A Fast, Efficient, and Tissue-Culture-Independent Genetic Transformation Method for Panax notoginseng and Lilium regale
Abstract
:1. Introduction
2. Results
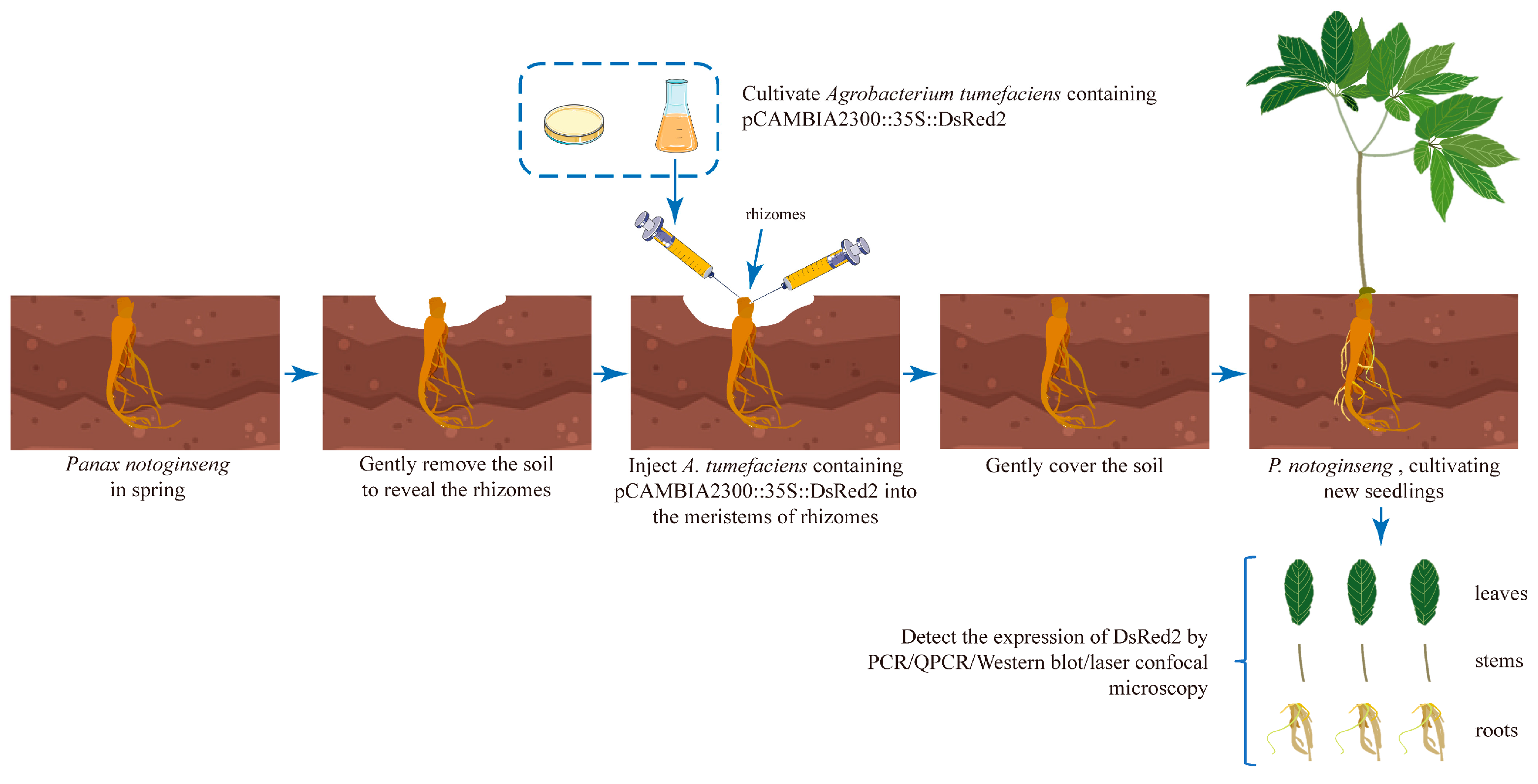
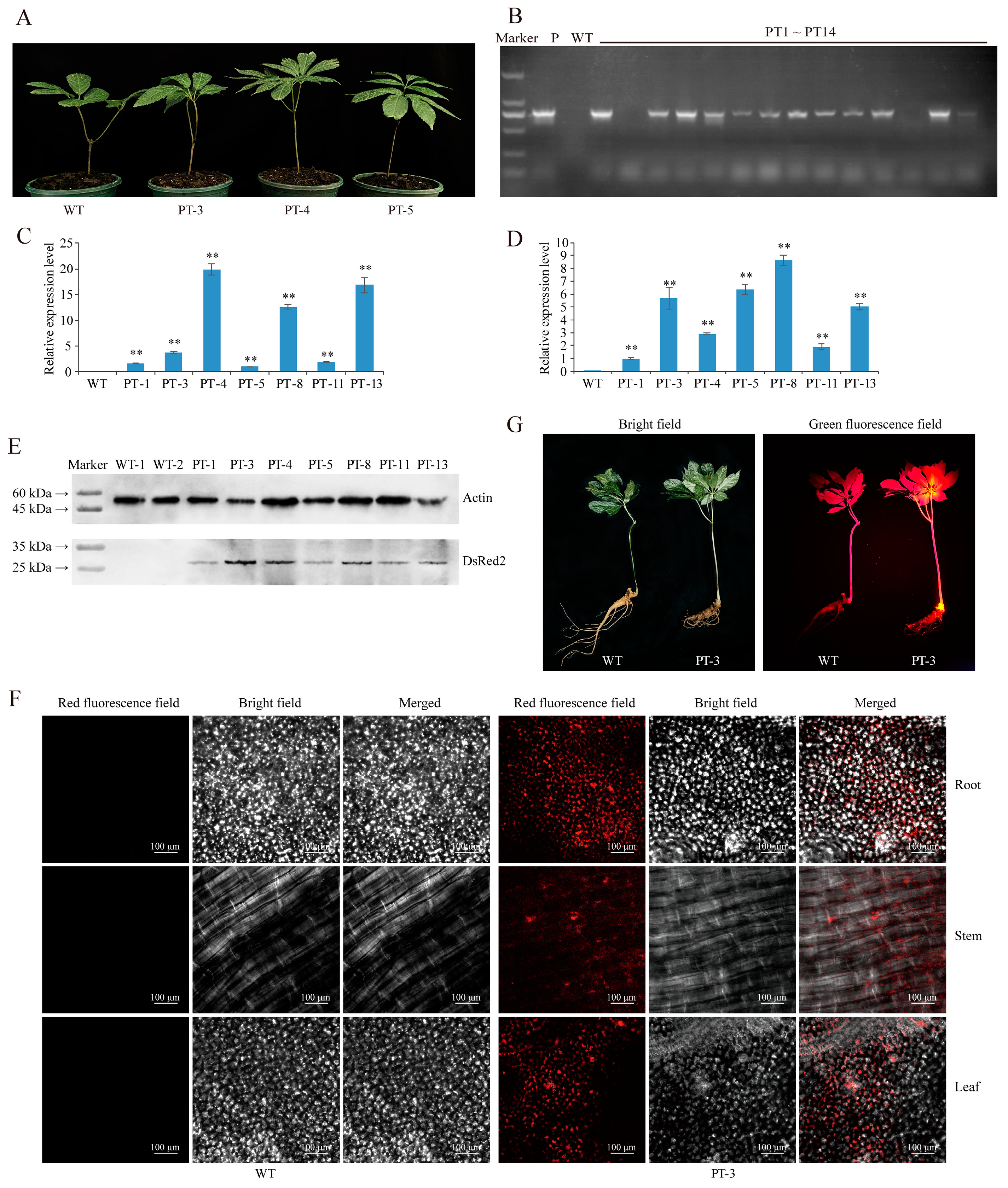
2.1. Establishment of Agrobacterium-Based Injection Rhizome Transformation Method in P. notoginseng
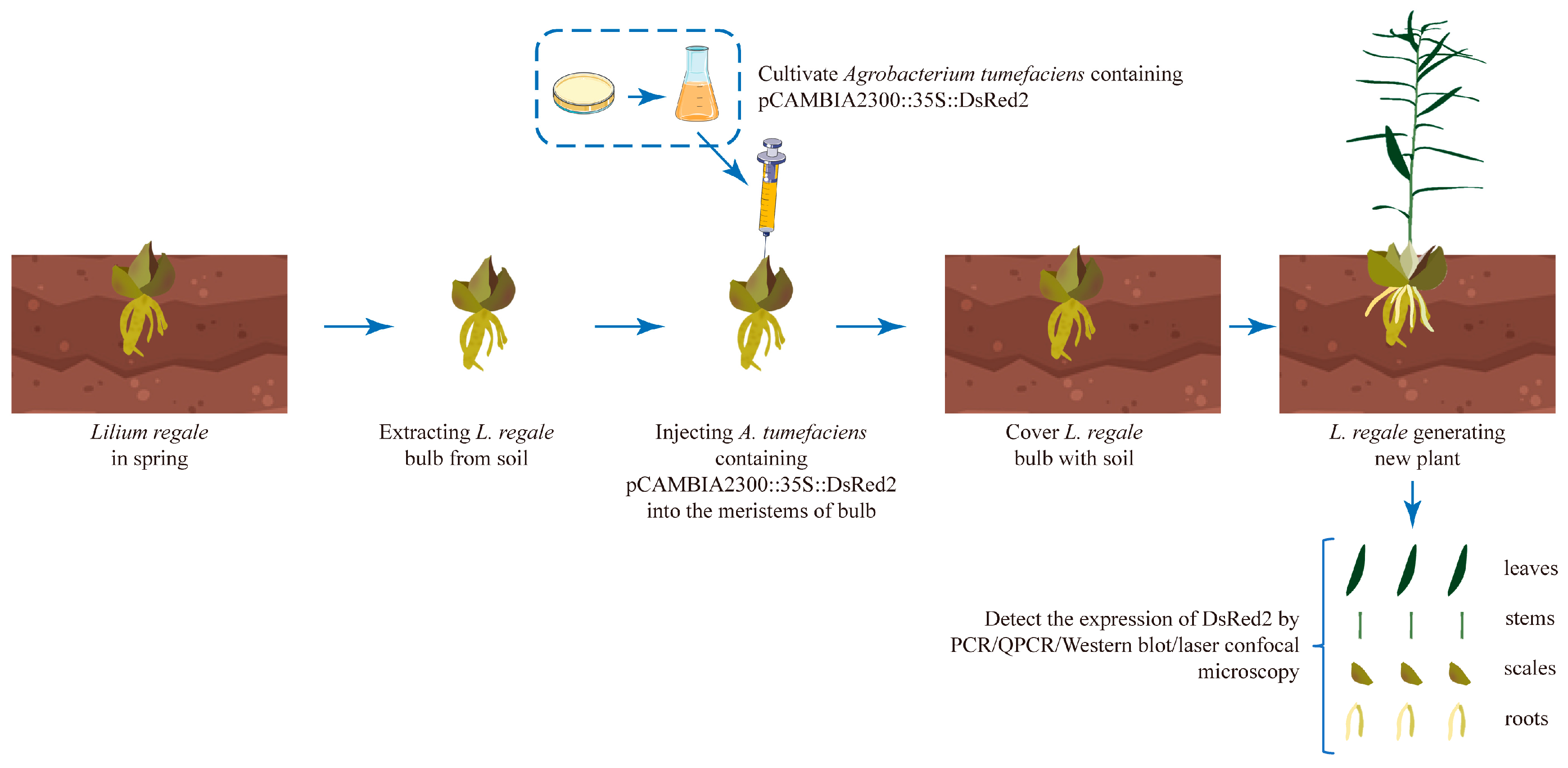
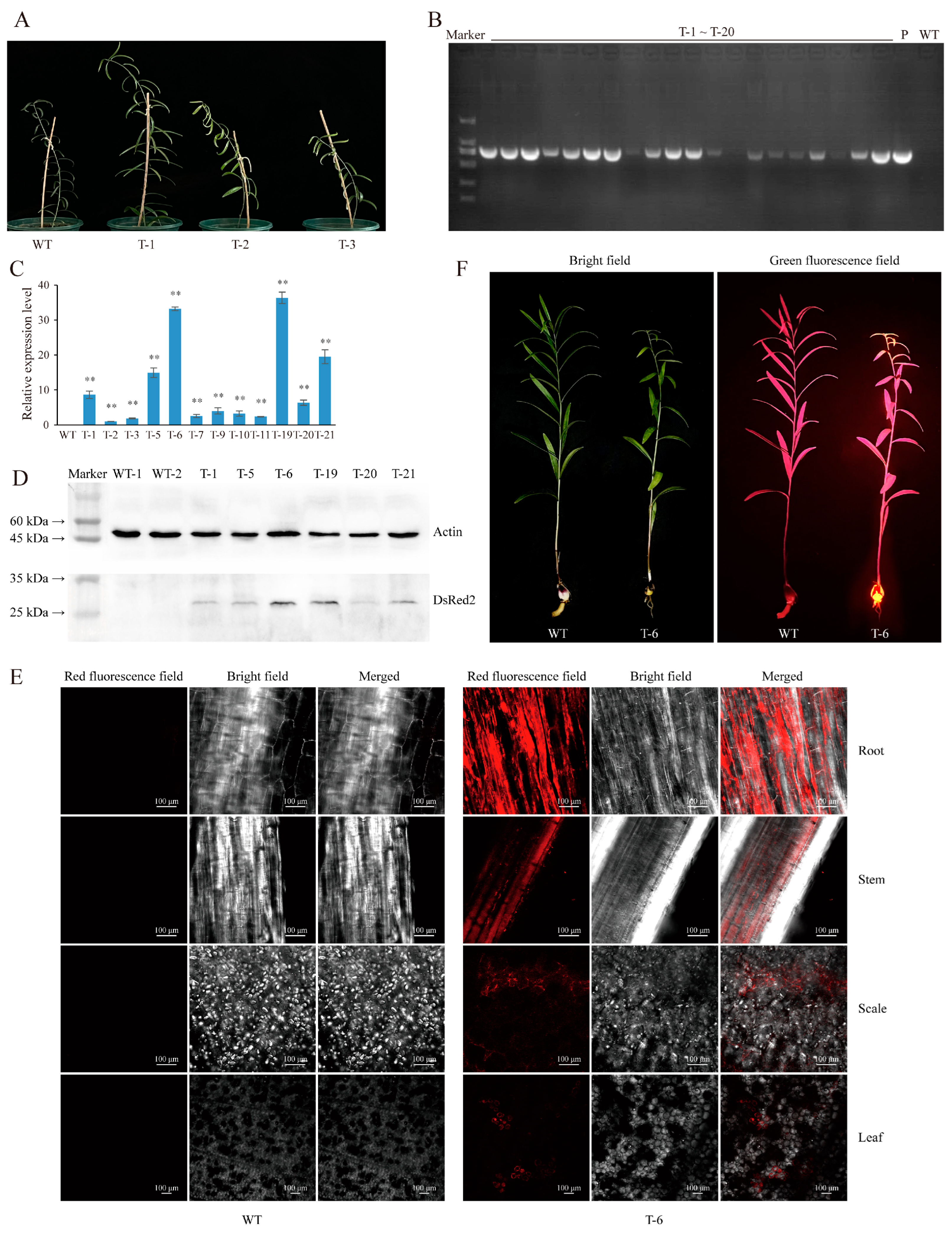
2.2. Agrobacterium-Based Injection in the Bulb Meristems Is a Fast and Efficient L. regale Genetic Transformation Method
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. A. tumefaciens Strain, Binary Vector, and Culture Preparation
4.3. Plant Preparation and Infection
4.4. PCR Screening of Positive Transgenic P. notoginseng and L. regale Plants
4.5. qRT-PCR-Based Detection of DsRed2 Expression
4.6. Analysis of Red Fluorescent Protein Expression through Western Blot
4.7. Observation of Specific Red Fluorescence in Different Organs of DsRed2 Transgenic Plants Using Laser Confocal Microscopy
4.8. Observing Red Fluorescence in Transgenic Plants Using a Handheld Fluorescence Detector
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krenek, P.; Samajova, O.; Luptovciak, I.; Doskocilova, A.; Komis, G.; Samaj, J. Transient plant transformation mediated by Agrobacterium tumefaciens: Principles, methods and applications. Biotechnol. Adv. 2015, 33, 1024–1042. [Google Scholar] [CrossRef]
- Azizi-Dargahlou, S.; Pouresmaeil, M. Agrobacterium tumefaciens-mediated plant transformation: A review. Mol. Biotechnol. 2024, 66, 1563–1580. [Google Scholar] [CrossRef] [PubMed]
- Shrawat, A.K.; Lörz, H. Agrobacterium-mediated transformation of cereals: A promising approach crossing barriers. Plant Biotechnol. J. 2006, 4, 575–603. [Google Scholar] [CrossRef]
- Rizwan, H.M.; Yang, Q.; Yousef, A.F.; Zhang, X.; Sharif, Y.; Kaijie, J.; Shi, M.; Li, H.; Munir, N.; Yang, X.; et al. Establishment of a novel and efficient Agrobacterium-Mediated in planta transformation system for passion fruit (Passiflora edulis). Plants 2021, 10, 2459. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.R.; Sun, X.; Xie, Y.M.; Du, A.; Chen, M.; Lai, S.S.; Wei, X.H.; Ji, L.L.; Wang, C.H. Panax notoginseng saponins alleviate diabetic retinopathy by inhibiting retinal inflammation: Association with the NF-κB signaling pathway. J. Ethnopharmacol. 2024, 319, 117135. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Zhang, D.; Yang, D.C. Biosynthesis and biotechnological production of ginsenosides. Biotechnol. Adv. 2015, 33, 717–735. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Koo, H.B.; Jeon, S.W.; Han, J.Y.; Kim, J.S.; Jun, K.M.; Choi, Y.E. Modification of ginsenoside saponin composition via the CRISPR/Cas9-mediated knockout of protopanaxadiol 6-hydroxylase gene in Panax ginseng. J. Ginseng Res. 2022, 46, 505–514. [Google Scholar] [CrossRef]
- Liu, S.Z.; Chen, X.X.; Zhao, T.Q.; Yu, J.H.; Chen, P.; Wang, Y.F.; Wang, K.Y.; Zhao, M.Z.; Jiang, Y.; Wang, Y.; et al. Identification of PgRg1-3 gene for ginsenoside Rg1 biosynthesis as revealed by combining genome-wide association study and gene co-expression network analysis of jilin ginseng core collection. Plants 2024, 13, 1784. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, Q.; Zeng, Z.X.; Wang, Y.; Zhao, M.Z.; Wang, K.G.; Zhang, M.P. The AP2/ERF transcription factor PgERF120 regulates ginsenoside biosynthesis in ginseng. Biomolecules 2024, 14, 345. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, Y.L.; Jiang, S.; Yu, H.; Xiang, Y.Y.; Liu, D.Q.; Qu, Y.; Cui, X.M.; Ge, F. Oleanane-type saponins biosynthesis in Panax notoginseng via transformation of β-Amyrin synthase gene from Panax japonicus. J. Agric. Food Chem. 2019, 67, 1982–1989. [Google Scholar] [CrossRef]
- Shi, Y.; Man, J.H.; Huang, Y.Y.; Zhang, J.H.; Zhang, Z.F.; Yin, G.Y.; Wang, X.; Liu, S.H.; Chen, Y.; Wang, X.H.; et al. Overexpression of PnMYB2 from Panax notoginseng induces cellulose and lignin biosynthesis during cell wall formation. Planta 2022, 255, 107. [Google Scholar] [CrossRef]
- Sui, J.J.; Jia, W.J.; Xin, Y.; Zhang, Y.Y. Transcriptomics-based identification of genes related to tapetum degradation and microspore development in lily. Genes 2022, 13, 366. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Wang, Z.P.; Ren, Y.M.; Li, H.Y.; Liu, N.; Sun, H.M. Establishment of efficient genetic transformation systems and application of CRISPR/Cas9 genome editing technology in Lilium pumilum DC. Fisch. and Lilium longiflorum White Heaven. Int. J. Mol. Sci. 2019, 20, 2920. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Cui, Q.; Yuan, L.; Jia, G.X. Efficient Agrobacterium tumefaciens (Smith & Towns.) conn-mediated transformation of Lilium ‘Sorbonne’ with genes encoding anthocyanin regulators. Can. J. Plant Sci. 2017, 97, 796–807. [Google Scholar]
- Fu, Y.Y.; Shu, L.L.; Li, H.Y.; Zhang, X.M.; Liu, X.; Ou, Z.Y.; Liang, X.M.; Qi, X.Y.; Yang, L.P. Establishment of highly efficient plant regeneration, callus transformation and analysis of Botrytis cinerea-responsive PR promoters in Lilium brownii var. viridulum. Plants 2023, 12, 1992. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.F.; Ma, X.; Jin, G.; Han, D.Y.; Xue, J.; Du, Y.P.; Chen, X.Q.; Yang, F.P.; Zhao, C.N.; Zhang, X.H. A modified method for transient transformation via pollen magnetofection in Lilium germplasm. Int. J. Mol. Sci. 2023, 24, 15304. [Google Scholar] [CrossRef]
- Sun, L.; Alariqi, M.; Zhu, Y.; Li, J.Y.; Li, Z.L.; Wang, Q.; Li, Y.J.; Rui, H.P.; Zhang, X.L.; Jin, S.X. Red fluorescent protein (DsRed2), an ideal reporter for cotton genetic transformation and molecular breeding. Crop J. 2018, 6, 366–376. [Google Scholar] [CrossRef]
- Huai, D.X.; Wu, J.; Xue, X.M.; Hu, M.L.; Zhi, C.Y.; Pandey, M.K.; Liu, N.; Huang, L.; Bai, D.M.; Yan, L.Y.; et al. Red fluorescence protein (DsRed2) promotes the screening efficiency in peanut genetic transformation. Front. Plant Sci. 2023, 14, 1123644. [Google Scholar] [CrossRef] [PubMed]
- Nandy, D.; Maity, A.; Mitra, A.K. Target-specific gene delivery in plant systems and their expression: Insights into recent developments. J. Biosci. 2020, 45, 30. [Google Scholar] [CrossRef]
- Filipecki, M.; Malepszy, S. Unintended consequences of plant transformation: A molecular insight. J. Appl. Genet. 2006, 47, 277–286. [Google Scholar] [CrossRef]
- Dong, O.X.; Ronald, P.C. Targeted DNA insertion in plants. Proc. Natl. Acad. Sci. USA 2021, 118, e2004834117. [Google Scholar] [CrossRef]
- Su, W.B.; Xu, M.Y.; Radani, Y.; Yang, L.M. Technological development and application of plant genetic transformation. Int. J. Mol. Sci. 2023, 24, 10646. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.S.; Xie, H.T.; Song, M.L.; Lu, J.H.; Ma, P.; Huang, B.Y.; Wang, M.G.; Tian, Y.F.; Chen, F.; Peng, J.; et al. Cut-dip-budding delivery system enables genetic modifications in plants without tissue culture. Innovation 2022, 4, 100345. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.H.; Li, S.S.; Deng, S.A.; Wang, M.G.; Wu, Y.H.; Li, M.; Dong, J.S.; Lu, S.H.; Su, C.L.; Li, G.F.; et al. A method of genetic transformation and gene editing of succulents without tissue culture. Plant Biotechnol. J. 2024, 22, 1981–1988. [Google Scholar] [CrossRef]
- Mei, G.G.; Chen, A.; Wang, Y.R.; Li, S.Q.; Wu, M.Y.; Hu, Y.L.; Liu, X.; Hou, X.L. A simple and efficient in planta transformation method based on the active regeneration capacity of plants. Plant Commun. 2024, 5, 100822. [Google Scholar] [CrossRef]
- Shah, T.; Andleeb, T.; Lateef, S.; Noor, M.A. Genome editing in plants: Advancing crop transformation and overview of tools. Plant Physiol. Biochem. 2018, 131, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Q.; Zhang, Z.Y.; Unver, T.; Zhang, B.H. CRISPR/Cas: A powerful tool for gene function study and crop improvement. J. Adv. Res. 2020, 29, 207–221. [Google Scholar] [CrossRef]
- Song, S.L.; Yan, R.; Wang, C.X.; Wang, J.X.; Sun, H.M. Improvement of a genetic transformation system and preliminary study on the function of LpABCB21 and LpPILS7 based on somatic embryogenesis in Lilium pumilum DC. Fisch. Int. J. Mol. Sci. 2020, 21, 6784. [Google Scholar] [CrossRef]
- Su, L.L.; Li, W.Y.; Chen, X.H.; Wang, P.C.; Liu, D.Q. Proline-rich protein PRPL1 enhances Panax notoginseng defence against Fusarium solani by regulating reactive oxygen species balance and strengthening the cell wall barrier. Plant Cell Environ. 2024, 47, 2377–2395. [Google Scholar] [CrossRef]
- Rao, X.Y.; Huang, X.L.; Zhou, Z.C.; Lin, X. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, J.; Li, W.; Li, X.; Liu, D.; Liu, G. A Fast, Efficient, and Tissue-Culture-Independent Genetic Transformation Method for Panax notoginseng and Lilium regale. Plants 2024, 13, 2509. https://doi.org/10.3390/plants13172509
Deng J, Li W, Li X, Liu D, Liu G. A Fast, Efficient, and Tissue-Culture-Independent Genetic Transformation Method for Panax notoginseng and Lilium regale. Plants. 2024; 13(17):2509. https://doi.org/10.3390/plants13172509
Chicago/Turabian StyleDeng, Jie, Wenyun Li, Xiaomin Li, Diqiu Liu, and Guanze Liu. 2024. "A Fast, Efficient, and Tissue-Culture-Independent Genetic Transformation Method for Panax notoginseng and Lilium regale" Plants 13, no. 17: 2509. https://doi.org/10.3390/plants13172509
APA StyleDeng, J., Li, W., Li, X., Liu, D., & Liu, G. (2024). A Fast, Efficient, and Tissue-Culture-Independent Genetic Transformation Method for Panax notoginseng and Lilium regale. Plants, 13(17), 2509. https://doi.org/10.3390/plants13172509






