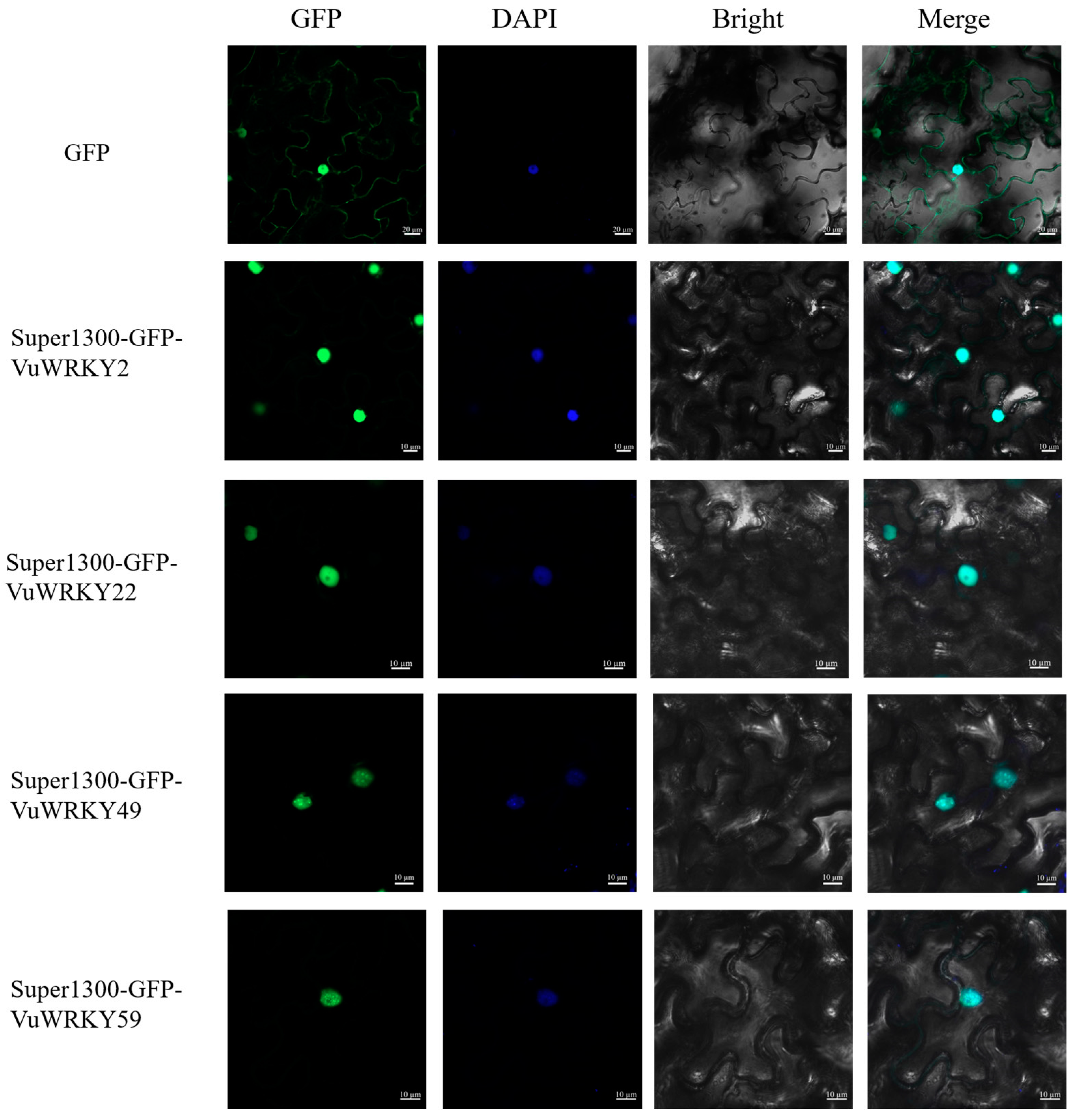
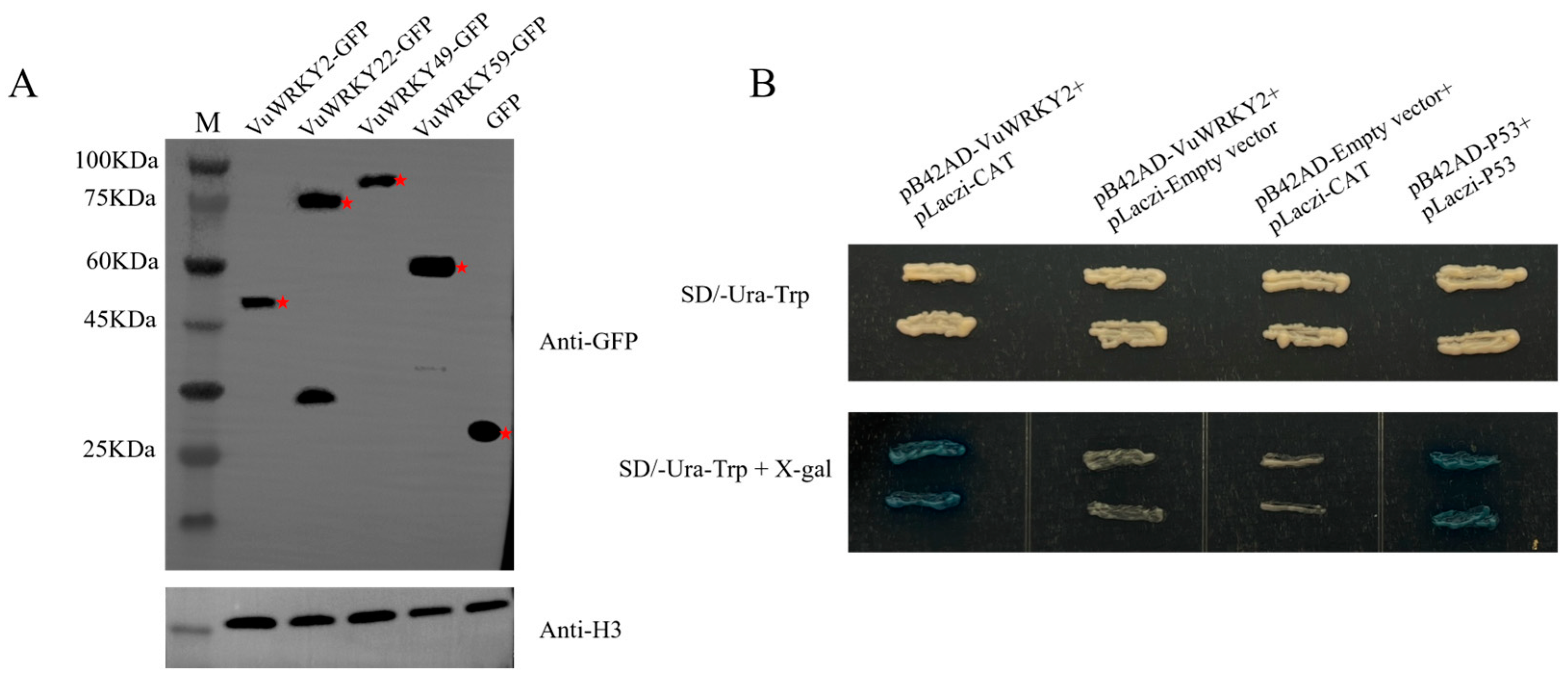
2.6. Subcellular Localization of VuWRKY Proteins
2.7. Binding of VuWRKY2 to the CAT Promoter
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of VuWRKY Genes in Cowpea
4.2. Gene Structure, Conserved Motifs, and Phylogenetic Tree Analysis
4.3. Chromosomal Localization, Gene Duplication, and Synteny Analysis of VuWRKY
4.4. Plant Material and Cowpea Fusarium Wilt Pathogen Inoculation
4.5. RT-qPCR Transcriptome Analysis, Identification of Differentially Expressed Genes, and Expression Validation
4.6. Subcellular Localization of VuWRKY and Western Blotting
4.7. Yeast One-Hybrid
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, Y.J.; Wu, J.; Wang, Y.P.; Sun, Q.F. Advances in the function of WRKY transcription factor in plant stress response. Mol. Plant Breed. 2020, 18, 7413–7422. [Google Scholar]
- Rushton, P.J.; Macdonald, H.; Huttly, A.K.; Lazarus, C.M.; Hooley, R. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol. Biol. 1995, 29, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, M.; Oelmüller, R. WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY Transcription Factor Family in Model Plants and Crops. Crit. Rev. Plant Sci. 2017, 5, 311–335. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 2, 86–101. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. MGG Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Ülker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 2007, 6, 827–842. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Liu, J.J.; Ekramoddoullah, A.K.M. Identification and characterization of the WRKY transcription factor family in Pinus monticola. Genome 2009, 52, 77–88. [Google Scholar] [CrossRef]
- Fang, Y.J.; Zheng, Y.Q.; Lu, W.; Li, J.; Duan, Y.J.; Zhang, S. WYP Roles of miR319-regulated TCPs in plant development and response to abiotic stress. Crop J. 2021, 9, 17–28. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Diezel, C.; Somssich, I.E. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 2012, 159, 266–285. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Li, Y.; Wang, F.; Cheng, Y.; Fan, B.; Yu, J.Q.; Chen, Z. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 2011, 23, 3824–3841. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.F.; Wang, Y.N.; Yu, L.; Eulgem, T.; Lai, Y.; Liu, Z.Q.; Wang, X.U.; Qiu, A.L.; Zhang, T.X.; Lin, J.; et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Liu, H.; Deng, Y.; Xiao, J.; Li, X.; Wang, S. The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 2015, 167, 1087–1099. [Google Scholar] [CrossRef]
- Li, C. Molecular Mechanism of WRKY26-bHLH3 Co-Promoting Red Pear Anthocyanin Synthesis and Anthocyanin Inhibition of Aspergillus niger Infection; Hefei University of Technology: Hefei, China, 2020. [Google Scholar]
- Li, S.; Liu, G.; Pu, L.; Liu, X.; Wang, Z.; Zhao, Q.; Chen, H.; Ge, F.; Liu, D. WRKY Transcription Factors Actively Respond to Fusarium oxysporum in Lilium regale. Phytopathology 2021, 111, 1625–1637. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Deng, J.; Liang, T.; Su, L.; Ge, F.; Liu, Q. Cloning and functional analysis of WRKY transcription factor gene from Lilium regale Wilso. Northwest Bot. J. 2021, 41, 1983–1993. [Google Scholar]
- Chakraborty, J.; Ghosh, P.; Sen, S.; Nandi, A.K.; Das, S. CaMPK9 increases the stability of CaWRKY40 transcription factor which triggers defense response in chickpea upon Fusarium oxysporum f. sp. ciceri Race1 infection. Plant Mol. Biol. 2019, 100, 411–431. [Google Scholar] [CrossRef]
- Markulin, L.; Corbin, C.; Renouard, S.; Drouet, S.; Durpoix, C.; Mathieu, C.; Lopez, T.; Auguin, D.; Hano, C.; Lainé, É. Characterization of LuWRKY36, a flax transcription factor promoting secoisolariciresinol biosynthesis in response to Fusarium oxysporum elicitors in Linum usitatissimum L. hairy roots. Planta 2019, 250, 347–366. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.R.P.; Ndeve, A.D.; Huynh, B.L.; Matthews, W.C.; Roberts, P.A. QTL mapping and transcriptome analysis of cowpea reveals candidate genes for root-knot nematode resistance. PLoS ONE 2018, 13, e0189185. [Google Scholar] [CrossRef]
- Tan, H.; Huang, H.; Tie, M.; Tang, Y.; Lai, Y.; Li, H. Transcriptome profiling of two asparagus bean (Vigna unguiculata subsp. sesquipedalis) cultivars differing in chilling tolerance under cold stress. PLoS ONE 2016, 11, e0151105. [Google Scholar] [CrossRef]
- Li, Q.; Fu, Q.; Wang, S.; Huang, G.; Liu, Y.; Ye, Q.; Wu, Q.; Sun, X. Pathogenic Identification and Indoor Screening of Cowpea Fusarium Wilt in Sanya, Hainan Province. Trop. Agric. Sci. 2017, 37, 38–42. [Google Scholar]
- Zhao, X.; Zhang, Y. Effects of p-hydroxybenzoic acid on cowpea Fusarium wilt fungus. Chin. Veg. J. 2009, 18, 38–40. [Google Scholar]
- Pottorff, M.O.; Li, G.; Ehlers, J.D.; Close, T.J.; Roberts, P.A. Genetic mapping, synteny, and physical location of two loci for Fusarium oxysporum f. sp. tracheiphilum race 4 resistance in cowpea [Vigna unguiculata (L.) Walp]. Mol. Breed. 2014, 33, 779–791. [Google Scholar] [CrossRef] [PubMed]
- da Silva Matos, M.K.; Benko-Iseppon, A.M.; Bezerra-Neto, J.P.; Ferreira-Neto, J.R.C.; Wang, Y.; Liu, H.; Pandolfi, V.; Amorim, L.L.B.; Willadino, L.; do Vale Amorim, T.C.; et al. The WRKY transcription factor family in cowpea: Genomic characterization and transcriptomic profiling under root dehydration. Gene 2022, 823, 146377. [Google Scholar] [CrossRef]
- Du, C. Function and application of the WRKY transcription factor superfamily in plant response to stresses. Grassl. Sci. 2021, 38, 1287–1300. [Google Scholar]
- Wang, B.; Chen, M.; Lin, L.; Ye, R.; Zhu, H.; Wen, Q. Signal pathways and related transcription factors of drought stress in plant. Northwest Bot. J. 2020, 40, 1792–1806. [Google Scholar]
- Sun, C.; Wang, C.; Liang, Y.; Li, C.; Zhang, Z.; Jiao, X.; Han, C. Research progress on plant WRKY transcription factors. Shandong For. Sci. Technol. 2020, 50, 79–84. [Google Scholar]
- Shen, D.; Yang, L.; Hu, W.; Kuang, L.; Guo, W.; Lu, T.; Liu, D.; Liu, Y. Cloning and expression analysis of stress response gene WRKY47 in citrus. Jiangsu Agric. J. 2021, 37, 129–138. [Google Scholar]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Ren, Q.; Chen, Y.; Xu, G.; Qian, Y. Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 2021, 21, 427. [Google Scholar] [CrossRef]
- Bencke-Malato, M.; Cabreira, C.; Wiebke-Strohm, B.; Bücker-Neto, L.; Mancini, E.; Osorio, M.B.; Homrich, M.S.; Turchetto-Zolet, A.C.; De Carvalho, M.C.; Stolf, R.; et al. Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol. 2014, 14, 236. [Google Scholar] [CrossRef]
- Li, X.Y.; Liu, H.; He, F.; Wang, X.; Li, M.N.; Long, R.C.; Kang, J.M.; Yang, Q.C.; Chen, L. Identification and expression pattern of the WRKY transcription factor family in Medicago sativa. Acta Prataculturae Sin. 2024, 33, 154–170. [Google Scholar]
- Chen, P.; Liu, Q. Genome-wide characterization of the WRKY gene family in cultivated strawberry (Fragaria × ananassa Duch.) and the importance of several group III members in continuous cropping. Sci. Rep. 2019, 9, 8423. [Google Scholar] [CrossRef]
- Lin, S.; Liu, J.; Zhang, X.; Bao, M.; Fu, X. Genome-wide identification and expression analysis of WRKY gene family in Dianthus caryophyllus. Acta Hortic. Sin. 2021, 4, 1768–1784. [Google Scholar]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef]
- Mangelsen, E.; Kilian, J.; Berendzen, K.W.; Kolukisaoglu, Ü.H.; Harter, K.; Jansson, C.; Wanke, D. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genom. 2008, 9, 194. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Chang, X.; Zhi, Y.; Wang, L.; Xing, G.; Song, W.; Nie, X. Evolution and identification of the WRKY gene family in Quinoa (Chenopodium quinoa). Genes 2019, 10, 131. [Google Scholar] [CrossRef]
- Ding, X.; Hu, M.; Xiang, S.; Lai, X.; Le, Y.; Liu, J.; Zhou, S. Bioinformatics analysis of the WRKY gene family in autumn eggplants. Jiangsu Agric. Sci. 2022, 50, 50–60. [Google Scholar]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–5036. [Google Scholar] [CrossRef] [PubMed]
- Van Verk, M.C.; Pappaioannou, D.; Neeleman, L.; Bol, J.F.; Linthorst, H.J.M. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008, 146, 1983–2199. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Xiang, S.; Chen, Y.; Zhang, H.; Yu, D. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 2021, 72, 1473–1489. [Google Scholar] [CrossRef]
- Xing, D.H.; Lai, Z.B.; Zheng, Z.Y.; Vinod, K.M.; Fan, B.F.; Chen, Z.X. Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol. Plant 2008, 1, 459–470. [Google Scholar] [CrossRef]
- Gao, Q.M.; Venugopal, S.; Navarre, D.; Kachroo, A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 2011, 155, 464–476. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Guo, D.; Zhao, G.; Wang, J.; Zhang, S.; Wang, C.; Guo, X. Group IIc WRKY transcription factors regulate cotton resistance to Fusarium oxysporum by promoting GhMKK2-mediated flavonoid biosynthesis. New Phytologist. 2022, 236, 249–265. [Google Scholar] [CrossRef] [PubMed]
- Encinas-Villarejo, S.; Maldonado, A.M.; Amil-Ruiz, F.; de los Santos, B.; Romero, F.; Pliego-Alfaro, F.; Muñoz-Blanco, J.; Caballero, J.L. Evidence for a positive regulatory role of strawberry (Fragaria × ananassa) Fa WRKY1 and Arabidopsis AtWRKY75 proteins in resistance. J. Exp. Bot. 2009, 60, 3043–3065. [Google Scholar] [CrossRef]
- Lai, Z.B.; Vinod, K.M.; Zheng, Z.Y.; Fan, B.F.; Chen, Z.X. Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 2008, 8, 68. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.Y.; Mosher, S.L.; Fan, B.F.; Klessig, D.F.; Chen, Z.X. Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 2007, 7, 2. [Google Scholar] [CrossRef]
- Xu, X.; Chen, C.; Fan, B.; Chen, Z. Physical and func tional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 2006, 18, 1310–1326. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lv, H.; Fang, Z.; Yang, L.; Xie, B.; Liu, Y.; Zhuang, M.; Zhang, Y.; Yang, Y. Research on screening of resistant resources to Fusarium wilt and inheritance of the resistant gene in cabbage. Acta Hortic. Sin. 2011, 38, 875–885. [Google Scholar]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar]
- Da Silva, H.A.P.; Nardeli, S.M.; Alves-Ferreira, M.; Simões-Araújo, J.L. Evaluation of Reference Genes for RT-qPCR Normalization in Cowpea under Drought Stress during Biological Nitrogen Fixation. Crop Sci. 2015, 55, 1660. [Google Scholar] [CrossRef]
- Kong, D.; Li, C.; Xue, W.; Wei, H.; Ding, H.; Hu, G.; Zhang, X.; Zhang, G.; Zou, T.; Xian, Y.; et al. UB2/UB3/TSH4-anchored transcriptional networks regulate early maize inflorescence development in response to simulated shade. Plant Cell 2023, 35, 717–737. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Y.; Liu, R.; Mao, Z.; Yang, Q.; Zheng, S.; Lu, X.; Yang, Y.; Xie, B.; Zhao, J.; Li, Y.; et al. Identification and Analysis of WRKY Transcription Factors in Response to Cowpea Fusarium Wilt in Cowpea. Plants 2024, 13, 2273. https://doi.org/10.3390/plants13162273
Hao Y, Liu R, Mao Z, Yang Q, Zheng S, Lu X, Yang Y, Xie B, Zhao J, Li Y, et al. Identification and Analysis of WRKY Transcription Factors in Response to Cowpea Fusarium Wilt in Cowpea. Plants. 2024; 13(16):2273. https://doi.org/10.3390/plants13162273
Chicago/Turabian StyleHao, Yali, Rui Liu, Zhenchuan Mao, Qihong Yang, Shijie Zheng, Xiaofei Lu, Yuhong Yang, Bingyan Xie, Jianlong Zhao, Yan Li, and et al. 2024. "Identification and Analysis of WRKY Transcription Factors in Response to Cowpea Fusarium Wilt in Cowpea" Plants 13, no. 16: 2273. https://doi.org/10.3390/plants13162273
APA StyleHao, Y., Liu, R., Mao, Z., Yang, Q., Zheng, S., Lu, X., Yang, Y., Xie, B., Zhao, J., Li, Y., Chen, G., & Ling, J. (2024). Identification and Analysis of WRKY Transcription Factors in Response to Cowpea Fusarium Wilt in Cowpea. Plants, 13(16), 2273. https://doi.org/10.3390/plants13162273





