Metabolome and Mycobiome of Aegilops tauschii Subspecies Differing in Susceptibility to Brown Rust and Powdery Mildew Are Diverse
Abstract
1. Introduction
2. Results
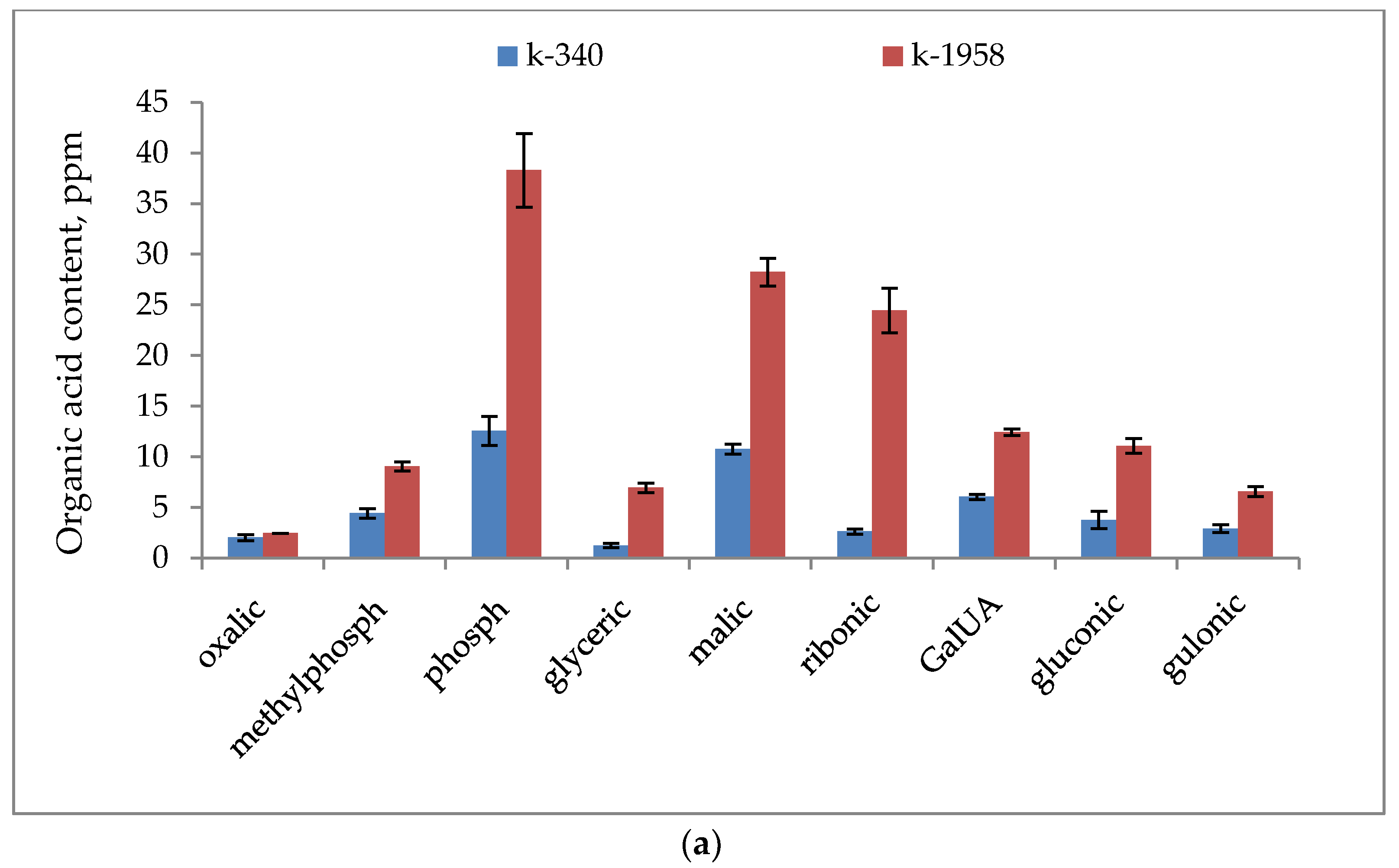
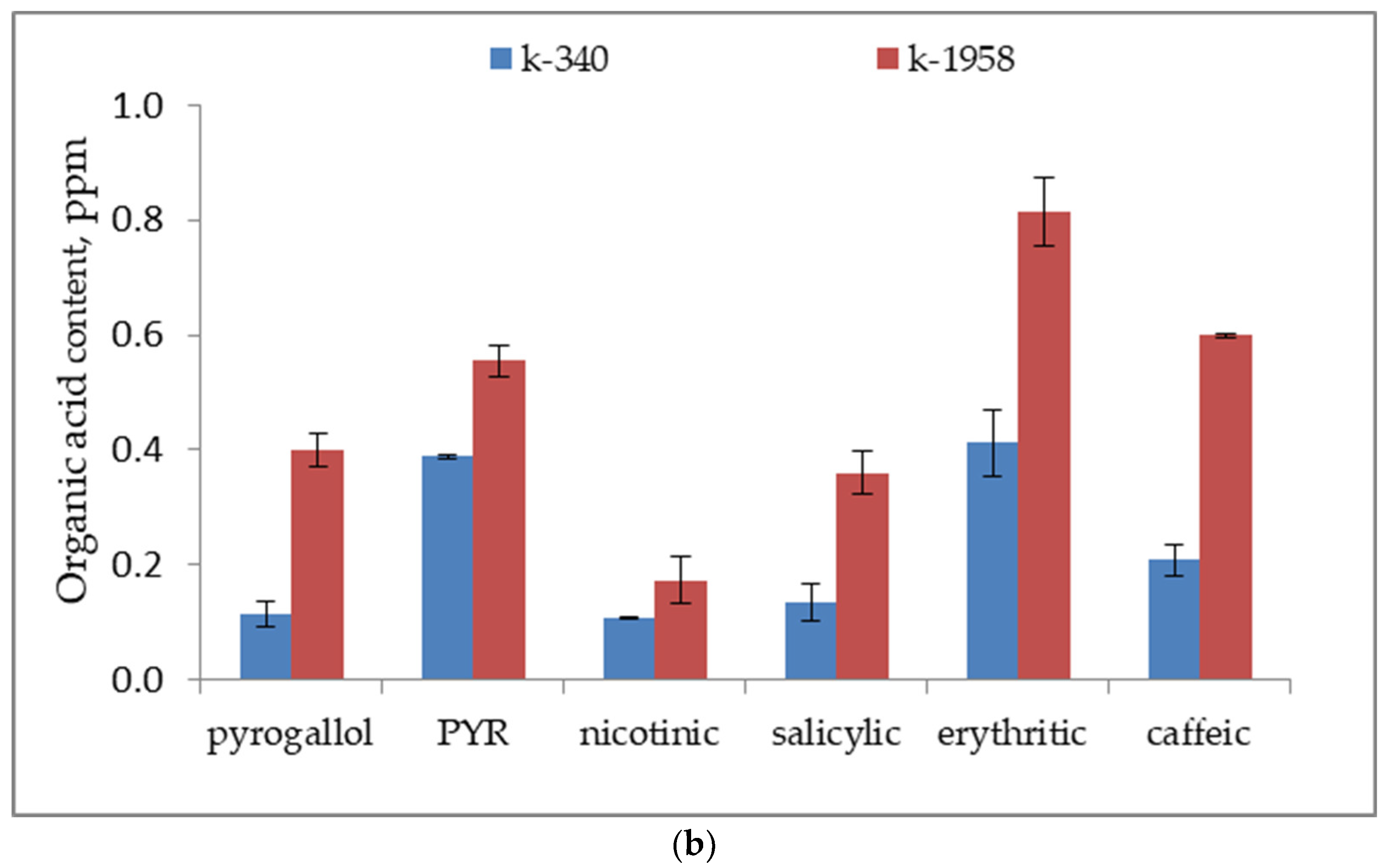
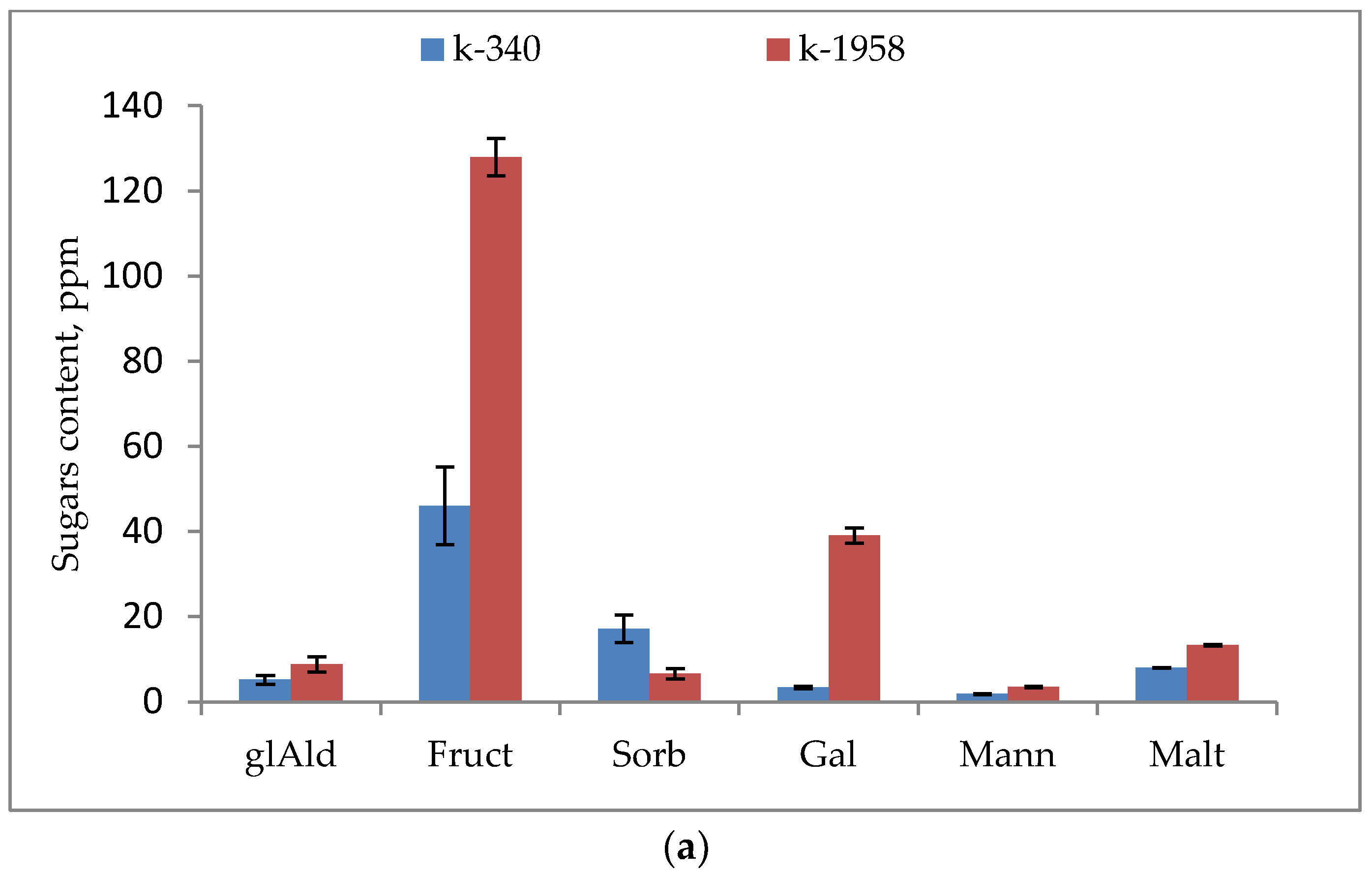
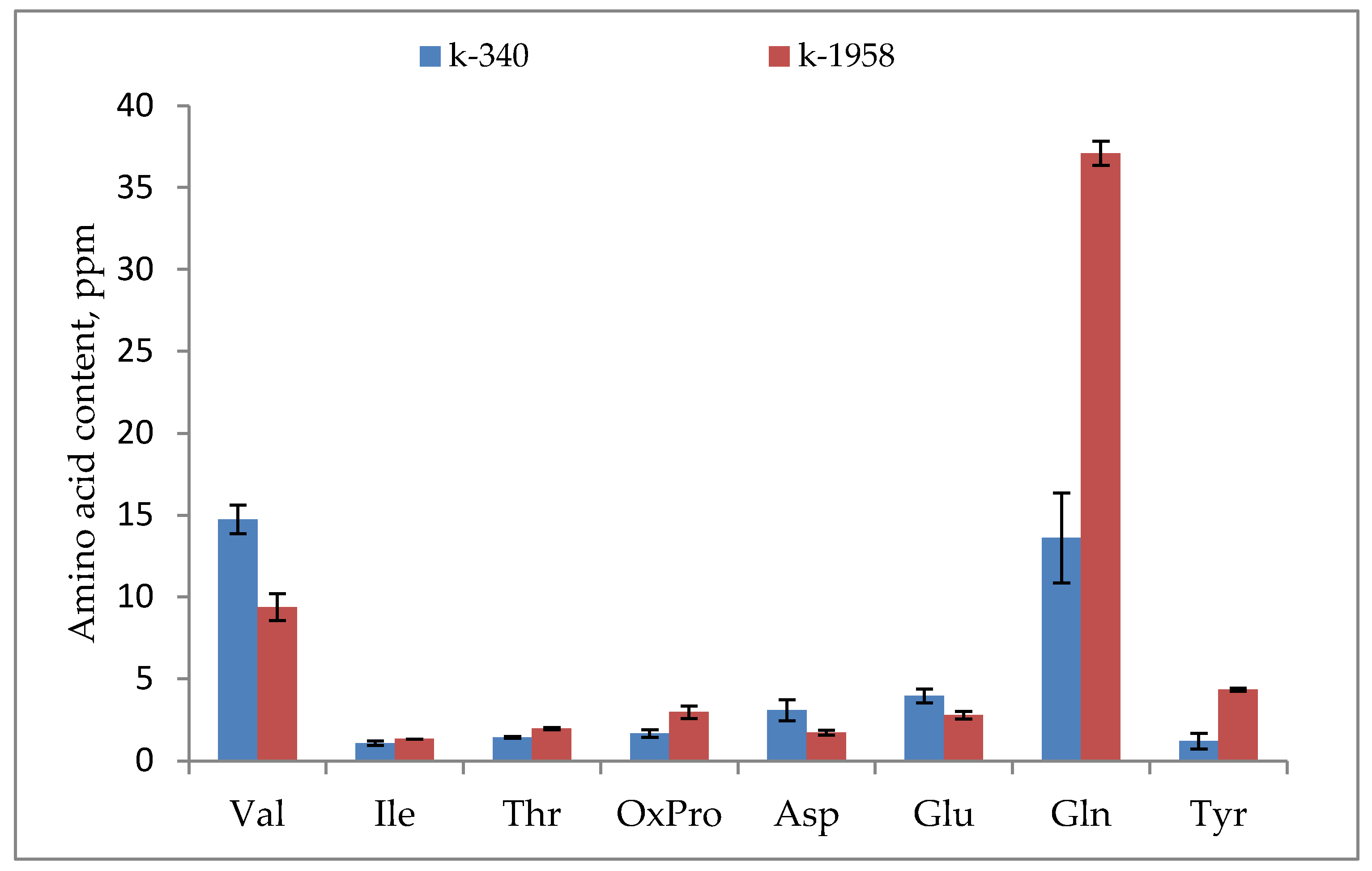
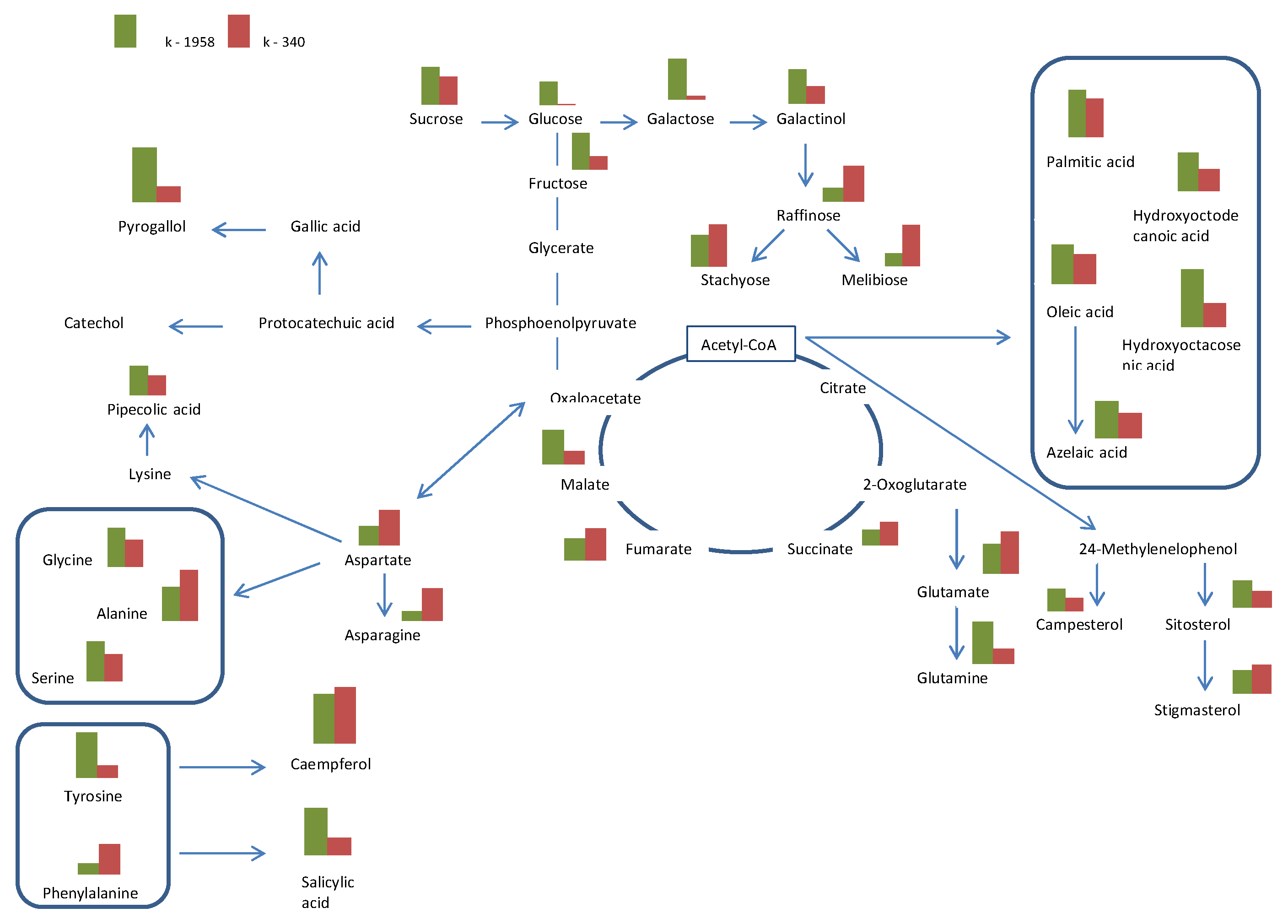
2.1. Metabolite Profiles of Ae. tauschii Seeds
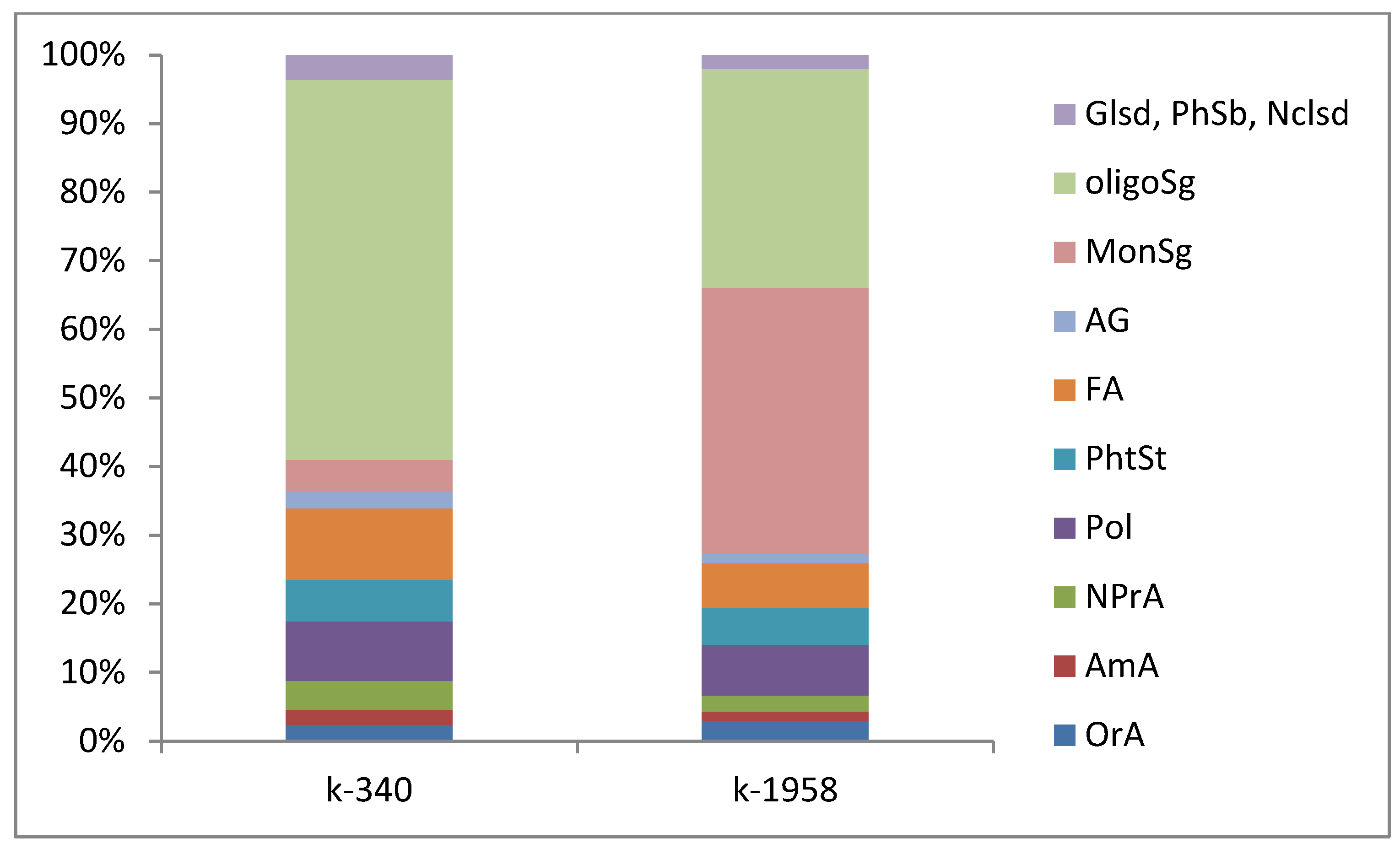
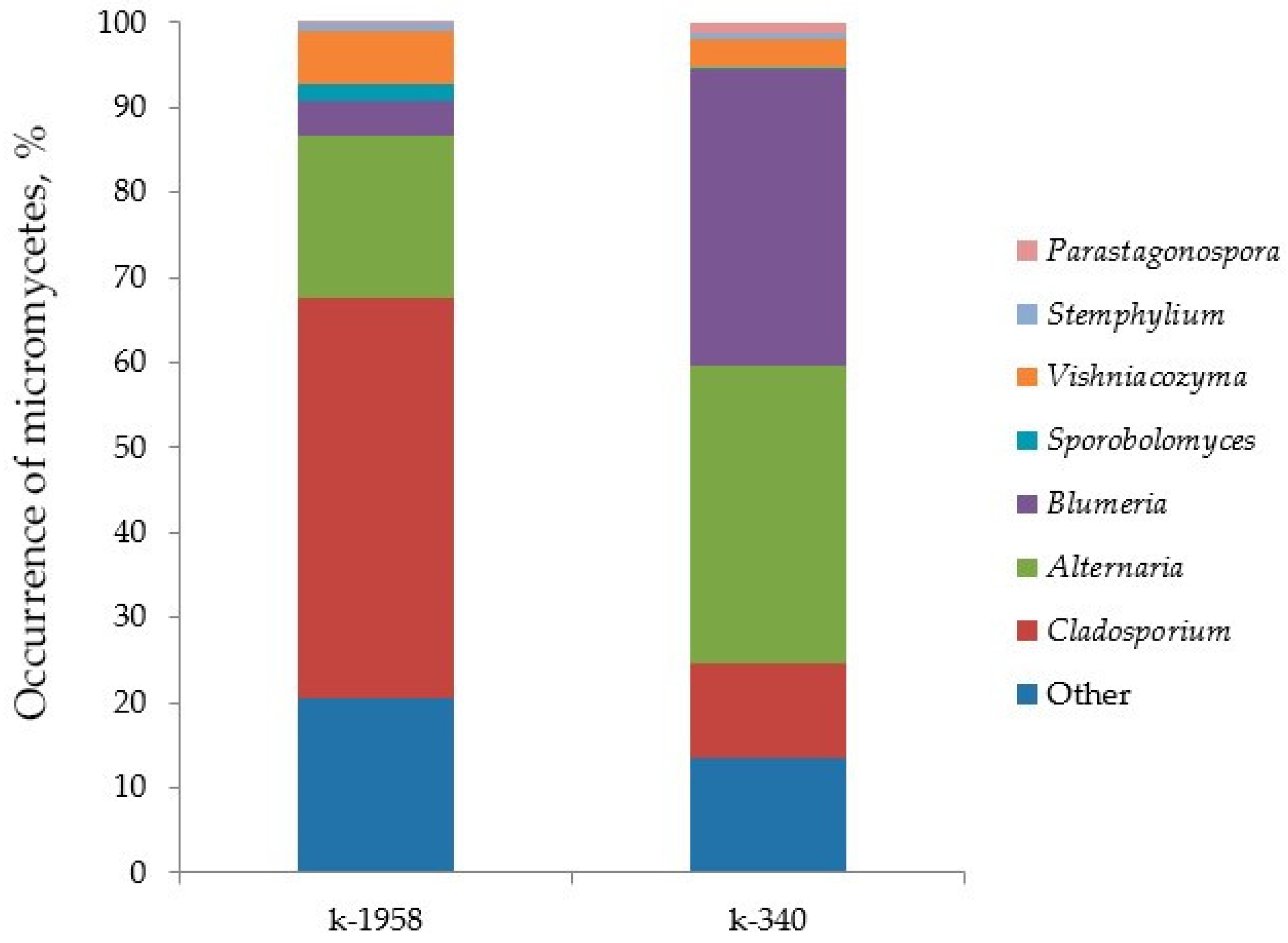
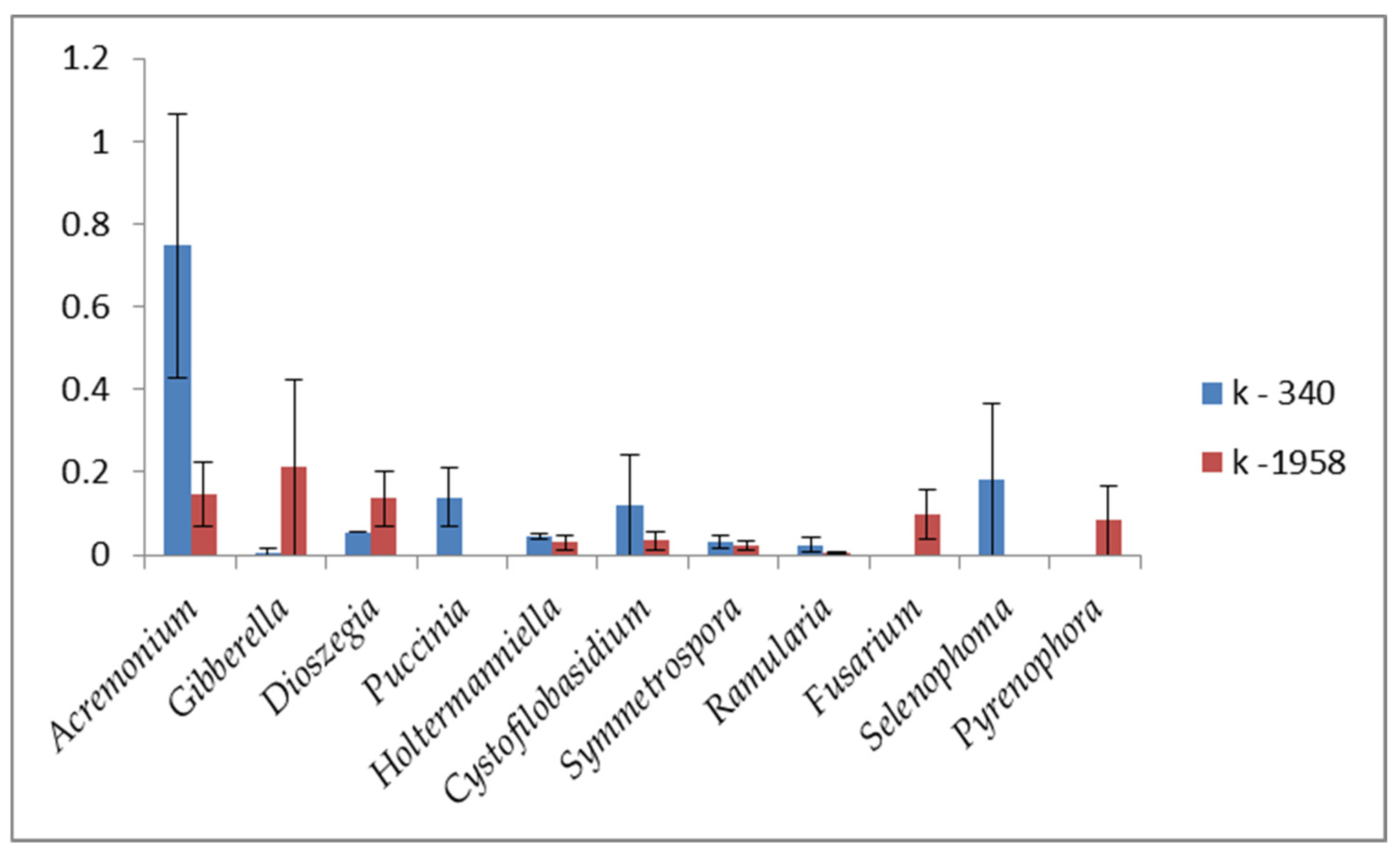
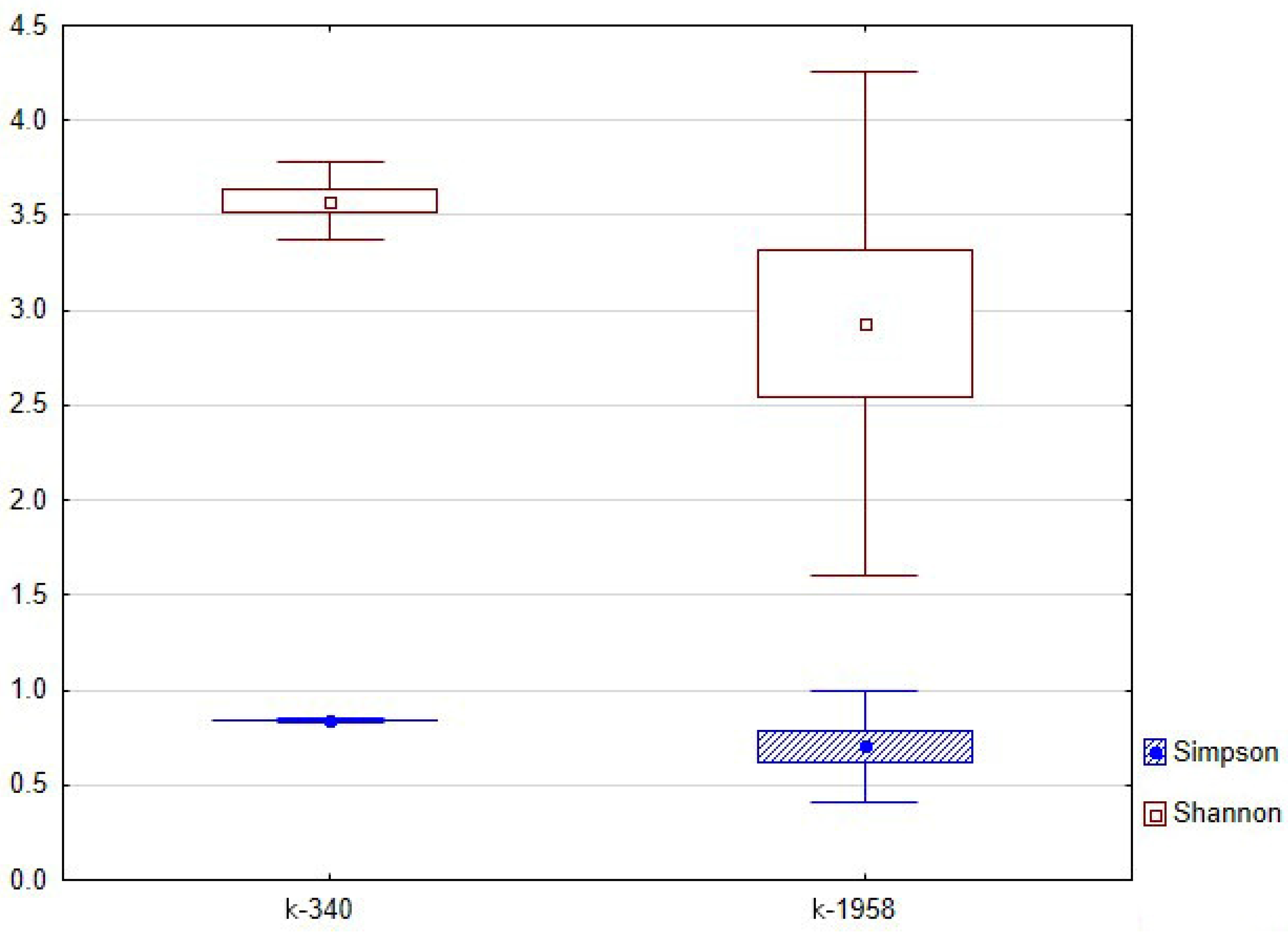
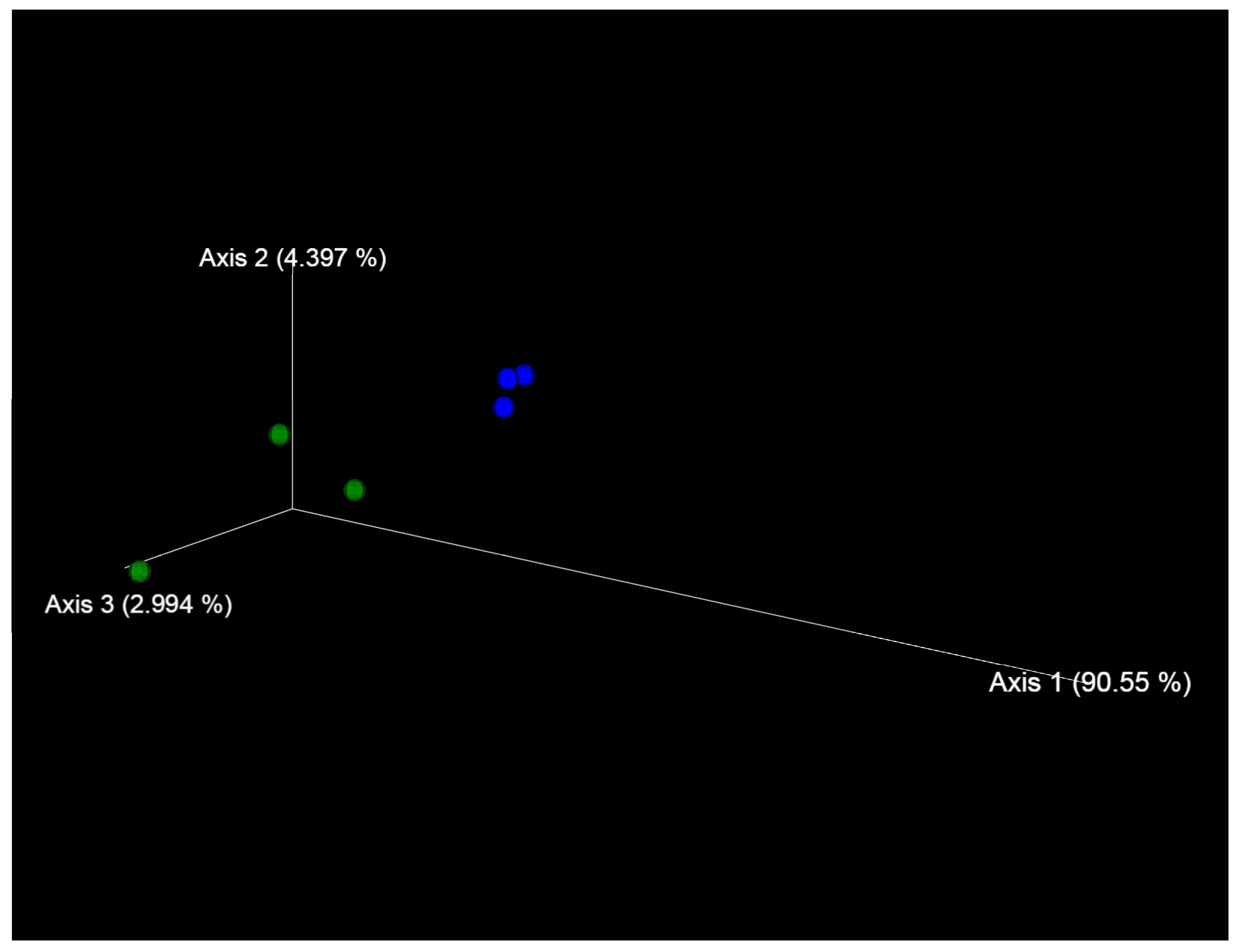
2.2. Fungal Microbiome (Mycobiome)
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Metabolome Analysis
4.3. Extraction of DNA, PCR and Sequencing
4.3.1. Isolation of Epiphytic Fungi
4.3.2. Sequencing ITS
4.3.3. Quantitative PCR
4.4. Systemic Analysis of Fungal Community and Metabolic Pathways
4.5. Statistical Analysis
5. Conclusions
- Our studies confirmed the difference in the seed metabolomic profiles of the studied accessions of Ae. tauschii which differed in field conditions in regard to their resistance to leaf rust and powdery mildew fungi. The resistant accession of Ae. tauschii ssp. strangulata k-1958 had a higher content of the main metabolites of glycolysis—glucose and pyruvic acid. The higher content of sucrose and stachyose in the grains of the resistant accession Ae tauschii k-1958 probably indicates the participation of maltose and stachyose in the formation of resistance of Ae tauschii to fungal pathogens. The content of plant metabolites acting as key components of systemic resistance induction, including salicylic, azelaic, and pipecolic acids, as well as galactinol, glycerol, and sitosterol was also significantly higher in the seeds of the resistant accession k-1958.
- Differences in the metabolome of Ae. tauschii seeds provided a different mycobiome of epiphytic micromycetes. The genera Alternaria, Blumeria, and Cladosporium dominated on the seeds of the mycobiome of susceptible accession Ae. tauschii ssp. meyeri k-340. The genera Cladosporium and Alternaria dominated on the seeds of resistant accession k-1958. Pathogens causing leaf diseases of plants were also found in the mycobiome of only the susceptible accession Ae. tauschii ssp. meyeri k-340, including Parastogonospora (1.15%) and Puccinia (0.14%). The mycobiome of the resistant accession k-1958 is characterized by a higher occurrence of saprotrophic microorganisms, many of which can be defined as potential biocontrol agents. These results on the composition of epiphytic micromycetes in the seed mycobiome of Ae. tauschii are preliminary and should be confirmed using a large number of seeds from diverse accessions differing in susceptibility to fungal diseases.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chebotar, V.K.; Chizhevskaya, E.P.; Andronov, E.E.; Vorobyov, N.I.; Keleinikova, O.V.; Baganova, M.E.; Konovalov, S.N.; Filippova, P.S.; Pishchik, V.N. Assessment of the rhizosphere bacterial community under maize growth using various agricultural technologies with biomodified mineral fertilizers. Agronomy 2023, 13, 1855. [Google Scholar] [CrossRef]
- Cole, B.J.; Feltcher, M.E.; Waters, R.J.; Wetmore, K.M.; Mucyn, T.S.; Ryan, E.M.; Wang, G.; Ul-Hasan, S.; McDonald, M.; Yoshikuni, Y.; et al. Genome-wide identification of bacterial plant colonization genes. PLoS Biol. 2017, 15, e2002860. [Google Scholar] [CrossRef]
- Su, Y.; Wang, J.; Gao, W.; Wang, R.; Yang, W.; Zhang, H.; Huang, L.; Guo, L. Dynamic metabolites: A bridge between plants and microbes. Sci. Total Environ. 2023, 899, 165612. [Google Scholar] [CrossRef] [PubMed]
- Broeckling, C.D.; Broz, A.K.; Bergelson, J.; Manter, D.K.; Jorge, M.; Vivanco, J.M. Root Exudates Regulate Soil Fungal Community Composition and Diversity. Appl. Environ. Microbiol. 2008, 74, 738–744. [Google Scholar] [CrossRef]
- Liu, F.; Hewezi, T.; Lebeis, S.L.; Pantalone, V.; Grewal, P.S.; Staton, M.E. Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 2019, 19, 201. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Zhang, T.; Wang, Z.; Li, G.; Zhao, W.; Lv, X.; Zhuang, L. Succession of endophytic fungi and arbuscular mycorrhizal fungi associated with the growth of plant and their correlation with secondary metabolites in the roots of plants. BMC Plant Biol. 2021, 21, 165. [Google Scholar] [CrossRef] [PubMed]
- Tiziani, R.; Miras-Moreno, B.; Malacrinò, A.; Vescio, R.; Lucini, L.; Mimmo, T.; Cesco, S.; Sorgona, A. Drought, heat, and their combination impact the root exudation patterns and rhizosphere microbiome in maize roots. Environ. Exp. Bot. 2022, 203, 105071. [Google Scholar] [CrossRef]
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef]
- Mastan, A.; Bharadwaj, R.; Kushwaha, R.K.; Vivek Babu, C.S. Functional fungal endophytes in Coleus forskohlii regulate labdane diterpene biosynthesis for elevated forskolin accumulation in roots. Microb. Ecol. 2019, 78, 914–926. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Koprivova, A.; Kopriva, S. Pinpointing secondary metabolites that shape the composition and function of the plant microbiome. J. Exp. Bot. 2021, 72, 57–69. [Google Scholar] [CrossRef]
- Pang, Z.; Chen, J.; Wang, T.; Gao, C.; Li, Z.; Guo, L.; Xu, J.; Cheng, Y. Linking plant secondary metabolites and plant microbiomes: A Review. Front. Plant Sci. 2021, 12, 621276. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Pánková, H.; Münzbergová, Z.; Rydlová, J.; Vosátka, M. Co-adaptation of plants and communities of arbuscular mycorrhizal fungi to their soil conditions. Folia Geobot. 2014, 49, 521–540. [Google Scholar] [CrossRef]
- Leroy, C.; Maes, A.Q.; Louisanna, E.; Schimann, H.; Séjalon-Delmas, N. Taxonomic, phylogenetic and functional diversity of root-associated fungi in bromeliads: Effects of host identity, life forms and nutritional modes. New Phytol. 2021, 231, 1195–1209. [Google Scholar] [CrossRef]
- Suryanarayanan, T.S.; Shaanker, R.U. Can fungal endophytes fast-track plant adaptations to climate change? Fungal Ecol. 2021, 50, 101039. [Google Scholar] [CrossRef]
- Carlström, C.I.; Field, C.M.; Bortfeld-Miller, M.; Müller, B.; Sunagawa, S.; Vorholt, J.A. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat. Ecol. Evol. 2019, 3, 1445–1454. [Google Scholar] [CrossRef]
- Michl, K.; Berg, G.; Cernava, T. The microbiome of cereal plants: The current state of knowledge and the potential for future applications. Environ. Microbiome 2023, 18, 28. [Google Scholar] [CrossRef]
- Hammer, K. Vorarbeiten zur monographischen Darstellung von Wildpflanzensortimenten: Aegilops L. Kulturpflanze 1980, 28, 33–180. (In German) [Google Scholar] [CrossRef]
- Mizuno, N.; Yamasaki, M.; Matsuoka, Y.; Kawahara, T.; Takumi, S. Population structure of wild wheat D-genome progenitor Aegilops tauschii Coss.: Implications for intraspecific lineage diversification and evolution of common wheat. Mol. Ecol. 2010, 19, 999–1013. [Google Scholar] [CrossRef]
- Wiersma, A.T.; Pulman, J.A.; Brown, L.K.; Cowger, C.; Olson, E.L. Identification of Pm58 from Aegilops tauschii. Theor. Appl. Genet. 2017, 130, 1123–1133. [Google Scholar] [CrossRef]
- Tyryshkin, L.G.; Kolesova, M.A. The use of molecular-genetic and phytopathological methods to identify genes for effective leaf rust resistance in Aegilops accessions. Proc. Appl. Bot. Genet. Breed. 2020, 181, 87–95. [Google Scholar] [CrossRef]
- Lin, G.; Chen, H.; Tian, B.; Sehgal, S.K.; Singh, L.; Xie, J.; Rawat, N.; Juliana, P.; Singh, N.; Shrestha, S.; et al. Cloning of the broadly effective wheat leaf rust resistance gene Lr42 transferred from Aegilops tauschii. Nat. Commun. 2022, 13, 3044. [Google Scholar] [CrossRef]
- Luo, M.; Xie, L.; Chakraborty, S.; Wang, A.; Matny, O.; Jugovich, M.A.; Kolmer, J.A.; Richardson, T.; Bhatt, D.; Hoque, M.; et al. A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nat. Biotechnol. 2021, 39, 561–566. [Google Scholar] [CrossRef]
- Huerta-Espino, J.; Singh, R.P.; Germán, S.; McCallum, B.D.; Park, R.F.; Chen, W.Q.; Bhardwaj, S.C.; Goyeau, H. Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 2011, 179, 143–160. [Google Scholar] [CrossRef]
- Markovska, O.; Dudchenko, V.; Grechishkina, T.T.; Stetsenko, I. Prevalence and harmfulness of winter wheat brown leaf rust (Puccinia recondita Rob. ex desm. f. sp. tritici) in the Southern Steppe of Ukraine. Ukr. J. Ecol. 2020, 10, 69–74. [Google Scholar] [CrossRef]
- Różewicz, M.; Wyzińska, M.; Grabiński, J. The most important fungal diseases of cereals—Problems and possible solutions. Agronomy 2021, 11, 714. [Google Scholar] [CrossRef]
- Mwale, V.M.; Chilembwe, H.C.; Uluko, H.C. Wheat powdery mildew (Blumeria graminis f. sp. tritici): Damage effects and genetic resistance developed in wheat (Triticum aestivum). Plant Sci. 2014, 5, 1–16. [Google Scholar]
- Shelenga, T.V.; Malyshev, L.L.; Kerv, Y.A.; Diubenko, T.V.; Konarev, A.V.; Horeva, V.I.; Belousova, M.K.; Kolesova, M.A.; Chikida, N.N. Metabolomic approach to search for fungal resistant forms of Aegilops tauschii Coss. from the VIR collection. Vavilovskii Zhurnal Genet. Sel. 2020, 24, 252–258. [Google Scholar] [CrossRef] [PubMed]
- KEGG PATHWAY Database. Available online: https://www.kegg.jp/kegg/pathway.html (accessed on 25 May 2024).
- The Americal Phytopathological Society (APS). Available online: https://www.apsnet.org/edcenter/resources/commonnames/Pages/Wheat.aspx (accessed on 25 May 2024).
- VENN DIAGRAM. Available online: https://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 25 May 2024).
- Kovanda, L.; Zhang, W.; Wei, X.; Luo, J.; Wu, X.; Atwill, E.R.; Vaessen, S.; Li, X.; Liu, Y. In vitro antimicrobial activities of organic acids and their derivatives on several species of gram-negative and gram-positive Bacteria. Molecules 2019, 24, 3770. [Google Scholar] [CrossRef]
- Ramírez-Gómez, X.; Jimenez Garcia, S.; Beltran-Campos, V.; Campos, M. Plant metabolites in plant defense against pathogens. In Plant Diseases—Current Threats and Management Trends; BoD: Norderstedt, Germany, 2019. [Google Scholar] [CrossRef]
- Khan, F.; Bamunuarachchi, N.I.; Tabassum, N.; Kim, Y.M. Caffeic acid and its derivatives: Antimicrobial drugs toward microbial pathogens. J. Agric. Food Chem. 2021, 69, 2979–3004. [Google Scholar] [CrossRef]
- Chizhevskaya, E.P.; Lapenko, N.G.; Chebotar, V.K. Antibacterial and antifungal activity of Chenopodium album L. Russ. J. Plant Physiol. 2023, 70, 194. [Google Scholar] [CrossRef]
- Shahzad, M.; Sherry, L.; Rajendran, R.; Edwards, C.A.; Combet, E.; Ramage, G. Utilising polyphenols for the clinical management of Candida albicans biofilms. Int. J. Antimicrob. Agents 2014, 44, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Qiu, Y.; Li, M.; Song, F.; Jiang, M. Functions of pipecolic acid on induced resistance against Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000 in tomato plants. J. Phytopathol. 2020, 168, 591–600. [Google Scholar] [CrossRef]
- Yao, D.; Zhang, G.; Chen, W.; Chen, J.; Li, Z.; Zheng, X.; Yin, H.; Hu, X. Pyrogallol and fluconazole interact synergistically in vitro against Candida glabrata through an efflux-associated mechanism. Antimicrob. Agents Chemother. 2021, 65, e0010021. [Google Scholar] [CrossRef]
- Mason, T.L.; Wasserman, B.P. Inactivation of red beet beta-glucan synthase by native and oxidized phenolic compounds. Phytochemistry 1987, 26, 2197–2202. [Google Scholar] [CrossRef]
- Kocaçalişkan, I.; Talan, I.; Terzi, I. Antimicrobial activity of catechol and pyrogallol as allelochemicals. Z. Naturforsch. C. J. Biosci. 2006, 61, 639–642. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, M.; Takada, K. Multiple effects of green tea catechin on the antifungal activity of antimycotics against Candida albicans. J. Antimicrob. Chemother. 2004, 53, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Ribnicky, D.M.; Shulaev, V.; Raskin, I. Intermediates of salicylic acid biosynthesis in tobacco. Plant Physiol. 1998, 118, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Zeier, J. N-hydroxypipecolic acid and salicylic acid: A metabolic duo for systemic acquired resistance. Curr. Opin. Plant Biol. 2019, 50, 44–57. [Google Scholar] [CrossRef]
- Huang, W.; Wang, Y.; Li, X.; Zhang, Y. Biosynthesis and regulation of salicylic acid and N-hydroxypipecolic acid in plant immunity. Mol. Plant 2020, 13, 31–41. [Google Scholar] [CrossRef]
- Dempsey, D.A.; Vlot, A.C.; Wildermuth, M.C.; Klessig, D.F. Salicylic acid biosynthesis and metabolism. Arab. Book 2011, 9, e0156. [Google Scholar] [CrossRef]
- Lim, G.-H.; Shine, M.; de Lorenzo, L.; Yu, K.; Cui, W.; Navarre, D.; Hunt, A.G.; Lee, J.-Y.; Kachroo, A.; Kachroo, P. Plasmodesmata localizing proteins regulate transport and signaling during systemic acquired immunity in plants. Cell Host Microbe 2016, 19, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Shah, L.; Rahman, S.; Riaz, M.W.; Yahya, M.; Xu, Y.J.; Liu, F.; Si, W.; Jiang, H.; Cheng, B. Plant defense mechanism and current understanding of salicylic acid and NPRs in activating SAR. Physiol. Mol. Plant Pathol. 2018, 104, 15–22. [Google Scholar] [CrossRef]
- da Rocha Neto, A.C.; Maraschin, M.; Di Piero, R.M. Antifungal activity of salicylic acid against Penicillium expansum and its possible mechanisms of action. Int. J. Food Microbiol. 2015, 215, 64–70. [Google Scholar] [CrossRef]
- Kachroo, A.; Liu, H.; Yuan, X.; Kurokawa, T.; Kachroo, P. Systemic acquired resistance-associated transport and metabolic regulation of salicylic acid and glycerol-3-phosphate. Essays Biochem. 2022, 66, 673–681. [Google Scholar] [CrossRef]
- Lim, G.H. Regulation of salicylic acid and n-hydroxy-pipecolic acid in systemic acquired resistance. Plant Pathol. J. 2023, 39, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Han, X.; Kahmann, R. Microbial effectors target multiple steps in the salicylic acid production and signaling pathway. Front. Plant Sci. 2015, 6, 349. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, H.; Sami, F.; Hayat, S. Glucose: Sweet or bitter effects in plants-a review on current and future perspective. Carbohydr. Res. 2020, 487, 107884. [Google Scholar] [CrossRef]
- Plaxton, W.C. The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 185–214. [Google Scholar] [CrossRef]
- Janse Van Rensburg, H.C.; Van den Ende, W.A. Potential signaling molecule in plants? Front. Plant Sci. 2018, 8, 2230. [Google Scholar] [CrossRef]
- Morkunas, I.; Ratajczak, L. The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol. Plant 2014, 36, 1607–1619. [Google Scholar] [CrossRef]
- Genga, A.; Mattana, M.; Coraggio, I.; Locatelli, F.; Piffanelli, P.; Consonni, R. Plant metabolomics: A characterisation of plant responses to abiotic stresses. In Abiotic Stress Plants—Mechanisms and Adaptations; Shanker, A., Venkateswarlu, S., Eds.; InTech: Houston, TX, USA, 2011. [Google Scholar] [CrossRef]
- Ibrahim, H.A.; Abdellatif, Y.M.R. Effect of maltose and trehalose on growth, yield and some biochemical components of wheat plant under water stress. Ann. Agric. Sci. 2016, 61, 267–274. [Google Scholar] [CrossRef]
- Lü, J.; Sui, X.; Ma, S.; Li, X.; Liu, H.; Zhang, Z. Suppression of cucumber stachyose synthase gene (CsSTS) inhibits phloem loading and reduces low temperature stress tolerance. Plant Mol. Biol. 2017, 95, 1–15. [Google Scholar] [CrossRef]
- Chibbar, R.N.; Jaiswal, S.; Gangola, M.; Båga, M. Carbohydrate Metabolism, Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar] [CrossRef]
- Trouvelot, S.; Héloir, M.-C.; Poinssot, B.; Gauthier, A.; Paris, F.; Guillier, C.; Combier, M.; Trdá, L.; Daire, X.; Adrian, M. Carbohydrates in plant immunity and plant protection: Roles and potential application as foliar sprays. Front. Plant Sci. 2014, 5, 592. [Google Scholar] [CrossRef]
- Vera, J.; Castro, J.; Gonzalez, A.; Moenne, A. Seaweed polysaccharides and derived oligosaccharides stimulate defense responses and protection against pathogens in plants. Mar. Drugs 2011, 9, 2514–2525. [Google Scholar] [CrossRef]
- Jung, H.W.; Tschaplinski, T.J.; Wang, L.; Glazebrook, J.; Greenberg, J.T. Priming in systemic plant immunity. Science 2009, 324, 89–91. [Google Scholar] [CrossRef]
- Chanda, B.; Xia, Y.; Mandal, M.K.; Yu, K.; Sekine, K.-T.; Gao, O.-M.; Selote, D.; Hu, Y.; Stromberg, A.; Navarre, D.; et al. Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 2011, 43, 421–427. [Google Scholar] [CrossRef]
- Návarová, H.; Bernsdorff, F.; Döring, A.-C.; Zeier, J. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 2012, 24, 5123–5141. [Google Scholar] [CrossRef]
- Zeier, J. Metabolic regulation of systemic acquired resistance. Curr. Opin. Plant Biol. 2021, 62, 102050. [Google Scholar] [CrossRef]
- Cho, S.M.; Kang, E.Y.; Kim, M.S.; Yoo, S.J.; Im, Y.J.; Kim, Y.C.; Yang, K.Y.; Kim, K.Y.; Kim, K.S.; Choi, Y.S.; et al. Jasmonate-dependent expression of a galactinol synthase gene is involved in priming of systemic fungal resistance in Arabidopsis thaliana. Botany 2010, 88, 452–461. [Google Scholar] [CrossRef]
- Dorostkar, S.; Dadkhodaie, A.; Ebrahimie, E.; Heidari, B.; Ahmadi-Kordshooli, M. Comparative transcriptome analysis of two contrasting resistant and susceptible Aegilops tauschii accessions to wheat leaf rust (Puccinia triticina) using RNA-sequencing. Sci. Rep. 2022, 12, 821. [Google Scholar] [CrossRef]
- Trovato, M.; Funck, D.; Forlani, G.; Okumoto, S.; Amir, R. Editorial: Amino acids in plants: Regulation and functions in development and stress defense. Front. Plant Sci. 2021, 12, 772810. [Google Scholar] [CrossRef]
- Celenza, J.L. Metabolism of tyrosine and tryptophan--new genes for old pathways. Curr. Opin. Plant Biol. 2001, 4, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Rachidi, F.; Benhima, R.; Kasmi, Y.; Sbabou, L.; Arroussi, H.E. Evaluation of microalgae polysaccharides as biostimulants of tomato plant defense using metabolomics and biochemical approaches. Sci. Rep. 2021, 11, 930. [Google Scholar] [CrossRef] [PubMed]
- Ziv, C.; Zhao, Z.; Gao, Y.G.; Xia, Y. Multifunctional roles of plant cuticle during plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1088. [Google Scholar] [CrossRef]
- Kaur, S.; Samota, M.K.; Choudhary, M.; Choudhary, M.; Pandey, A.; Sharma, A.; Thakur, J. How do plants defend themselves against pathogens-biochemical mechanisms and genetic interventions. Physiol. Mol. Biol. Plants 2022, 28, 485–504. [Google Scholar] [CrossRef] [PubMed]
- Conde, A.; Regalado, A.; Rodrigues, D.; Costa, J.M.; Blumwald, E.; Chaves, M.M.; Gerós, H. Polyols in grape berry: Transport and metabolic adjustments as a physiological strategy for water-deficit stress tolerance in grapevine. J. Exp. Bot. 2015, 66, 889–906. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Yu, H.; Fan, J.; Zhang, J.; Su, L.; Li, D.; Pan, H.; Zhang, X. Crucial Roles of the High-Osmolarity Glycerol Pathway in the Antifungal Activity of Isothiocyanates against Cochliobolus heterostrophus. J. Agric. Food Chem. 2023, 71, 15466–15475. [Google Scholar] [CrossRef]
- Tatebayashi, K.; Yamamoto, K.; Tomida, T.; Nishimura, A.; Takayama, T.; Oyama, M.; Kozuka-Hata, H.; Adachi-Akahane, S.; Tokunaga, Y.; Saito, H. Osmostress enhances activating phosphorylation of Hog1 MAP kinase by mono-phosphorylated Pbs2 MAP2K. EMBO J. 2020, 39, e103444. [Google Scholar] [CrossRef]
- Fagard, M.; Launay, A.; Clément, G.; Courtial, J.; Dellagi, A.; Farjad, M.; Krapp, A.; Soulié, M.-C.; Masclaux-Daubresse, C. Nitrogen metabolism meets phytopathology. J. Exp. Bot. 2014, 65, 5643–5656. [Google Scholar] [CrossRef]
- Qiu, X.M.; Sun, Y.Y.; Ye, X.Y.; Li, Z.G. Signaling role of glutamate. Front. Plant Sci. 2020, 10, 1743. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Yang, F.; He, X.; Du, X.; Mu, P.; Ma, W. Advances in the functional study of glutamine synthetase in plant abiotic stress tolerance response. Crop J. 2022, 10, 917–923. [Google Scholar] [CrossRef]
- Shitan, N. Secondary metabolites in plants: Transport and self-tolerance mechanisms. Biosci. Biotechnol. Biochem. 2016, 80, 1283–1293. [Google Scholar] [CrossRef]
- Monreal, C.M.; Schnitzer, M. The chemistry and biochemistry of organic components in the soil solutions of wheat rhizospheres. In Advances in Agronomy; Chapter 4; Sparks, D.L., Ed.; Elsevier: Amsterdam, The Netherlands, 2013; Volume 121, pp. 179–251. [Google Scholar] [CrossRef]
- Edreva, A.M.; Velikova, V.; Tsonev, T. Phenylamides in plants. Russ. J. Plant Physiol. 2007, 54, 287–301. [Google Scholar] [CrossRef]
- Noecker, C.; Chiu, H.C.; McNally, C.P.; Borenstein, E. Defining and evaluating microbial contributions to metabolite variation in microbiome-metabolome association studies. mSystems 2019, 4, e00579-19. [Google Scholar] [CrossRef]
- Wang, H.; Guo, Y.; Luo, Z.; Gao, L.; Li, R.; Zhang, Y.; Kalaji, H.M.; Qiang, S.; Chen, S. Recent advances in Alternaria phytotoxins: A review of their occurrence, structure, bioactivity, and biosynthesis. J. Fungi 2022, 8, 168. [Google Scholar] [CrossRef] [PubMed]
- Gannibal, P.B.; Orina, A.S.; Kononenko, G.P.; Burkin, A.A. Distinction of Alternaria Sect. Pseudoalternaria strains among other Alternaria fungi from cereals. J. Fungi 2022, 8, 423. [Google Scholar] [CrossRef]
- Perelló, A.; Moreno, M.; Sisterna, M. Alternaria infectoria species-group associated with black point of wheat in Argentina. Plant Pathol. 2008, 57, 379. [Google Scholar] [CrossRef]
- Somma, S.; Amatulli, M.T.; Masiello, M.; Moretti, A.; Logrieco, A.F. Alternaria species associated to wheat black point identified through a multilocus sequence approach. Int. J. Food Microbiol. 2019, 293, 34–43. [Google Scholar] [CrossRef]
- Jankovics, T.; Komáromi, J.; Fábián, A.; Jäger, K.; Vida, G.; Kiss, L. New insights into the life cycle of the wheat powdery mildew: Direct observation of ascosporic infection in Blumeria graminis f. sp. tritici. Phytopathology 2015, 105, 797–804. [Google Scholar] [CrossRef]
- Zhao, J.; Fang, Y.; Chu, G.; Yan, H.; Hu, L.; Huang, L. Identification of leaf-scale wheat powdery mildew (Blumeria graminis f. sp. tritici) combining hyperspectral imaging and an SVM classifier. Plants 2020, 9, 936. [Google Scholar] [CrossRef]
- Li, M.; Yang, Z.; Liu, J.; Chang, C. Wheat susceptibility genes TaCAMTA2 and TaCAMTA3 negatively regulate post-penetration resistance against Blumeria graminis forma sp. tritici. Int. J. Mol. Sci. 2023, 24, 10224. [Google Scholar] [CrossRef]
- Barkat, E.; Hardy, G.; Ren, Y.; Calver, M.; Bayliss, K. Fungal contaminants of stored wheat vary between Australian states. Australas. Plant Pathol. 2016, 45, 621–628. [Google Scholar] [CrossRef]
- Bensch, K.; Braun, U.; Groenewald, J.Z.; Crous, P.W. The genus Cladosporium. Stud. Mycol. 2012, 72, 1–401. [Google Scholar] [CrossRef] [PubMed]
- Golosna, L. Mycobiota of wheat seeds with signs of “Black Point” under conditions of forest-steppe and forest zones of Ukraine. Chem. Proc. 2022, 10, 93. [Google Scholar] [CrossRef]
- Hernández, G.; Ramos, B.; Sultani, H.N.; Ortiz, Y.; Spengler, I.; Castañeda, R.F.; Rivera, D.G.; Arnold, N.; Westermann, B.; Mirabal, Y. Cultural characterization and antagonistic activity of Cladobotryum virescens against some phytopathogenic fungi and oomycetes. Agronomy 2023, 13, 389. [Google Scholar] [CrossRef]
- Khan, A.M.; Khan, M.; Salman, H.M.; Ghazali, H.M.Z.U.; Ali, R.I.; Hussain, M.; Yousaf, M.M.; Hafeez, Z.; Khawja, M.S.; Alharbi, S.A.; et al. Detection of seed-borne fungal pathogens associated with wheat (Triticum aestivum L.) seeds collected from farmer fields and grain market. J. King Saud Univ. Sci. 2023, 35, 102590. [Google Scholar] [CrossRef]
- Knorr, K.; Jørgensen, L.N.; Nicolaisen, M. Fungicides have complex effects on the wheat phyllosphere mycobiome. PLoS ONE 2019, 14, e0213176. [Google Scholar] [CrossRef]
- Bartosiak, S.F.; Arseniuk, E.; Szechyńska-Hebda, M.; Bartosiak, E. Monitoring of natural occurrence and severity of leaf and glume blotch diseases of winter wheat and winter triticale incited by necrotrophic fungi Parastagonospora spp. and Zymoseptoria tritici. Agronomy 2021, 11, 967. [Google Scholar] [CrossRef]
- Croll, D.; Crous, P.W.; Pereira, D.; Mordecai, E.A.; McDonald, B.A.; Brunner, P.C. Genome-scale phylogenies reveal relationships among Parastagonospora species infecting domesticated and wild grasses. Persoonia 2021, 46, 116–128. [Google Scholar] [CrossRef]
- Kushnirenko, I.; Shreyder, E.; Bondarenko, N.; Shaydayuk, E.; Kovalenko, N.; Titova, J.; Gultyaeva, E. Genetic protection of soft wheat from diseases in the southern ural of Russia and virulence variability of foliar pathogens. Agriculture 2021, 11, 703. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Hernández-Restrepo, M.; Iturrieta-González, I.; García, D.; Gené, J.; Groenewald, J.Z.; Cai, L.; Chen, Q.; Quaedvlieg, W.; Schumacher, R.K.; et al. Genera of phytopathogenic fungi: GOPHY 3. Stud. Mycol. 2019, 94, 1–124. [Google Scholar] [CrossRef]
- Kim, M.; Choi, G.; Lee, H. Property of Curcuma longa L. rhizome-derived curcumin against phytopathogenic fungi in a greenhouse. J. Agric. Food Chem. 2003, 51, 1578–1581. [Google Scholar] [CrossRef]
- Chen, W.; Wellings, C.; Chen, X.; Kang, Z.; Liu, T. Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 2014, 15, 433–446. [Google Scholar] [CrossRef]
- Jalli, M.; Laitinen, P.; Latvala, S. The emergence of cereal fungal diseases and the incidence of leaf spot diseases in Finland. Agric. Food Sci. 2011, 20, 62–73. [Google Scholar] [CrossRef]
- Palágyi, A.; Bakonyi, J.; Tar, M.; Cséplő, M.; Csősz, M. Isolation and identification of Pyrenophora chaetomioides from winter oat in Hungary. Cereal Res. Commun. 2020, 48, 57–63. [Google Scholar] [CrossRef]
- Strelkov, S.E.; Lamari, L. Host–parasite interactions in tan spot (Pyrenophora tritici-repentis) of wheat. Can. J. Plant Pathol. 2003, 25, 339–349. Available online: http://hdl.handle.net/1993/2776 (accessed on 15 August 2024). [CrossRef]
- Francioli, D.; Cid, G.; Hajirezaei, M.-R.; Kolb, S. Response of the wheat mycobiota to flooding revealed substantial shifts towards plant pathogens. Front. Plant Sci. 2022, 13, 1028153. [Google Scholar] [CrossRef]
- Imathiu, S.M.; Ray, R.V.; Back, M.; Hare, M.C.; Edwards, S.G. Fusarium langsethiae pathogenicity and aggressiveness towards oats and wheat in wounded and unwounded in vitro detached leaf assays. Eur. J. Plant Pathol. 2009, 124, 117–126. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; De Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Vujanovic, V.; Mavragani, D.; Hamel, C. Fungal communities associated with durum wheat production system: A characterization by growth stage, plant organ and preceding crop. Crop Prot. 2012, 37, 26–34. [Google Scholar] [CrossRef]
- Fokkema, N.J. Competition for endogenous and exogenous nutrients between Sporobolomyces roseus and Cochliobolus sativus. Can. J. Bot. 1984, 62, 2463–2468. [Google Scholar] [CrossRef]
- Sanzani, S.M.; Sgaramella, M.; Mosca, S.; Solfrizzo, M.; Ippolito, A. control of Penicillium expansum by an epiphytic basidiomycetous yeast. Horticulturae 2021, 7, 473. [Google Scholar] [CrossRef]
- Latz, M.A.C.; Jensen, B.; David, B.; Collinge, D.B.; Jørgensen, H.J.L. Identification of two endophytic fungi that control Septoria tritici blotch in the field, using a structured screening approach. Biol. Control 2019, 141, 104128. [Google Scholar] [CrossRef]
- Rojas, E.C.; Jensen, B.; Jørgensen, H.J.L.; Latz, M.A.C.; Esteban, P.; Ding, Y.; Collinge, D.B. Selection of fungal endophytes with biocontrol potential against Fusarium head blight in wheat. Biol. Control 2020, 144, 104222. [Google Scholar] [CrossRef]
- Ownley, B.H.; Griffin, M.R.; Klingeman, W.E.; Gwinn, K.D.; Moulton, J.K.; Pereira, R.M. Beauveria bassiana: Endophytic colonization and plant disease control. J. Invertebr. Pathol. 2008, 98, 267–270. [Google Scholar] [CrossRef]
- Volkova, G.V.; Kremneva, O.Y.; Shumilov, Y.V.; Gladkova, E.V.; Vaganova, O.F.; Mitrofanova, O.P.; Lysenko, N.S.; Chikida, N.N.; Khakimova, A.G.; Zuev, E.V. Immunological assessment of wheat accessions, its rare species, aegilops from the collection federal research center “Vavilov all-russian institute of genetic resources” and selection of sources with group resistance. Plant Prot. News 2016, 3, 38–39. (In Russian) [Google Scholar]
- Batasheva, B.A.; Abdullaev, R.A.; Kovaleva, O.N.; Zveinek, I.A.; Radchenko, E.E. Powdery mildew resistance of barley in Southern Dagestan. Proc. Appl. Bot. Genet. Breed 2021, 182, 153–156. [Google Scholar] [CrossRef]
- Loskutov, I.G.; Shelenga, T.V.; Konarev, A.V.; Shavarda, A.L.; Blinova, E.V.; Dzubenko, N.I. The metabolomic approach to the comparative analysis of wild and cultivated species of oats (Avena L.). Russ. J. Genet. Appl. Res. 2017, 7, 501–508. [Google Scholar] [CrossRef]
- Solanki, M.K.; Abdelfattah, A.; Sadhasivam, S.; Zakin, V.; Wisniewski, M.; Droby, S.; Sionov, E. Analysis of stored wheat grain-associated microbiota reveals biocontrol activity among microorganisms against mycotoxigenic fungi. J. Fungi 2021, 7, 781. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, R.B. Asessment of soil microbial community structure by use of taxon specific quantitative PCR assays. Appl. Environ. Microb. 2005, 71, 4117–4120. [Google Scholar] [CrossRef] [PubMed]


| Metabolites | Metabolites Contents, ppm | |
|---|---|---|
| k-340 | k-1958 | |
| Salicylic acid * | 0.13 ± 0.03 | 0.36 ± 0.04 |
| Pyrogallol * | 0.11 ± 0.02 | 0.40 ± 0.03 |
| Azelaic acid * | 1.86 ± 0.20 | 2.73 ± 0.17 |
| Pipecolic acid * | 0.34 ± 0.01 | 0.51 ± 0.03 |
| Glycerol * | 59.68 ± 3.98 | 76.71 ± 3.21 |
| Galactinol * | 77.58 ± 2.80 | 151.05 ± 8.82 |
| Sitosterol | 186.34 ± 26.21 | 304.23 ± 10.27 |
| Blast ID | Phylum | Genus Anamorph/Teleomorph | Disease in Wheat | Relative Abundance, % | |
|---|---|---|---|---|---|
| k-1958 | k-340 | ||||
| Cladosporium | Ascomycota | Cladosporium/Davidiella | Non-pathogenic/black head mold/black point smudge | 47.14 ± 22.11 | 12.35 ± 3.07 |
| Alternaria infectoria | Ascomycota | Alternaria | Black point | 15.53 ± 5.19 | 30.67 ± 2.43 |
| Blumeria graminis | Ascomycota | Blumeria | Powdery mildew | 0 | 7.30 ± 1.99 |
| Vishniacozyma | Basidiomycota | Vishniacozyma | Non-pathogenic | 6.15 ± 3.35 | 3.58 ± 0.49 |
| Sporobolomyces roseus | Basidiomycota | Sporobolomyces | Non-pathogenic | 2.06 ± 1.55 | 0.40 ± 0.04 |
| Parastogonospora | Ascomycota | Parastogonospora/Phaeosphaeria | Spot blotch | 0 | 1.28 ± 0.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pishchik, V.N.; Chizhevskaya, E.P.; Kichko, A.A.; Aksenova, T.S.; Andronov, E.E.; Chebotar, V.K.; Filippova, P.S.; Shelenga, T.V.; Belousova, M.H.; Chikida, N.N. Metabolome and Mycobiome of Aegilops tauschii Subspecies Differing in Susceptibility to Brown Rust and Powdery Mildew Are Diverse. Plants 2024, 13, 2343. https://doi.org/10.3390/plants13172343
Pishchik VN, Chizhevskaya EP, Kichko AA, Aksenova TS, Andronov EE, Chebotar VK, Filippova PS, Shelenga TV, Belousova MH, Chikida NN. Metabolome and Mycobiome of Aegilops tauschii Subspecies Differing in Susceptibility to Brown Rust and Powdery Mildew Are Diverse. Plants. 2024; 13(17):2343. https://doi.org/10.3390/plants13172343
Chicago/Turabian StylePishchik, Veronika N., Elena P. Chizhevskaya, Arina A. Kichko, Tatiana S. Aksenova, Evgeny E. Andronov, Vladimir K. Chebotar, Polina S. Filippova, Tatiana V. Shelenga, Maria H. Belousova, and Nadezhda N. Chikida. 2024. "Metabolome and Mycobiome of Aegilops tauschii Subspecies Differing in Susceptibility to Brown Rust and Powdery Mildew Are Diverse" Plants 13, no. 17: 2343. https://doi.org/10.3390/plants13172343
APA StylePishchik, V. N., Chizhevskaya, E. P., Kichko, A. A., Aksenova, T. S., Andronov, E. E., Chebotar, V. K., Filippova, P. S., Shelenga, T. V., Belousova, M. H., & Chikida, N. N. (2024). Metabolome and Mycobiome of Aegilops tauschii Subspecies Differing in Susceptibility to Brown Rust and Powdery Mildew Are Diverse. Plants, 13(17), 2343. https://doi.org/10.3390/plants13172343








