Isolation and Selection of Protein-Rich Mutants of Chlorella vulgaris by Fluorescence-Activated Cell Sorting with Enhanced Biostimulant Activity to Germinate Garden Cress Seeds
Abstract
:1. Introduction
2. Results
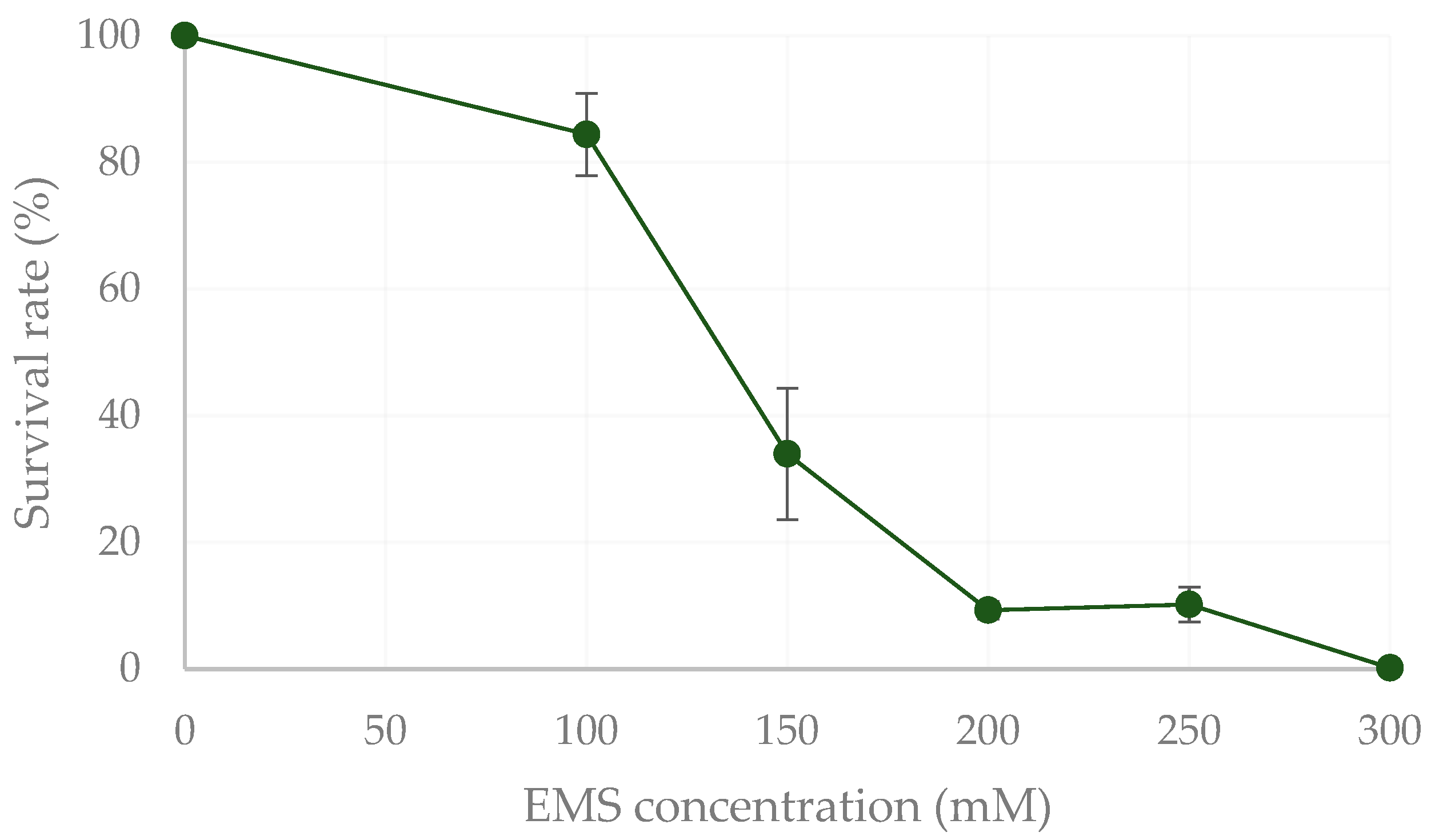
2.1. Mutagenesis and Isolation of Protein-Rich Mutants by FACS
2.2. Screening of Mutants
2.2.1. Growth Performance and Protein Content
2.2.2. Chlorophyll and Carotenoid Profiles
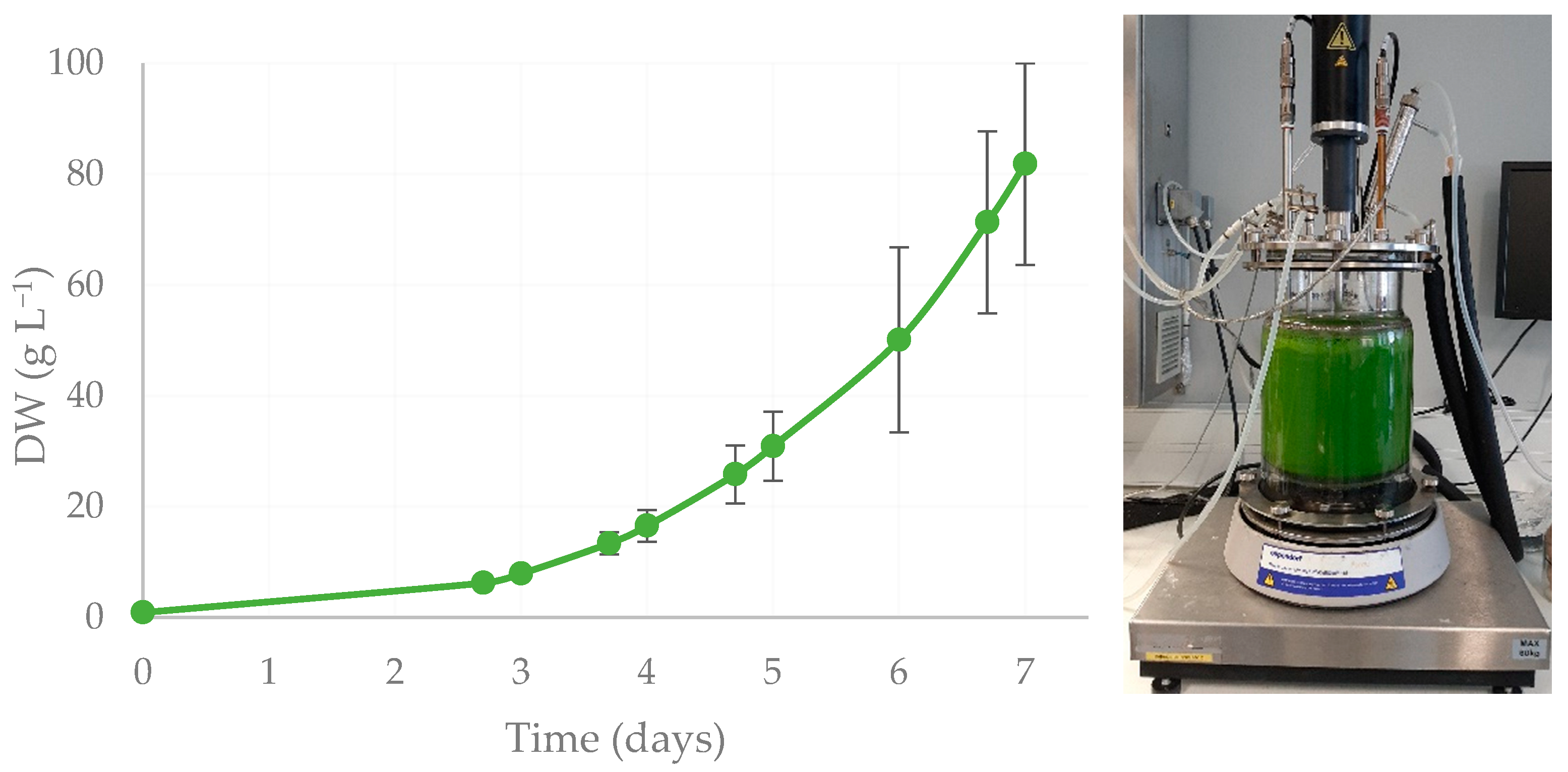
2.3. Biomass Growth and Protein Production
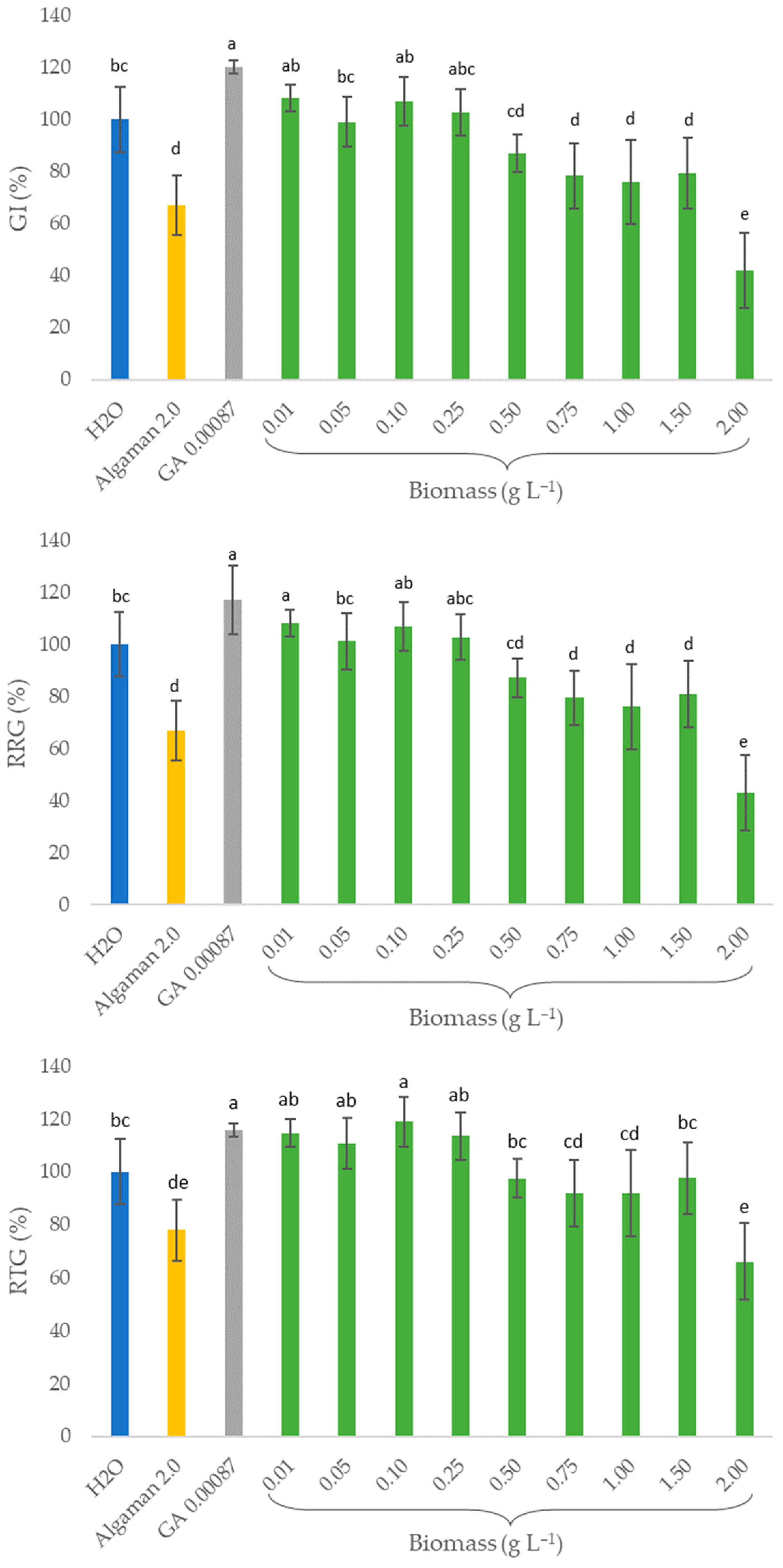
2.4. Biostimulant Activity (In Vitro Assays)
3. Discussion
4. Materials and Methods
4.1. Microalgae Strain and Mutagenesis
4.2. Isolation of Protein-Rich Strains by FACS
4.3. Screening of FACS Mutants
4.3.1. Growth Performance
4.3.2. Biochemical Analysis of the Biomass
Protein Content
Extraction and Quantification of Chlorophyll and Carotenoids
4.4. Biomass Growth and Protein Production (7 L Fermenter)
4.5. Biostimulant Activity (In Vitro Assays)
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- United Nations|Department of Economic and Social Affairs. Goal 2—End Hunger, Achieve Food Security and Improved Nutrition and Promote Sustainable Agriculture. Available online: https://sdgs.un.org/goals/goal2 (accessed on 20 May 2024).
- Mannino, G. A New Era of Sustainability: Plant Biostimulants. Int. J. Mol. Sci. 2023, 24, 16329. [Google Scholar] [CrossRef]
- Landrigan, P.J.; Stegeman, J.J.; Fleming, L.E.; Allemand, D.; Anderson, D.M.; Backer, L.C.; Brucker-Davis, F.; Chevalier, N.; Corra, L.; Czerucka, D.; et al. Human Health and Ocean Pollution. Ann. Glob. Health 2020, 86, 151. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Song, S.; Wang, R.; Liu, Z.; Meng, J.; Sweetman, A.J.; Jenkins, A.; Ferrier, R.C.; Li, H.; Luo, W.; et al. Impacts of soil and water pollution on food safety and health risks in China. Environ. Int. 2015, 77, 5–15. [Google Scholar] [CrossRef]
- Ahmad, W.; Iqbal, J.; Nasir, M.J.; Ahmad, B.; Khan, M.T.; Khan, S.N.; Adnan, S. Impact of land use/land cover changes on water quality and human health in district Peshawar Pakistan. Sci. Rep. 2021, 11, 16526. [Google Scholar] [CrossRef] [PubMed]
- Agriculture and Food System. Available online: https://www.eea.europa.eu/en/topics/in-depth/agriculture-and-food (accessed on 20 May 2024).
- Greene, C.; Scott-Buechler, C.; Hausner, A.; Johnson, Z.; Lei, X.G.; Huntley, M. Transforming the Future of Marine Aquaculture: A Circular Economy Approach. Oceanography 2022, 35, 26–34. [Google Scholar] [CrossRef]
- Williamson, E.; Ross, I.L.; Wall, B.T.; Hankamer, B. Microalgae: Potential novel protein for sustainable human nutrition. Trends Plant Sci. 2024, 29, 370–382. [Google Scholar] [CrossRef]
- Enzing, C.; Ploeg, M.; Barbosa, M.; Sijtsma, L. Microalgae-Based Products for the Food and Feed Sector: An Outlook for Europe; JRC Scientific and Policy Reports; Publications Office of the EU: Luxembourg, 2014; pp. 19–37. [Google Scholar]
- Wells, M.L.; Potin, P.; Craigie, J.S.; Raven, J.A.; Merchant, S.S.; Helliwell, K.E.; Smith, A.G.; Camire, M.E.; Brawley, S.H. Algae as nutritional and functional food sources: Revisiting our understanding. J. Appl. Phycol. 2017, 29, 949–982. [Google Scholar] [CrossRef]
- Navarro-López, E.; Cerón-García, M.d.C.; López-Rodríguez, M.; Acién-Fernández, F.G.; Molina-Grima, E. Biostimulants obtained after pilot-scale high-pressure homogenization of Scenedesmus sp. grown in pig manure. Algal Res. 2020, 52, 102123. [Google Scholar] [CrossRef]
- Colla, G.; Rouphael, Y. Microalgae: New Source of Plant Biostimulants. Agronomy 2020, 10, 1240. [Google Scholar] [CrossRef]
- Ronga, D.; Biazzi, E.; Parati, K.; Carminati, D.; Carminati, E.; Tava, A. Microalgal Biostimulants and Biofertilisers in Crop Productions. Agronomy 2019, 9, 192. [Google Scholar] [CrossRef]
- Gitau, M.M.; Farkas, A.; Ördög, V.; Maróti, G. Evaluation of the biostimulant effects of two Chlorophyta microalgae on tomato (Solanum lycopersicum). J. Clean. Prod. 2022, 364, 132689. [Google Scholar] [CrossRef]
- Villaró-Cos, S.; Franco, M.C.; García-Vaquero, M.; Morán, L.; Alarcón, F.J.; Lafarga, T. Composition of microalgae produced using different types of water and nutrient sources. Algal Res. 2024, 78, 103394. [Google Scholar] [CrossRef]
- Farid, R.; Mutale-Joan, C.; Redouane, B.; Najib, E.M.; Abderahime, A.; Laila, S.; Hicham, E.A. Effect of Microalgae Polysaccharides on Biochemical and Metabolomics Pathways Related to Plant Defense in Solanum lycopersicum. Appl. Biochem. Biotechnol. 2018, 188, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Shahrajabian, M.H.; Kuang, Y.; Wang, N. Amino Acids Biostimulants and Protein Hydrolysates in Agricultural Sciences. Plants 2024, 13, 210. [Google Scholar] [CrossRef] [PubMed]
- de Queiroz, A.R.; Hines, C.; Brown, J.; Sahay, S.; Vijayan, J.; Stone, J.M.; Bickford, N.; Wuellner, M.; Glowacka, K.; Buan, N.R.; et al. The effects of exogenously applied antioxidants on plant growth and resilience. Phytochem. Rev. 2023, 22, 407–447. [Google Scholar] [CrossRef]
- Trovão, M.; Schüler, L.M.; Machado, A.; Bombo, G.; Navalho, S.; Barros, A.; Pereira, H.; Silva, J.; Freitas, F.; Varela, J. Random Mutagenesis as a Promising Tool for Microalgal Strain Improvement towards Industrial Production. Mar. Drugs 2022, 20, 440. [Google Scholar] [CrossRef] [PubMed]
- Schüler, L.; de Morais, E.G.; Trovão, M.; Machado, A.; Carvalho, B.; Carneiro, M.; Maia, I.; Soares, M.; Duarte, P.; Barros, A.; et al. Isolation and Characterization of Novel Chlorella Vulgaris Mutants with Low Chlorophyll and Improved Protein Contents for Food Applications. Front. Bioeng. Biotechnol. 2020, 8, 469. [Google Scholar] [CrossRef]
- Spicer, A.; Molnar, A. Gene Editing of Microalgae: Scientific Progress and Regulatory Challenges in Europe. Biology 2018, 7, 21. [Google Scholar] [CrossRef]
- Manandhar-Shrestha, K.; Hildebrand, M. Development of flow cytometric procedures for the efficient isolation of improved lipid accumulation mutants in a Chlorella sp. microalga. J. Appl. Phycol. 2013, 25, 1643–1651. [Google Scholar] [CrossRef]
- Cazzaniga, S.; Dall’Osto, L.; Szaub, J.; Scibilia, L.; Ballottari, M.; Purton, S.; Bassi, R. Domestication of the green alga Chlorella sorokiniana: Reduction of antenna size improves light-use efficiency in a photobioreactor. Biotechnol. Biofuels 2014, 7, 157. [Google Scholar] [CrossRef]
- Pereira, H.; Schulze, P.S.; Schüler, L.M.; Santos, T.; Barreira, L.; Varela, J. Fluorescence activated cell-sorting principles and applications in microalgal biotechnology. Algal Res. 2018, 30, 113–120. [Google Scholar] [CrossRef]
- Gao, F.; Sá, M.; Cabanelas, I.T.D.; Wijffels, R.H.; Barbosa, M.J. Improved fucoxanthin and docosahexaenoic acid productivities of a sorted self-settling Tisochrysis lutea phenotype at pilot scale. Bioresour. Technol. 2021, 325, 124725. [Google Scholar] [CrossRef]
- Nayak, M.; Suh, W.I.; Oh, Y.T.; Ryu, A.J.; Jeong, K.J.; Kim, M.; Mohapatra, R.K.; Lee, B.; Chang, Y.K. Directed evolution of Chlorella sp. HS2 towards enhanced lipid accumulation by ethyl methanesulfonate mutagenesis in conjunction with fluorescence-activated cell sorting based screening. Fuel 2022, 316, 123410. [Google Scholar] [CrossRef]
- Malerba, M.E.; Connolly, S.R.; Heimann, K. Standard flow cytometry as a rapid and non-destructive proxy for cell nitrogen quota. J. Appl. Phycol. 2015, 28, 1085–1095. [Google Scholar] [CrossRef]
- Gerashchenko, B.I. Fluorescence-Activated Cell Sorting (FACS)-Based Characterization of Microalgae. Mar. Ecol. Curr. Future Dev. 2020, 2, 148–160. [Google Scholar] [CrossRef]
- Doucha, J.; Lívanský, K. Production of high-density Chlorella culture grown in fermenters. J. Appl. Phycol. 2011, 24, 35–43. [Google Scholar] [CrossRef]
- Barros, A.; Pereira, H.; Campos, J.; Marques, A.; Varela, J.; Silva, J. Heterotrophy as a tool to overcome the long and costly autotrophic scale-up process for large scale production of microalgae. Sci. Rep. 2019, 9, 13935. [Google Scholar] [CrossRef] [PubMed]
- Smetana, S.; Sandmann, M.; Rohn, S.; Pleissner, D.; Heinz, V. Autotrophic and heterotrophic microalgae and cyanobacteria cultivation for food and feed: Life cycle assessment. Bioresour. Technol. 2017, 245, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.; Wijffels, R.H.; Dominguez, M.; Barbosa, M.J. Heterotrophic vs autotrophic production of microalgae: Bringing some light into the everlasting cost controversy. Algal Res. 2022, 64, 102698. [Google Scholar] [CrossRef]
- Diaz, C.J.; Douglas, K.J.; Kang, K.; Kolarik, A.L.; Malinovski, R.; Torres-Tiji, Y.; Molino, J.V.; Badary, A.; Mayfield, S.P. Developing algae as a sustainable food source. Front. Nutr. 2023, 9, 1029841. [Google Scholar] [CrossRef]
- Schüler, L.M.; Santos, T.; Pereira, H.; Duarte, P.; Katkam, N.G.; Florindo, C.; Schulze, P.S.; Barreira, L.; Varela, J.C. Improved production of lutein and β-carotene by thermal and light intensity upshifts in the marine microalga Tetraselmis sp. CTP4. Algal Res. 2020, 45, 101732. [Google Scholar] [CrossRef]
- Lau, K.Y.; Pleissner, D.; Lin, C.S.K. Recycling of food waste as nutrients in Chlorella vulgaris cultivation. Bioresour. Technol. 2014, 170, 144–151. [Google Scholar] [CrossRef]
- Cai, Y.; Zhai, L.; Wu, K.; Li, Z.; Gu, Z.; Wang, Y.; Cui, X.; Zhou, T.; Ruan, R.; Liu, T.; et al. Mechanisms of promotion in the heterotrophic growth of Chlorella vulgaris by the combination of sodium acetate and hydrolysate of broken rice. Bioresour. Technol. 2022, 364, 127965. [Google Scholar] [CrossRef]
- Hyka, P.; Lickova, S.; Přibyl, P.; Melzoch, K.; Kovar, K. Flow cytometry for the development of biotechnological processes with microalgae. Biotechnol. Adv. 2013, 31, 2–16. [Google Scholar] [CrossRef]
- Krul, E.S. Calculation of Nitrogen-to-Protein Conversion Factors: A Review with a Focus on Soy Protein. J. Am. Oil Chem. Soc. 2019, 96, 339–364. [Google Scholar] [CrossRef]
- Kjeldahl, J. Neue Methode zur Bestimmung des Stickstoffs in organischen Körpern. Z. Anal. Chem. 1883, 22, 366–382. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pereira, H.; Gangadhar, K.N.; Schulze, P.S.C.; Santos, T.; de Sousa, C.B.; Schueler, L.M.; Custódio, L.; Malcata, F.X.; Gouveia, L.; Varela, J.C.S.; et al. Isolation of a euryhaline microalgal strain, Tetraselmis sp. CTP4, as a robust feedstock for biodiesel production. Sci. Rep. 2016, 6, 35663. [Google Scholar] [CrossRef]
- Sakarika, M.; Kornaros, M. Kinetics of growth and lipids accumulation in Chlorella vulgaris during batch heterotrophic cultivation: Effect of different nutrient limitation strategies. Bioresour. Technol. 2017, 45, 674–691. [Google Scholar] [CrossRef]
- Yun, H.-S.; Kim, Y.-S.; Yoon, H.-S. Effect of Different Cultivation Modes (Photoautotrophic, Mixotrophic, and Heterotrophic) on the Growth of Chlorella sp. and Biocompositions. Front. Bioeng. Biotechnol. 2021, 9, 774143. [Google Scholar] [CrossRef]
- Trovão, M.; Cardoso, L.; Schüler, L.; Machado, A.; Santo, G.E.; Pedroso, H.; Reis, A.; Barros, A.; Correia, N.; Costa, M.; et al. Oxyfluorfen: A novel metabolic inhibitor to select microalgal chlorophyll-deficient mutant strains for nutritional applications. Algal Res. 2024, 81, 103572. [Google Scholar] [CrossRef]
- Kim, J.; Kim, M.; Lee, S.; Jin, E. Development of a Chlorella vulgaris mutant by chemical mutagenesis as a producer for natural violaxanthin. Algal Res. 2020, 46, 101790. [Google Scholar] [CrossRef]
- Eregie, S.B.; Sanusi, I.A.; Kana, G.E.B.; Olaniran, A.O. Effect of ultra-violet light radiation on Scenedesmus vacuolatus growth kinetics, metabolic performance, and preliminary biodegradation study. Biodegradation 2024, 35, 71–86. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Wei, D.; Xing, X. Breeding a novel chlorophyll-deficient mutant of Auxenochlorella pyrenoidosa for high-quality protein production by atmospheric room temperature plasma mutagenesis. Bioresour. Technol. 2023, 390, 129907. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.-S.; Lee, B.; Jeong, B.-R.; Chang, Y.K.; Kwon, J.-H. Truncated light-harvesting chlorophyll antenna size in Chlorella vulgaris improves biomass productivity. J. Appl. Phycol. 2016, 28, 3193–3202. [Google Scholar] [CrossRef]
- Corcoran, A.A.; Saunders, M.A.; Hanley, A.P.; Lee, P.A.; Lopez, S.; Ryan, R.; Yohn, C.B. Iterative screening of an evolutionary engineered Desmodesmus generates robust field strains with pesticide tolerance. Algal Res. 2018, 31, 443–453. [Google Scholar] [CrossRef]
- Dall’osto, L.; Cazzaniga, S.; Guardini, Z.; Barera, S.; Benedetti, M.; Mannino, G.; Maffei, M.E.; Bassi, R. Combined resistance to oxidative stress and reduced antenna size enhance light-to-biomass conversion efficiency in Chlorella vulgaris cultures. Biotechnol. Biofuels 2019, 12, 221. [Google Scholar] [CrossRef]
- Guardini, Z.; Dall’Osto, L.; Barera, S.; Jaberi, M.; Cazzaniga, S.; Vitulo, N.; Bassi, R. High Carotenoid Mutants of Chlorella vulgaris Show Enhanced Biomass Yield under High Irradiance. Plants 2021, 10, 911. [Google Scholar] [CrossRef]
- Schüler, L.M.; Bombo, G.; Duarte, P.; Santos, T.F.; Maia, I.B.; Pinheiro, F.; Marques, J.; Jacinto, R.; Schulze, P.S.; Pereira, H.; et al. Carotenoid biosynthetic gene expression, pigment and n-3 fatty acid contents in carotenoid-rich Tetraselmis striata CTP4 strains under heat stress combined with high light. Bioresour. Technol. 2021, 337, 125385. [Google Scholar] [CrossRef]
- Tharek, A.; Yahya, A.; Salleh, M.M.; Jamaluddin, H.; Yoshizaki, S.; Hara, H.; Iwamoto, K.; Suzuki, I.; Mohamad, S.E. Improvement and screening of astaxanthin producing mutants of newly isolated Coelastrum sp. using ethyl methane sulfonate induced mutagenesis technique. Biotechnol. Rep. 2021, 32, e00673. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Prakash, G.; Lali, A.M. Reduced chlorophyll antenna mutants of Chlorella saccharophila for higher photosynthetic efficiency and biomass productivity under high light intensities. J. Appl. Phycol. 2020, 32, 1559–1567. [Google Scholar] [CrossRef]
- Nakanishi, K.; Deuchi, K. Culture of a high-chlorophyll-producing and halotolerant Chlorella vulgaris. J. Biosci. Bioeng. 2014, 117, 617–619. [Google Scholar] [CrossRef] [PubMed]
- Vigeolas, H.; Duby, F.; Kaymak, E.; Niessen, G.; Motte, P.; Franck, F.; Remacle, C. Isolation and partial characterization of mutants with elevated lipid content in Chlorella sorokiniana and Scenedesmus obliquus. J. Biotechnol. 2012, 162, 3–12. [Google Scholar] [CrossRef]
- Xi, Y.; Yin, L.; Chi, Z.Y.; Luo, G. Characterization and RNA-seq transcriptomic analysis of a Scenedesmus obliqnus mutant with enhanced photosynthesis efficiency and lipid productivity. Sci. Rep. 2021, 11, 11795. [Google Scholar] [CrossRef]
- Kim, H.S.; Park, W.-K.; Lee, B.; Seon, G.; Suh, W.I.; Moon, M.; Chang, Y.K. Optimization of heterotrophic cultivation of Chlorella sp. HS2 using screening, statistical assessment, and validation. Sci. Rep. 2019, 9, 19383. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Xia, Y.; Zeng, Y.; Li, X.; Zhang, Y. Nitrate concentration-shift cultivation to enhance protein content of heterotrophic microalga Chlorella vulgaris: Over-compensation strategy. Bioresour. Technol. 2017, 233, 247–255. [Google Scholar] [CrossRef]
- Santo, G.E.; Barros, A.; Costa, M.; Pereira, H.; Trovão, M.; Cardoso, H.; Carvalho, B.; Soares, M.; Correia, N.; Silva, J.T.; et al. Scenedesmus rubescens Heterotrophic Production Strategies for Added Value Biomass. Mar. Drugs 2023, 21, 411. [Google Scholar] [CrossRef]
- Morillas-España, A.; Ruiz-Nieto, Á.; Lafarga, T.; Acién, G.; Arbib, Z.; González-López, C.V. Biostimulant Capacity of Chlorella and Chlamydopodium Species Produced Using Wastewater and Centrate. Biology 2022, 11, 1086. [Google Scholar] [CrossRef]
- Gitau, M.M.; Farkas, A.; Balla, B.; Ördög, V.; Futó, Z.; Maróti, G. Strain-Specific Biostimulant Effects of Chlorella and Chlamydomonas Green Microalgae on Medicago truncatula. Plants 2021, 10, 1060. [Google Scholar] [CrossRef] [PubMed]
- Alling, T.; Funk, C.; Gentili, F.G. Nordic microalgae produce biostimulant for the germination of tomato and barley seeds. Sci. Rep. 2023, 13, 3509. [Google Scholar] [CrossRef] [PubMed]
- Martini, F.; Beghini, G.; Zanin, L.; Varanini, Z.; Zamboni, A.; Ballottari, M. The potential use of Chlamydomonas reinhardtii and Chlorella sorokiniana as biostimulants on maize plants. Algal Res. 2021, 60, 102515. [Google Scholar] [CrossRef]
- Gharib, F.A.E.L.; Osama, K.; El Sattar, A.M.A.; Ahmed, E.Z. Impact of Chlorella vulgaris, Nannochloropsis salina, and Arthrospira platensis as bio-stimulants on common bean plant growth, yield and antioxidant capacity. Sci. Rep. 2024, 14, 1398. [Google Scholar] [CrossRef] [PubMed]
- Viana, C.; Genevace, M.; Gama, F.; Coelho, L.; Pereira, H.; Varela, J.; Reis, M. Chlorella vulgaris and Tetradesmus obliquus Protect Spinach (Spinacia oleracea L.) against Fusarium oxysporum. Plants 2024, 13, 1697. [Google Scholar] [CrossRef]
- Mrid, R.B.; Benmrid, B.; Hafsa, J.; Boukcim, H.; Sobeh, M.; Yasri, A. Secondary metabolites as biostimulant and bioprotectant agents: A review. Sci. Total Environ. 2021, 777, 146204. [Google Scholar] [CrossRef]
- Guercio, A.M.; Palayam, M.; Shabek, N. Strigolactones: Diversity, perception, and hydrolysis. Phytochem. Rev. 2023, 22, 339–359. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, S.; Wei, P.; Guo, S.; Wu, J. A briefly overview of the research progress for the abscisic acid analogues. Front. Chem. 2022, 10, 967404. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.; Grande, P.M.; Blank, L.M.; Klose, H. Insights into cell wall disintegration of Chlorella vulgaris. PLoS ONE 2022, 17, e0262500. [Google Scholar] [CrossRef]
- Carullo, D.; Abera, B.D.; Scognamiglio, M.; Donsì, F.; Ferrari, G.; Pataro, G. Application of Pulsed Electric Fields and High-Pressure Homogenization in Biorefinery Cascade of C. vulgaris Microalgae. Foods 2022, 11, 471. [Google Scholar] [CrossRef]
- Ritchie, R.J. Universal chlorophyll equations for estimating chlorophylls a, b, c, and d and total chlorophylls in natural assemblages of photosynthetic organisms using acetone, methanol, or ethanol solvents. Photosynthetica 2008, 46, 115–126. [Google Scholar] [CrossRef]
- Couso, I.; Vila, M.; Vigara, J.; Cordero, B.F.; Vargas, M.; Rodríguez, H.; León, R. Synthesis of carotenoids and regulation of the carotenoid biosynthesis pathway in response to high light stress in the unicellular microalga Chlamydomonas reinhardtii. Eur. J. Phycol. 2012, 47, 223–232. [Google Scholar] [CrossRef]
- Zucconi, F.; Forte, M.; Monaco, A.; De Bertoldi, M. Biological evaluation of compost maturity. Biocycle 1981, 22, 27–29. [Google Scholar]

| Strain | P (g L−1 d−1) | µ (d−1) | Protein (% DW) | PP (g L−1 d−1) |
|---|---|---|---|---|
| WT | 1.51 ± 0.05 b | 1.05 ± 0.02 d | 32.1 ± 1.1 c | 0.56 ± 0.05 b |
| A6 | 1.54 ± 0.32 b | 1.05 ± 0.06 d | 31.8 ± 0.8 c,d | 0.51 ± 0.03 b,c,d |
| B4 | 1.26 ± 0.02 b,e | 0.99 ± 0.01 c,d | 29.8 ± 1.1 c,d | 0.46 ± 0.02 b,c,d |
| C1 | 1.39 ± 0.02 b,c | 1.05 ± 0.01 c | 29.9 ± 0.8 c,d | 0.47 ± 0.06 b,c,d |
| C3 | 1.54 ± 0.05 b | 1.07 ± 0.02 c | 29.2 ± 0.7 c,d | 0.53 ± 0.02 b,d |
| C4 | 1.52 ± 0.10 b | 1.04 ± 0.02 c | 31.0 ± 1.4 c,d | 0.68 ± 0.12 a,b,d |
| C5 | 1.88 ± 0.13 a,b | 1.12 ± 0.02 b,c | 38.2 ± 0.4 b | 0.66 ± 0.01 a,b,d |
| C10 | 1.49 ± 0.13 b | 1.07 ± 0.01 c | 30.8 ± 0.9 c,d | 0.50 ± 0.02 b,c,d |
| D3 | 1.40 ± 0.03 b,d | 1.05 ± 0.01 c | 33.3 ± 0.4 c | 0.50 ± 0.01 b,c,d |
| D4 | 1.35 ± 0.10 b,d | 1.05 ± 0.01 c | 30.3 ± 0.4 c,d | 0.46 ± 0.04 b,c,d |
| D5 | 1.36 ± 0.08 b,d | 1.05 ± 0.02 c | 28.0 ± 0.7 c,d | 0.44 ± 0.02 b,c,d |
| E2 | 0.84 ± 0.04 e | 0.95 ± 0.01 d | 39.1 ± 0.2 b | 0.43 ± 0.03 b,c,d |
| E4 | 1.24 ± 0.15 b,d | 1.02 ± 0.04 c,d | 27.9 ± 1.5 c,d | 0.39 ± 0.01 d |
| E5 | 1.22 ± 0.04 b,d | 1.02 ± 0.01 c,d | 30.3 ± 0.7 c,d | 0.43 ± 0.02 b,c,d |
| F4 | 2.08 ± 0.17 a | 1.28 ± 0.05 a | 38.1 ± 0.5 b | 0.64 ± 0.03 a,b |
| F5 | 1.88 ± 0.08 a,b | 1.26 ± 0.03 a | 38.2 ± 1.1 b | 0.63 ± 0.02 a,b |
| G2 | 1.67 ± 0.18 b | 1.23 ± 0.01 a | 43.0 ± 1.7 a | 0.60 ± 0.03 a,b |
| G3 | 1.55 ± 0.10 b | 1.05 ± 0.01 c | 35.6 ± 1.7 b,c | 0.60 ± 0.05 a,b |
| Chlorophyll a | Chlorophyll b | Total Chlorophyll | Neoxanthin | Violaxanthin | Lutein | β-Carotene | |
|---|---|---|---|---|---|---|---|
| WT | 0.30 ± 0.03 a | 0.18 ± 0.02 a | 0.48 ± 0.05 a | 0.36 ± 0.01 a | 0.17 ± 0.01 | 0.53 ± 0.01 a | 0.74 ± 0.02 a |
| F4 | 0.54 ± 0.01 b | 0.24 ± 0.01 c | 0.78 ± 0.01 c | 0.20 ± 0.01 b | <LOQ | 0.59 ± 0.04 a | 0.37 ± 0.05 b |
| G2 | 0.48 ± 0.03 b | 0.21 ± 0.01 e | 0.69 ± 0.04 c | 0.22 ± 0.03 b | n.d. | 0.82 ± 0.08 b | 0.67 ± 0.07 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trovão, M.; Schüler, L.; Pedroso, H.; Reis, A.; Santo, G.E.; Barros, A.; Correia, N.; Ribeiro, J.; Bombo, G.; Gama, F.; et al. Isolation and Selection of Protein-Rich Mutants of Chlorella vulgaris by Fluorescence-Activated Cell Sorting with Enhanced Biostimulant Activity to Germinate Garden Cress Seeds. Plants 2024, 13, 2441. https://doi.org/10.3390/plants13172441
Trovão M, Schüler L, Pedroso H, Reis A, Santo GE, Barros A, Correia N, Ribeiro J, Bombo G, Gama F, et al. Isolation and Selection of Protein-Rich Mutants of Chlorella vulgaris by Fluorescence-Activated Cell Sorting with Enhanced Biostimulant Activity to Germinate Garden Cress Seeds. Plants. 2024; 13(17):2441. https://doi.org/10.3390/plants13172441
Chicago/Turabian StyleTrovão, Mafalda, Lisa Schüler, Humberto Pedroso, Ana Reis, Gonçalo Espírito Santo, Ana Barros, Nádia Correia, Joana Ribeiro, Gabriel Bombo, Florinda Gama, and et al. 2024. "Isolation and Selection of Protein-Rich Mutants of Chlorella vulgaris by Fluorescence-Activated Cell Sorting with Enhanced Biostimulant Activity to Germinate Garden Cress Seeds" Plants 13, no. 17: 2441. https://doi.org/10.3390/plants13172441










