Physiological and Proteomic Analysis of Various Priming on Rice Seed under Chilling Stress
Abstract
:1. Introduction
2. Material and Methods
2.1. Experimental Design and Treatment Details
2.2. Seed Germination Measurement and Sampling
2.3. Total Protein Extraction
2.4. Protein Digestion and TMT Labeling
2.5. High pH RPLC Separation
2.6. LC-MS/MS Analysis
2.7. Identification of Proteins
2.8. RNA Extraction and Quantitative Real-Time Transcription Polymerase Chain Reaction (qRT-PCR)
2.9. Statistical Analysis
3. Results
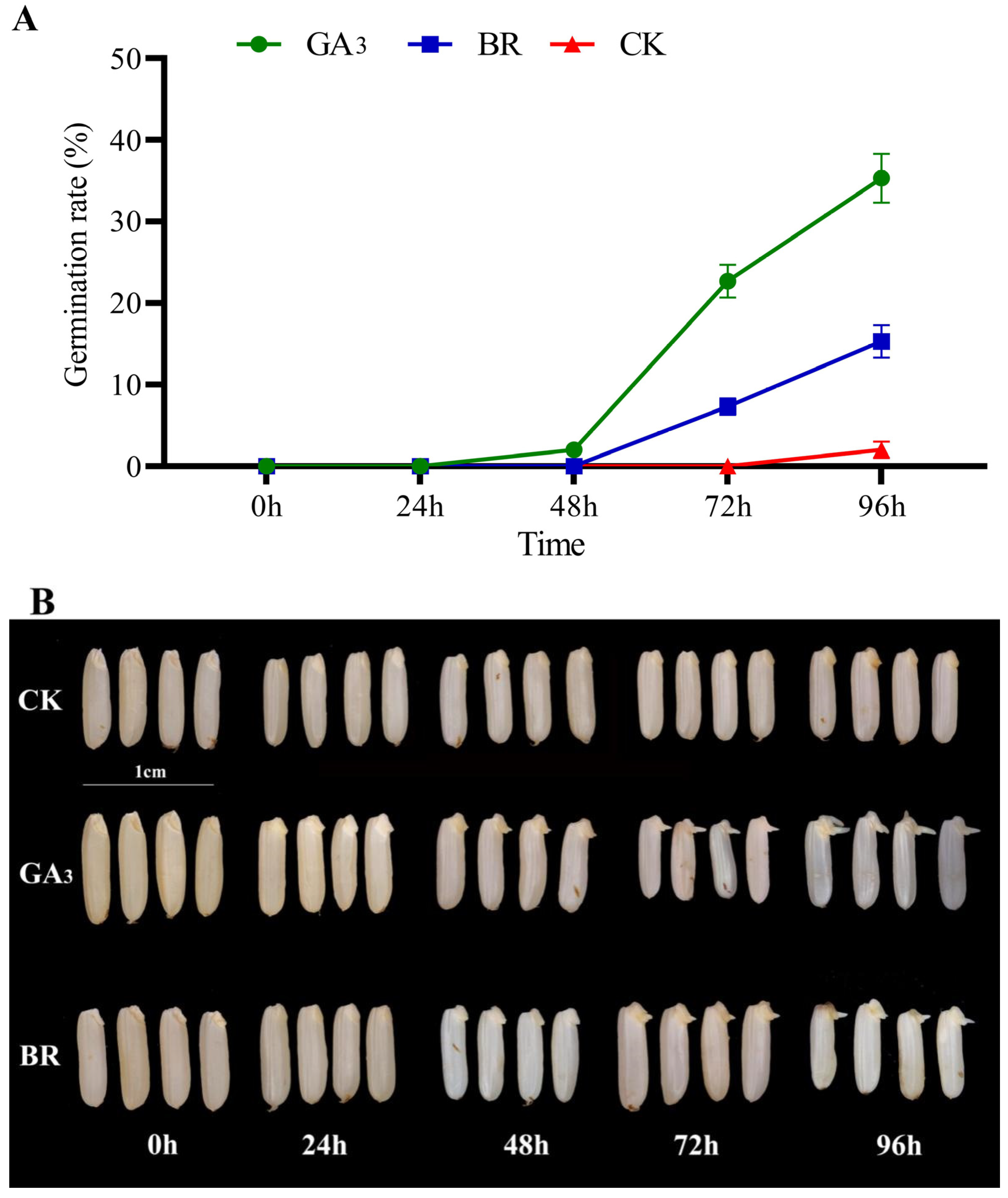
3.1. The Effect of Different Initiators on the Germination Rate of Rice Seeds under Chilling Stress
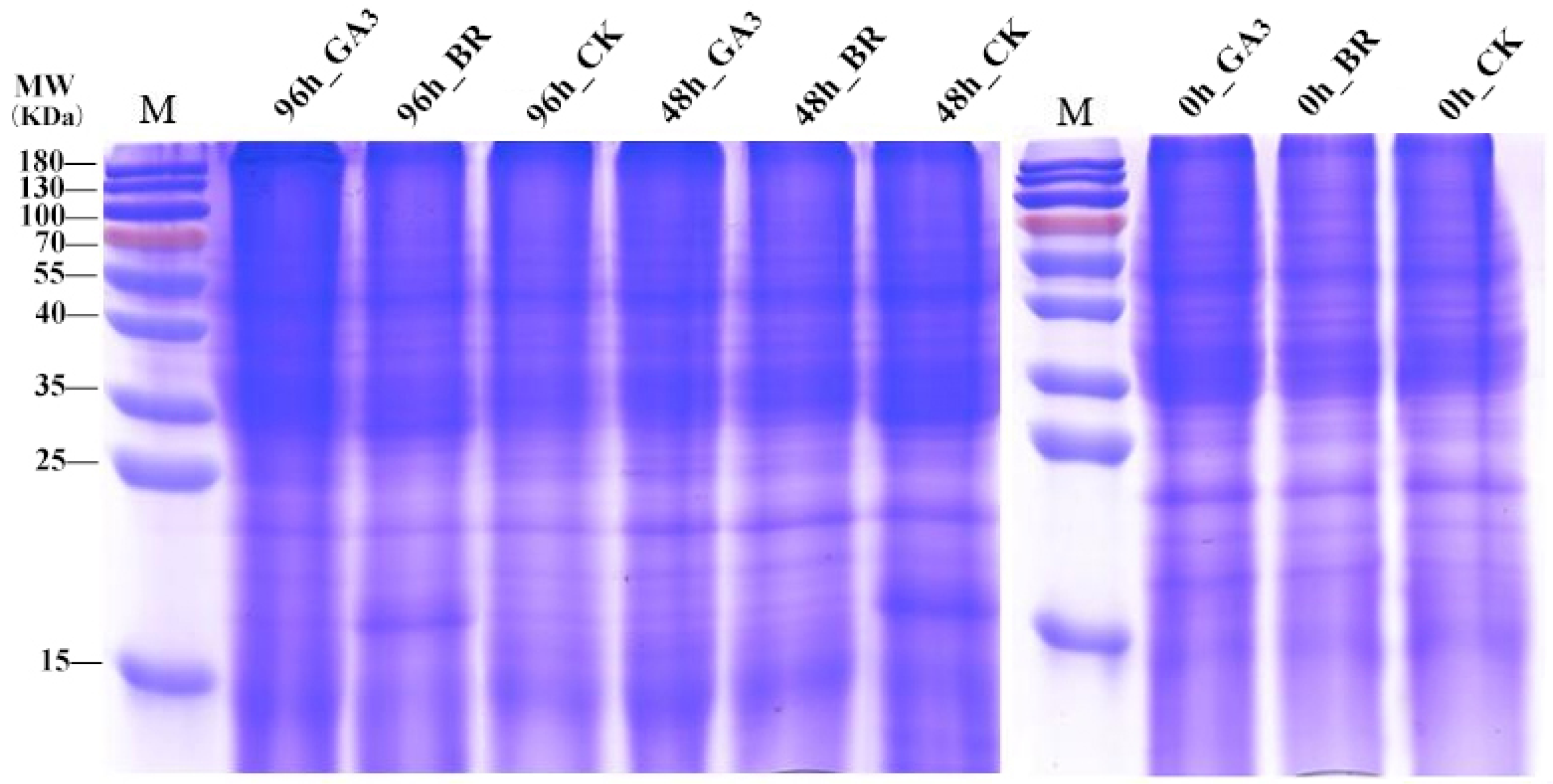
3.2. Protein Detection
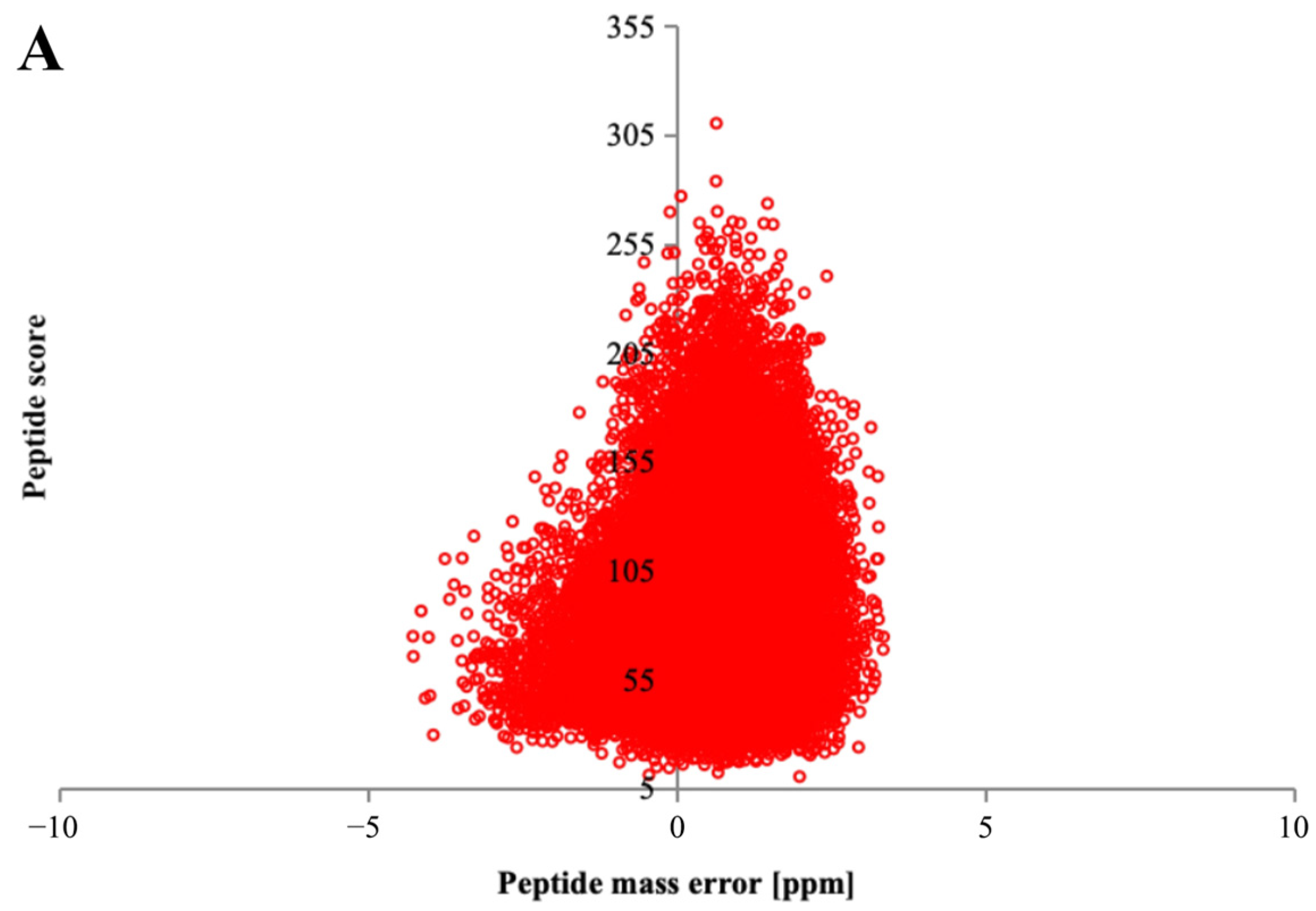
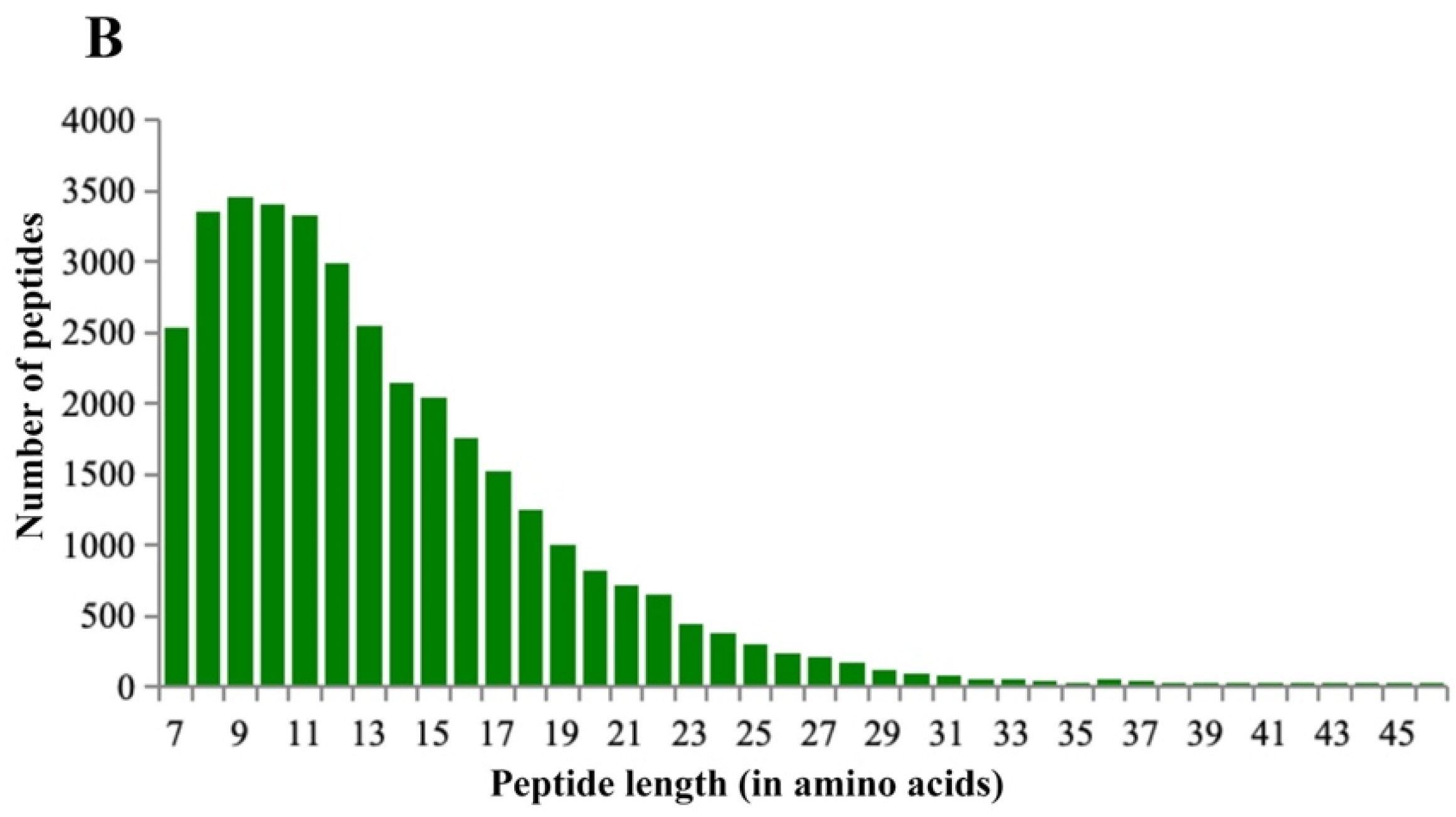
3.3. Protein Mass Spectrometry Identification
3.4. Identification of Rice Embryo Proteins Induced by GA3, BR, and CK
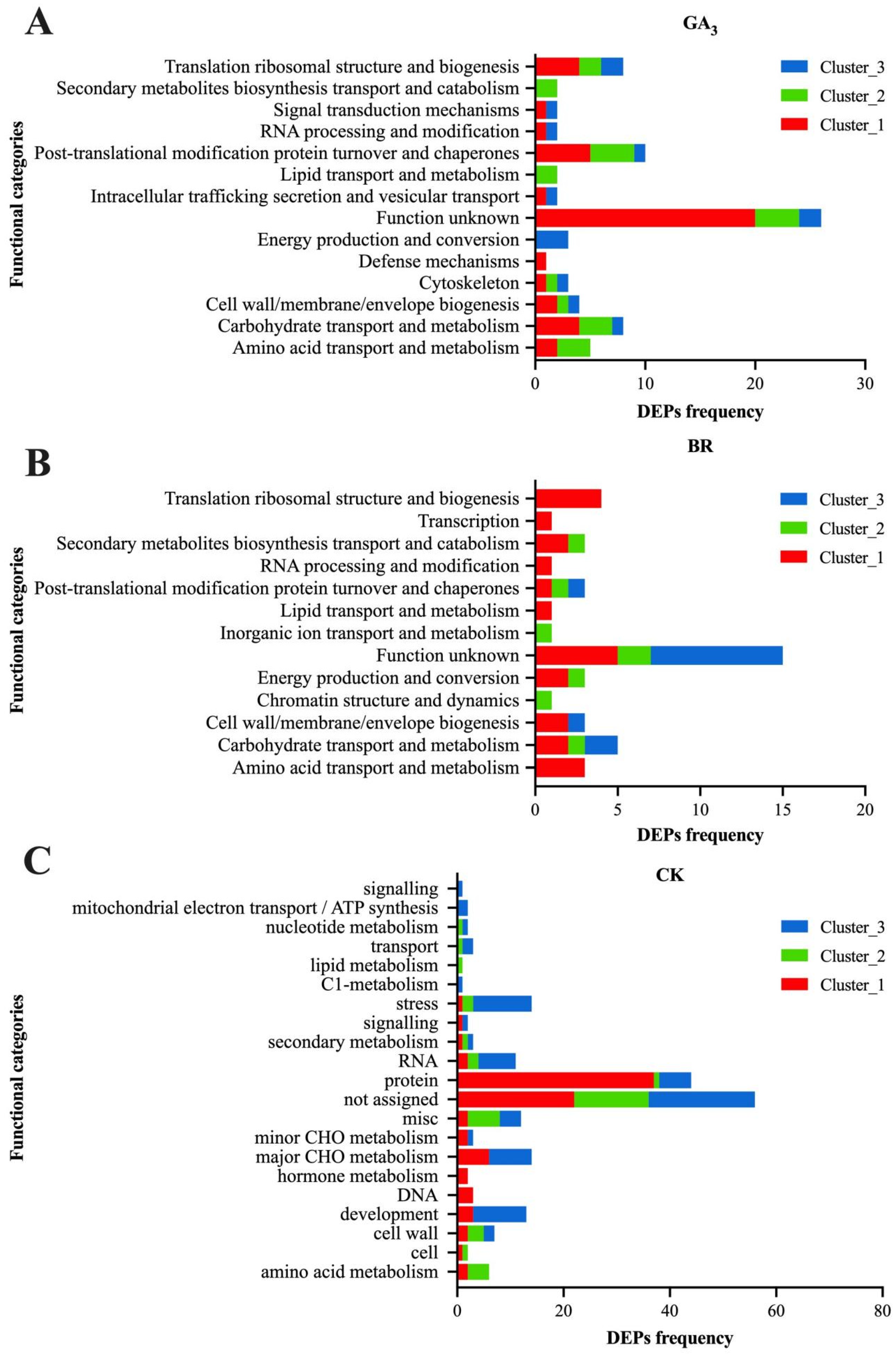
3.5. Fuzzy Clustering and COG Functional Classification of Differentially Expressed Proteins in Rice Embryos Induced by GA3, BR, and CK
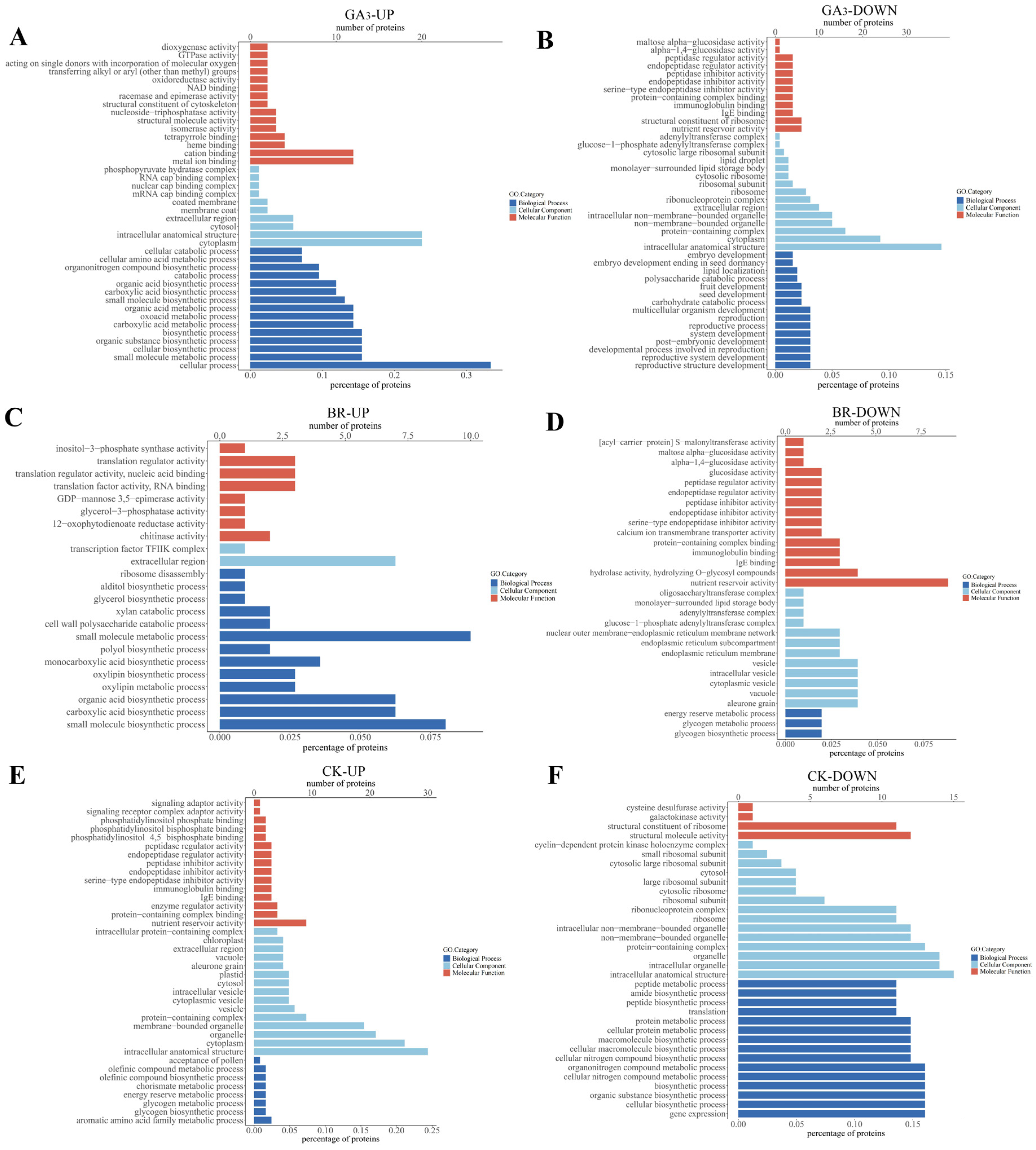
3.6. GO Analysis of DEPs in Rice Embryos Induced by GA3, BR, and CK
3.7. KEGG Analysis of DEPs in Rice Embryos Induced by GA3, BR, and CK
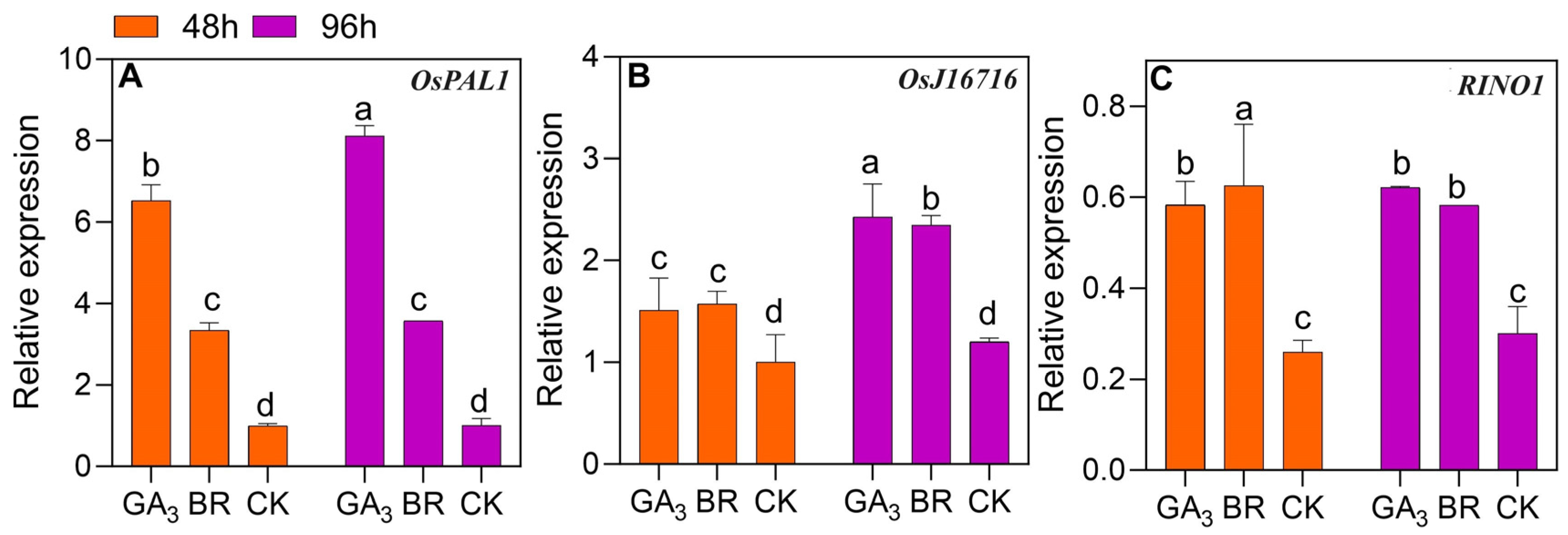
3.8. Expression Patterns of Genes Related to Observed Proteins
4. Discussion
4.1. Seed Germination
4.2. Effects of GA3 and BR Seed Initiation on Protein Content in Low-Temperature Germinated Rice Embryos
4.3. Genes Expression Patterns
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Zhang, X.; Gao, G.; Ali, I.; Wu, X.; Tang, M.; Chen, L.; Jiang, L.; Liang, T. Effects of various seed priming on morphological, physiological, and biochemical traits of rice under chilling stress. Front. Plant Sci. 2023, 14, 1146285. [Google Scholar] [CrossRef]
- Farooq, M.; Basra, S.; Wahid, A.; Khaliq, A.; Kobayashi, N. Rice seed invigoration: A review. In Organic Farming, Pest Control and Remediation of Soil Pollutants; Springer: Dordrecht, The Netherlands, 2010; Volume 1, pp. 137–175. [Google Scholar]
- Krishnan, P.; Ramakrishnan, B.; Reddy, K.R.; Reddy, V. High-temperature effects on rice growth, yield, and grain quality. Adv. Agron. 2011, 111, 87–206. [Google Scholar]
- Heydecker, W.; Higgins, J.; Gulliver, R. Accelerated germination by osmotic seed treatment. Nature 1973, 246, 42–44. [Google Scholar] [CrossRef]
- Mondal, S.; Bose, B. Seed priming: An interlinking technology between seeds, Seed germination and seedling establishment. In Plant Reproductive Ecology-Recent Advances; IntechOpen: London, Uk, 2021; pp. 107–122. [Google Scholar]
- Bhanuprakash, K.; Yogeesha, H. Seed priming for abiotic stress tolerance: An overview. Abiotic Stress Physiol. Hortic. Crops 2016, 35, 103–117. [Google Scholar]
- Nedunchezhiyan, V.; Velusamy, M.; Subburamu, K. Seed priming to mitigate the impact of elevated carbon dioxide associated temperature stress on germination in rice (Oryza sativa L.). Arch. Agron. Soil Sci. 2020, 66, 83–95. [Google Scholar] [CrossRef]
- Ya, W.; Yuetao, W.; Ruifang, Y.; Fuhua, W.; Jing, F.; Wenbo, Y.; Tao, B.; Shengxuan, W.; Haiqing, Y. Effects of gibberellin priming on seedling emergence and transcripts involved in mesocotyl elongation in rice under deep direct-seeding conditions. J. Zhejiang Univ. Sci. B 2021, 22, 1002. [Google Scholar]
- Zhang, K.; Khan, Z.; Wu, H.; Khan, M.N.; Hu, L. Gibberellic acid priming improved rapeseed drought tolerance by modulating root morphology, ROS homeostasis, and chloroplast autophagy. J. Plant Growth Regul. 2023, 42, 5977–5990. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, J.; Zhang, Y.; Xie, X.; Knapp, A. Seed priming with brassinolide improves lucerne (Medicago sativa L.) seed germination and seedling growth in relation to physiological changes under salinity stress. Aust. J. Agric. Res. 2007, 58, 811–815. [Google Scholar] [CrossRef]
- Neha; Twinkle; Mohapatra, S.; Sirhindi, G.; Dogra, V. Seed priming with brassinolides improves growth and reinforces antioxidative defenses under normal and heat stress conditions in seedlings of Brassica juncea. Physiol. Plant. 2022, 174, e13814. [Google Scholar] [CrossRef]
- Wang, R.; Anjum, S.A.; Niu, J.; Liu, M.; Li, J.; Zohaib, A.; Song, J.; Lv, J.; Wang, S.; Zong, X. Exogenous application of brassinolide ameliorate chilling stress in Leymus chinensis (Trin.) Tzvel. by modulating morphological, physiological and biochemical traits. Bangladesh J. Bot. 2016, 45, 143–150. [Google Scholar]
- He, D.; Yang, P. Proteomics of rice seed germination. Front. Plant Sci. 2013, 4, 246. [Google Scholar] [CrossRef]
- Zhang, Y.-X.; Xu, H.-H.; Liu, S.-J.; Li, N.; Wang, W.-Q.; Møller, I.M.; Song, S.-Q. Proteomic analysis reveals different involvement of embryo and endosperm proteins during aging of Yliangyou 2 hybrid rice seeds. Front. Plant Sci. 2016, 7, 1394. [Google Scholar] [CrossRef]
- Fu, Q.; Wang, B.-C.; Jin, X.; Li, H.-B.; Han, P.; Wei, K.-h.; Zhang, X.-m.; Zhu, Y.-X. Proteomic analysis and extensive protein identification from dry, germinating Arabidopsis seeds and young seedlings. BMB Rep. 2005, 38, 650–660. [Google Scholar] [CrossRef]
- Kim, S.T.; Wang, Y.; Kang, S.Y.; Kim, S.G.; Rakwal, R.; Kim, Y.C.; Kang, K.Y. Developing rice embryo proteomics reveals essential role for embryonic proteins in regulation of seed germination. J. Proteome Res. 2009, 8, 3598–3605. [Google Scholar] [CrossRef] [PubMed]
- Bønsager, B.C.; Finnie, C.; Roepstorff, P.; Svensson, B. Spatio-temporal changes in germination and radical elongation of barley seeds tracked by proteome analysis of dissected embryo, aleurone layer, and endosperm tissues. Proteomics 2007, 7, 4528–4540. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Komatsu, S. Proteomic analysis of rice seedlings during cold stress. Proteomics 2007, 7, 1293–1302. [Google Scholar] [CrossRef]
- Yoon, S.H.; Lee, J.H.; Kim, B.N. Chest CT Findings in Hospitalized Patients with SARS-CoV-2: Delta versus Omicron Variants. Radiology 2023, 306, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Pradeep Krishnan, S.; Rajendran, S.; Balamurugan, B.; Elangovan, D.; Ezhumalai, S.; Amooru, H.; Senthilkumar, S.; Nimmy, M.S.; Jha, S.K.; Sathee, L. Proteomic Responses to Salinity and Drought Stress in Plants. In Plant Proteomics: Implications in Growth, Quality Improvement, and Stress Resilience; CRC Press: Boca Raton, FL, USA, 2024. [Google Scholar]
- Tuan, P.A.; Kumar, R.; Rehal, P.K.; Toora, P.K.; Ayele, B.T. Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front. Plant Sci. 2018, 9, 362906. [Google Scholar] [CrossRef] [PubMed]
- Parwez, R.; Aftab, T.; Gill, S.S.; Naeem, M. Abscisic acid signaling and crosstalk with phytohormones in regulation of environmental stress responses. Environ. Exp. Bot. 2022, 199, 104885. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Xu, D.; Li, G.; Wu, L.; Zhou, J.; Xu, Y. Primegens: Robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics 2002, 18, 1432–1437. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A. Effect of low-temperature stress on germination, growth, and phenology of plants: A review. In Physiological Processes in Plants under Low Temperature Stress; Springer: Singapore, 2022; pp. 1–106. [Google Scholar]
- Yang, Y.; Zhu, W.; Dong, Z.; Chao, Y.; Xu, L.; Chen, M.; Liu, Z. 1D coordination polymer nanofibers for low-temperature photothermal therapy. Adv. Mater. 2017, 29, 1703588. [Google Scholar] [CrossRef] [PubMed]
- Aziz, T.; Pekşen, E. Seed priming with gibberellic acid rescues chickpea (Cicer arietinum L.) from chilling stress. Acta Physiol. Plant. 2020, 42, 139. [Google Scholar] [CrossRef]
- Pipinis, E.; Milios, E.; Kiamos, N.; Mavrokordopoulou, O.; Smiris, P. Effects of stratification and pre-treatment with gibberellic acid on seed germination of two Carpinus species. Seed Sci. Technol. 2012, 40, 21–31. [Google Scholar] [CrossRef]
- Jin, S.-K.; Zhang, M.-Q.; Leng, Y.-J.; Xu, L.-N.; Jia, S.-W.; Wang, S.-L.; Song, T.; Wang, R.-A.; Yang, Q.-Q.; Tao, T. OsNAC129 regulates seed development and plant growth and participates in the brassinosteroid signaling pathway. Front. Plant Sci. 2022, 13, 905148. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Khan, T.A.; Shahzad, S.; Ullah, S.; Shahzadi, I.; Ali, A.; Akram, W.; Yasin, N.A.; Yusuf, M. BioClay nanosheets infused with GA3 ameliorate the combined stress of hexachlorobenzene and temperature extremes in Brassica alboglabra plants. Front. Plant Sci. 2022, 13, 964041. [Google Scholar] [CrossRef]
- Yu, H.; Teng, Z.; Liu, B.; Lv, J.; Chen, Y.; Qin, Z.; Peng, Y.; Meng, S.; He, Y.; Duan, M. Transcription factor OsMYB30 increases trehalose content to inhibit α-amylase and seed germination at low temperature. Plant Physiol. 2024, 194, 1815–1833. [Google Scholar] [CrossRef] [PubMed]
- Rubio, S.; Donoso, A.; Pérez, F.J. The dormancy-breaking stimuli “chilling, hypoxia and cyanamide exposure” up-regulate the expression of α-amylase genes in grapevine buds. J. Plant Physiol. 2014, 171, 373–381. [Google Scholar] [CrossRef]
- Chaudhuri, A.; Halder, K.; Abdin, M.Z.; Majee, M.; Datta, A. Abiotic stress tolerance in plants: Brassinosteroids navigate competently. Int. J. Mol. Sci. 2022, 23, 14577. [Google Scholar] [CrossRef]
- Ahmad, F.; Singh, A.; Kamal, A. Crosstalk of brassinosteroids with other phytohormones under various abiotic stresses. J. Appl. Biol. Biotechnol. 2018, 6, 56–62. [Google Scholar]
- Hafeez, M.B.; Zahra, N.; Zahra, K.; Raza, A.; Batool, A.; Shaukat, K.; Khan, S. Brassinosteroids: Molecular and physiological responses in plant growth and abiotic stresses. Plant Stress 2021, 2, 100029. [Google Scholar] [CrossRef]
- Dong, C.; Li, L.; Cao, N.; Shang, Q.; Zhang, Z. Roles of phenylalanine ammonia-lyase in low temperature tolerance in cucumber seedlings. Yingyong Shengtai Xuebao 2015, 26, 2041–2049. [Google Scholar] [PubMed]
- RayChaudhuri, A.; Hait, N.C.; DasGupta, S.; Bhaduri, T.J.; Deb, R.; Majumder, A.L. L-myo-lnositol 1-phosphate synthase from plant sources (characteristics of the chloroplastic and cytosolic enzymes). Plant Physiol. 1997, 115, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Unel, N.M.; Baloglu, M.C.; Altunoglu, Y.Ç. Comprehensive investigation of cucumber heat shock proteins under abiotic stress conditions: A multi-omics survey. J. Biotechnol. 2023, 374, 49–69. [Google Scholar] [CrossRef]
- Diogo-Jr, R.; de Resende Von Pinho, E.V.; Pinto, R.T.; Zhang, L.; Condori-Apfata, J.A.; Pereira, P.A.; Vilela, D.R. Maize heat shock proteins—Prospection, validation, categorization and in silico analysis of the different ZmHSP families. Stress Biol. 2023, 3, 37. [Google Scholar]
- Mondal, S.; Karmakar, S.; Panda, D.; Pramanik, K.; Bose, B.; Singhal, R.K. Crucial plant processes under heat stress and tolerance through heat shock proteins. Plant Stress 2023, 10, 100227. [Google Scholar] [CrossRef]
- Abdirad, S.; Wu, Y.; Ghorbanzadeh, Z.; Tazangi, S.E.; Amirkhani, A.; Fitzhenry, M.J.; Kazemi, M.; Ghaffari, M.R.; Koobaz, P.; Zeinalabedini, M. Proteomic analysis of the meristematic root zone in contrasting genotypes reveals new insights in drought tolerance in rice. Proteomics 2022, 22, 2200100. [Google Scholar] [CrossRef]
- Ma, L.; Wang, Y.; Liu, W.; Liu, Z. Overexpression of an alfalfa GDP-mannose 3, 5-epimerase gene enhances acid, drought and salt tolerance in transgenic Arabidopsis by increasing ascorbate accumulation. Biotechnol. Lett. 2014, 36, 2331–2341. [Google Scholar] [CrossRef]
- Gevaert, O.; Van Overtveldt, S.; Da Costa, M.; Beerens, K.; Desmet, T. GDP-altrose as novel product of GDP-mannose 3, 5-epimerase: Revisiting its reaction mechanism. Int. J. Biol. Macromol. 2020, 165, 1862–1868. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Xiao, N.; Zhang, G.; Wang, F.; Chen, X.; Fang, R. Rice Germin-like Protein 2-1 functions in seed dormancy under the control of abscisic acid and gibberellic acid signaling pathways. Plant Physiol. 2020, 183, 1157–1170. [Google Scholar] [CrossRef]
- Gevaert, O.; Van Overtveldt, S.; Beerens, K.; Desmet, T. Characterization of the first bacterial and thermostable GDP-mannose 3, 5-epimerase. Int. J. Mol. Sci. 2019, 20, 3530. [Google Scholar] [CrossRef] [PubMed]
- Beerens, K.; Gevaert, O.; Desmet, T. GDP-mannose 3, 5-epimerase: A view on structure, mechanism, and industrial potential. Front. Mol. Biosci. 2022, 8, 784142. [Google Scholar] [CrossRef] [PubMed]
- Silambarasan, S.; Logeswari, P.; Cornejo, P.; Kannan, V.R. Evaluation of the production of exopolysaccharide by plant growth promoting yeast Rhodotorula sp. strain CAH2 under abiotic stress conditions. Int. J. Biol. Macromol. 2019, 121, 55–62. [Google Scholar] [CrossRef]
- Zou, P.; Lu, X.; Zhao, H.; Yuan, Y.; Meng, L.; Zhang, C.; Li, Y. Polysaccharides derived from the brown algae Lessonia nigrescens enhance salt stress tolerance to wheat seedlings by enhancing the antioxidant system and modulating intracellular ion concentration. Front. Plant Sci. 2019, 10, 48. [Google Scholar] [CrossRef]
- Mariani, D.; Mathias, C.J.; da Silva, C.G.; Herdeiro, R.d.S.; Pereira, R.; Panek, A.D.; Eleutherio, E.C.; Pereira, M.D. Involvement of glutathione transferases, Gtt1and Gtt2, with oxidative stress response generated by H2O2 during growth of Saccharomyces cerevisiae. Redox Rep. 2008, 13, 246–254. [Google Scholar] [CrossRef]
- Chronopoulou, E.; Kontouri, K.; Chantzikonstantinou, M.; Pouliou, F.; Perperopoulou, F.; Voulgari, G.; Bosmali, E.; Axarli, I.; Nianiou-Obeidat, I.; Madesis, P. Plant glutathione transferases: Structure, antioxidant catalytic function and in planta protective role in biotic and abiotic stress. Curr. Chem. Biol. 2014, 8, 58–75. [Google Scholar] [CrossRef]
- Primavera, A.; Fustinoni, S.; Biroccio, A.; Ballerini, S.; Urbani, A.; Bernardini, S.; Federici, G.; Capuccil, E.; Manno, M.; Lo Bello, M. Glutathione transferases and glutathionyl haernoglobin as biomarkers of oxidative stress in subjects exposed to low doses of 1, 3-butadiene in a petrochemical plant. Toxicol. Lett. 2008, 180, S26–S27. [Google Scholar] [CrossRef]
- Zhang, J.; Yuan, H.; Yang, Y.; Fish, T.; Lyi, S.M.; Thannhauser, T.W.; Zhang, L.; Li, L. Plastid ribosomal protein S5 is involved in photosynthesis, plant development, and cold stress tolerance in Arabidopsis. J. Exp. Bot. 2016, 67, 2731–2744. [Google Scholar] [CrossRef]
- Robles, P.; Quesada, V. Transcriptional and post-transcriptional regulation of organellar gene expression (OGE) and its roles in plant salt tolerance. Int. J. Mol. Sci. 2019, 20, 1056. [Google Scholar] [CrossRef]
- Doddagoudar, S.; Nagaraja, M.; Lakshmikanth, M.; Srininvas, A.; Shakuntala, N.; Hiremath, U.; Mahanthshivayogayya, K. Improving the resilience of rice seedlings to low temperature stress through seed priming. S. Afr. J. Bot. 2023, 162, 183–192. [Google Scholar]



| Gene Names | Protein Accession | Description | Fold Change (FC) | Regulation | |
|---|---|---|---|---|---|
| 48/0 h | 96/0 h | ||||
| Os03g0821100 | Q84TA1 | Heat shock cognate 70 kDa protein 2 | 2.878 | 2.881 | UP |
| Os04g0688200 | A3AYV8 | Peroxidase | 4.537 | 6.489 | UP |
| Os03g0854100 | Q0DLN9 | Similar to ARF GAP-like zinc finger-containing protein ZIGA2 | 3.618 | 3.641 | UP |
| Os02g0612300 | Q84L14 | Nuclear cap-binding protein subunit 2 | 3.734 | 4.930 | UP |
| Os03g0192700 | O64437 | Myo-inositol 3-phosphate synthase 1 | 7.521 | 8.943 | UP |
| Os02g0739600 | Q6Z5N4 | Pyruvate dehydrogenase E1 component subunit | 6.098 | 4.130 | UP |
| Os10g0417600 | A3C4S4 | GDP-mannose 3,5-epimerase 1 | 6.235 | 6.112 | UP |
| Os12g0567200 | Q2QNF7 | Diaminopimelate epimerase | 6.682 | 4.608 | UP |
| Os03g0331700 | Q10LX4 | Probable calcium-binding protein | 8.361 | 7.654 | UP |
| Os02g0626100 | P14717 | Phenylalanine ammonia-lyase | 8.451 | 12.649 | UP |
| Os06g0730800 | Q5Z414 | mRNA splicing factor | 17.032 | 0.010 | UP |
| Os07g0693100 | Q0D3D2 | Pyruvate decarboxylase | 0.010 | 0.010 | Down |
| Os01g0231700 | Q5NB69 | Elongation step of protein synthesis. | 0.010 | 5.696 | Down |
| Os01g0374000 | Q93WM2 | Glutathione transferase | 0.133 | 0.170 | Down |
| Os06g0219600 | B9FS82 | Similar to Poly(A)-binding protein II-like. | 0.156 | 4.831 | Down |
| Os01g0500900 | A0A0P0V2Y9 | Tyrosine--tRNA ligase | 0.211 | 0.235 | Down |
| Os05g0595100 | Q8LNZ3 | UDP-glucose 4-epimerase | 0.238 | 0.144 | Down |
| Os06g0675700 | Q653V7 | Alpha-glucosidase in rice seeds | 0.29 | 0.397 | Down |
| Os07g0184300 | P31674 | 40S ribosomal protein S15 | 0.339 | 0.219 | Down |
| Os07g0675100 | Q6ZDX2 | Pectinesterase | 0.400 | 0.010 | Down |
| Gene Names | Protein Accession | Description | Fold Change (FC) | Regulation | |
|---|---|---|---|---|---|
| 48/0 h | 96/0 h | ||||
| Os04g0688200 | A3AYV8 | Peroxidase | 4.624 | 5.788 | UP |
| Os08g0558800 | B7F845 | 60S ribosomal protein | 2.476 | 3.373 | UP |
| Os03g0192700 | O64437 | Inositol-3-phosphate synthase | 7.191 | 7.754 | UP |
| Os02g0626100 | P14717 | Phenylalanine ammonia-lyase | 6.882 | 9.259 | UP |
| Os07g0622200 | Q75W16 | Phospho-2-dehydro-3-deoxyheptonate aldolase | 5.189 | 4.498 | UP |
| Os10g0517500 | Q7XCS3 | Cys/Met metabolism PLP-dependent enzyme family protein | 2.799 | 3.545 | UP |
| Os07g0681400 | Q7XHW4 | Probable calcium-binding protein | 3.629 | 3.103 | UP |
| Os02g0169900 | Q6H6B9 | Inositol-1-monophosphatase | 0.010 | 0.010 | Down |
| Os06g0708832 | Q5Z9H5 | Similar to arogenate dehydrogenase. | 0.010 | 0.010 | Down |
| Os02g0738900 | Q0DXR0 | Dynamin GTPase | 0.135 | 0.179 | Down |
| Os05g0595100 | Q8LNZ3 | UDP-glucose 4-epimerase | 0.190 | 0.186 | Down |
| Os01g0633100 | Q7G065 | Glucose-1-phosphate adenylyltransferase large subunit | 0.232 | 6.741 | Down |
| Os06g0320200 | Q5Z9Z0 | Beta-glucosidase | 0.245 | 0.028 | Down |
| Os06g0172600 | Q69SI5 | 40S ribosomal protein | 0.386 | 0.020 | Down |
| Os05g0461400 | Q6L500 | Probable histone H2A. | 0.492 | 0.021 | Down |
| Gene Names | Protein Accession | Description | Fold Change (FC) | Regulation | |
|---|---|---|---|---|---|
| 48/0 h | 96/0 h | ||||
| Os04g0688200 | A3AYV8 | Peroxidase | 2.530 | 4.491 | UP |
| Os10g0517500 | Q7XCS3 | Cys/Met metabolism PLP-dependent enzyme family protein | 3.420 | 6.565 | UP |
| Os07g0622200 | Q75W16 | Phospho-2-dehydro-3-deoxyheptonate aldolase | 3.558 | 6.192 | UP |
| Os02g0626100 | P14717 | Phenylalanine ammonia-lyase | 3.596 | 4.920 | UP |
| Os03g0192700 | O64437 | Inositol-3-phosphate synthase | 5.191 | 5.754 | UP |
| Os07g0681400 | Q7XHW4 | Probable calcium-binding protein | 5.468 | 7.842 | UP |
| Os06g0569500 | Q0DBF4 | Ent-sandaracopimaradiene 3-hydroxylase | 6.003 | 7.766 | UP |
| Os08g0536000 | Q6Z1G7 | Pyruvate dehydrogenase E1 component subunit | 0.010 | 0.063 | Down |
| Os11g0123400 | A0A0N7KSC9 | CTP:phosphoethanolamine cytidylyltransferase | 0.010 | 0.010 | Down |
| Os05g0595100 | Q8LNZ3 | UDP-glucose 4-epimerase | 0.296 | 0.314 | Down |
| Os02g0785800 | Q6K8U8 | Similar to Ribosomal protein L35A. | 0.448 | 0.299 | Down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Hui, G.; Gao, G.; Ali, I.; Tang, M.; Chen, L.; Zhong, X.; Jiang, L.; Liang, T.; Zhang, X. Physiological and Proteomic Analysis of Various Priming on Rice Seed under Chilling Stress. Plants 2024, 13, 2430. https://doi.org/10.3390/plants13172430
Zhang H, Hui G, Gao G, Ali I, Tang M, Chen L, Zhong X, Jiang L, Liang T, Zhang X. Physiological and Proteomic Analysis of Various Priming on Rice Seed under Chilling Stress. Plants. 2024; 13(17):2430. https://doi.org/10.3390/plants13172430
Chicago/Turabian StyleZhang, Hua, Guo Hui, Guoqing Gao, Izhar Ali, Maoyan Tang, Lei Chen, Xiaoyuan Zhong, Ligeng Jiang, Tianfeng Liang, and Xiaoli Zhang. 2024. "Physiological and Proteomic Analysis of Various Priming on Rice Seed under Chilling Stress" Plants 13, no. 17: 2430. https://doi.org/10.3390/plants13172430
APA StyleZhang, H., Hui, G., Gao, G., Ali, I., Tang, M., Chen, L., Zhong, X., Jiang, L., Liang, T., & Zhang, X. (2024). Physiological and Proteomic Analysis of Various Priming on Rice Seed under Chilling Stress. Plants, 13(17), 2430. https://doi.org/10.3390/plants13172430








