Multiomics Analysis of the Mechanism by Which Gibberellin Alleviates S-Metolachlor Toxicity in Rice Seedlings
Abstract
:1. Introduction
2. Results
2.1. GA Recovered S-Metolachlor Inhibition of Rice Seedlings
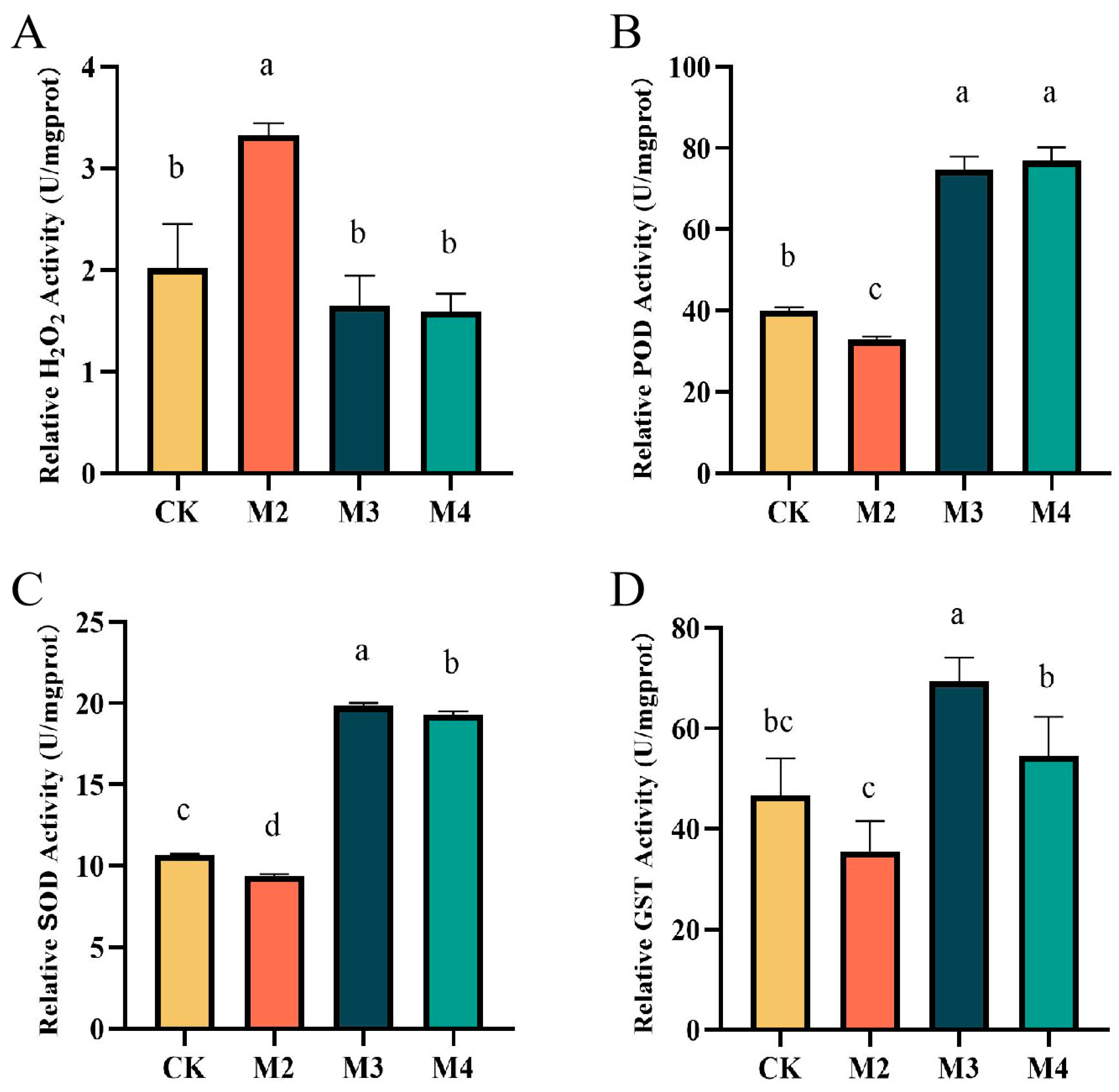
2.2. S-Metolachlor and GA3 Affect the Activity of Various Enzymes
2.3. GA3 Alleviates Toxicity by Affecting the Synthesis Pathway of Flavonoids
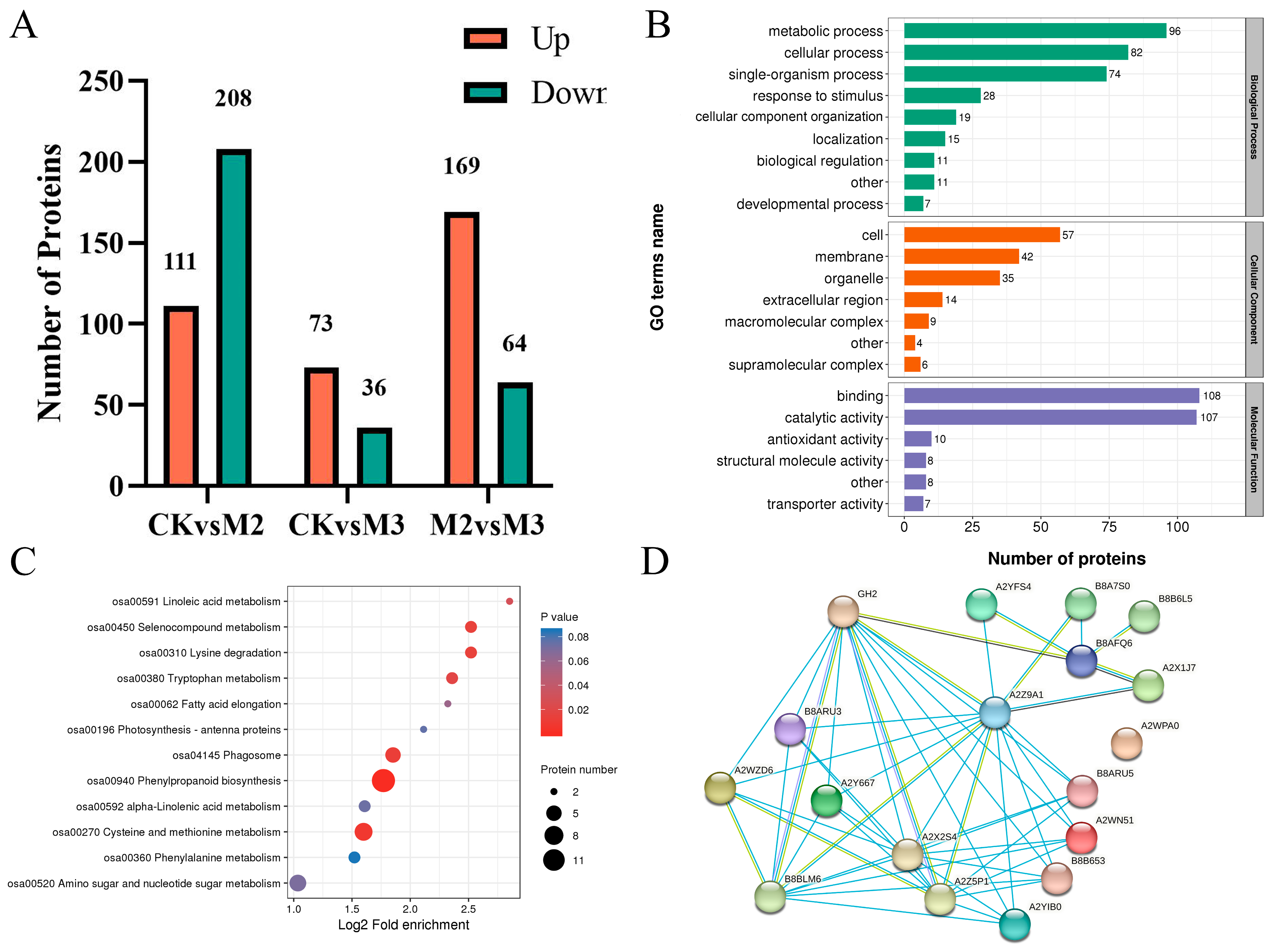
2.4. GA3 Affects Gene Expression on Multiple Pathways
2.5. GA3 Affects Multiple Protein Synthesis Pathways
2.6. Conjoint Analysis
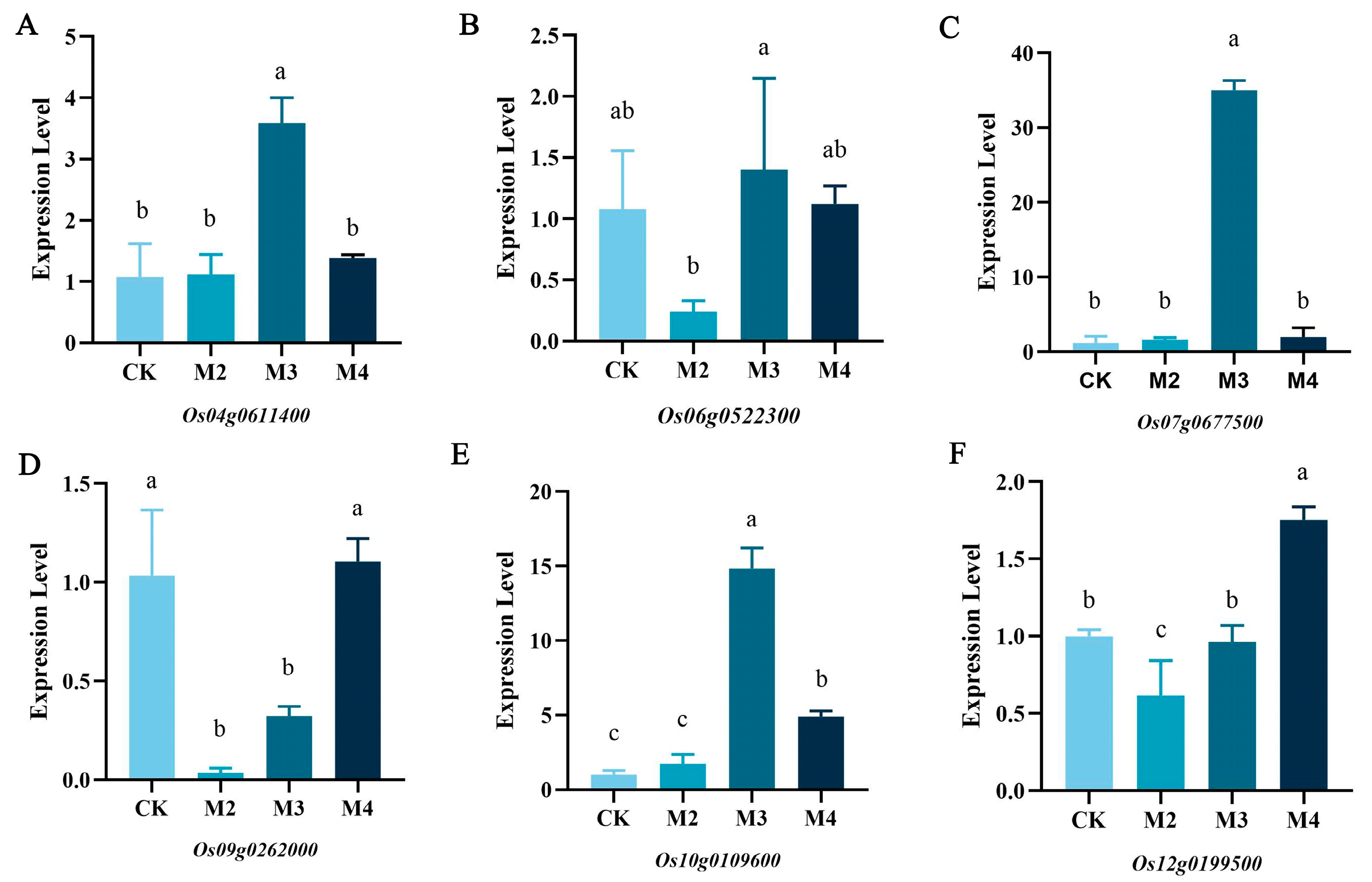
2.7. Q-PCR Validation of Candidate Genes
3. Discussion
3.1. GA3 Recovered S-Metolachor Inhibition of Rice Seedlings
3.2. GA3 Alleviates S-Metolachlor Toxicity by Affecting Enzyme Activity
3.3. Adding GA3 Affects the Synthesis of Flavonoids
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Determination of Enzyme Activities
4.3. Metabolome Analysis
4.4. RNA-Seq Analysis
4.5. Proteomics Analysis
4.6. Differential Genes and Proteins GO Analysis
4.7. Real Time Quantative PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xia, C.X.; Liang, G.H.; Chong, K.; Xu, Y.Y. The COG1-OsSERL2 complex senses cold to trigger signaling network for chilling tolerance in japonica rice. Nat. Commun. 2023, 14, 3104. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.Q.; Li, R.H.; Chu, S.H.; Zhu, W.D.; Chen, Q.Z. Weed communities and distribution characteristics in the paddy fields of main rice growing regions of Hubei Province. Chin. J. Eco-Agric. 2009, 17, 533–536. [Google Scholar] [CrossRef]
- Obiri, B.D.; Obeng, E.A.; Oduro, K.A. Farmers’ perceptions of herbicide usage in forest landscape restoration programs in Ghana. Sci. Afr. 2021, 11, e00672. [Google Scholar] [CrossRef]
- Qu, Q.; Li, Y.; Zhang, Z.Y.; Cui, H.Z.; Cui, A.; Zhao, Q.Q.; Liu, W.Y.; Lu, T.; Qian, H.F. Effects of s-metolachlor on wheat (Triticum aestivum L.) seedling root exudates and the rhizosphere microbiome. J. Hazard. Mater. 2021, 411, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.K.; Zhou, X.M.; Wu, J.; Li, J.B.; Bai, L.Y. Effect and mechanism of Sanshools on alleviating injury from metolachlor in rice. Pestic. Biochem. Physiol. 2014, 110, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Demaill, F.; Elfeky, I.; Malbezin, L.; Guédard, M.L.; Eon, M.; Bessoule, J.J.; Agnès, F.M.; Delmas, F.; Mazzella, N.; Gonzalez, P.; et al. Impact of diuron and S-metolachlor on the freshwater diatom Gomphonema gracile: Complementarity between fatty acid profiles and different kinds of ecotoxicological impact-endpoints. Sci. Total Environ. 2019, 688, 960–969. [Google Scholar] [CrossRef]
- Liu, H.J.; Huang, R.N.; Xie, F.; Zhang, S.X.; Shi, J. Enantioselective phytotoxicity of metolachlor against maize and rice roots. J. Hazard. Mater. 2012, 217, 330–337. [Google Scholar] [CrossRef]
- Liu, H.; Xiong, M. Comparative toxicity of racemic metolachlor and s-metolachlor to Chlorella pyrenoidosa. Aquat. Toxicol. 2009, 93, 100–106. [Google Scholar] [CrossRef]
- Kang, T.; Gao, S.; Zhao, L.X.; Zhai, Y.; Ye, F.; Fu, Y. Design, synthesis, and SAR of novel 1,3-disubstituted imidazolidine or hexahydropyrimidine derivatives as herbicide safeners. J. Agric. Food Chem. 2021, 69, 45–54. [Google Scholar] [CrossRef]
- Acharya, S.P.; Johnson, J.; Weidhaas, J. Adsorption kinetics of the herbicide safeners, benoxacor and furilazole, to activated carbon and agricultural soils. J. Environ. Sci. 2020, 89, 23–24. [Google Scholar] [CrossRef]
- Ye, F.; Zhai, Y.; Huo, K.L.; Liu, Y.X.; Li, N.; Gao, S.; Zhao, L.X.; Fu, Y. Safeners Improve Maize Tolerance under Herbicide Toxicity Stress by Increasing the Activity of Enzymes in vivo. J. Agric. Food Chem. 2019, 67, 11568–11576. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.G.; Rieu, I.; Steber, C.M. Gibberellin Metabolism and Signaling. Vitam. Horm. 2005, 72, 289–338. [Google Scholar]
- Richards, D.E.; King, K.E.; Ait-Ali, T.; Harberd, N.P. How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. 2001, 52, 67–88. [Google Scholar] [CrossRef] [PubMed]
- Dayan, J. Gibberellin Transport. Annu. Plant Rev. Gibberellins 2016, 49, 95–120. [Google Scholar]
- Wang, H.Z.; Ma, J.; Li, X.Y.; Zhang, R.P. Relationship between Physio-biochemical Characters and Drought Tolerance of Rice at Seedling Stage. Acta Agric. Boreali-Occident. Sin. 2009, 24, 174–178. [Google Scholar]
- Deng, X.L. A Mini Review on Natural Safeners: Chemistry, Uses, Modes of Action, and Limitations. Plants 2022, 11, 3509. [Google Scholar] [CrossRef] [PubMed]
- Busi, R. Resistance to herbicides inhibiting the biosynthesis of very-long-chain fatty acids. Pest Manag. Sci. 2014, 70, 1378–1384. [Google Scholar] [CrossRef] [PubMed]
- Kouame, B.J.; Mary, C.S.; Cammy, D.W.; Matthew, B.B.; Thomas, R.B.; Erin, G.; Nilda, R.B. S-metolachlor persistence in soil as influenced by within-season and inter-annual herbicide use. Environ. Adv. 2022, 9, 100318. [Google Scholar] [CrossRef]
- Tao, B.; Wang, Y.K.; Li, D.P.; Li, J.J. Study on detoxification effect of safeners AD-67 on S-metolachlor. J. Northeast. Agric. Univ. 2019, 50, 29–35. [Google Scholar]
- Liu, Y.Y.; Jing, W.; Ke, X.C.; Zhang, J.; Sun, X.D.; Wang, C.T.; Yang, Y.P. Gibberellin inhibition of taproot formation by modulation of DELLA-NAC complex activity in turnip (Brassica rapa var. rapa). Protoplasma 2021, 258, 925–934. [Google Scholar] [CrossRef]
- Marikovsky, M.; Ziv, V.; Nevo, N.; Catherine, H.C.; Mahler, O. Cu/Zn superoxide dismutase plays important role in immune response. J. Immunol. 2003, 170, 2993–3001. [Google Scholar] [CrossRef] [PubMed]
- Wu, R. Phytotoxicity of s-metolachlor to wheat and its safener. Plant Prot. 2017, 43, 99–105. [Google Scholar]
- Gray, S.B.; Brady, S.M. Plant developmental responses to climate change. Dev. Biol. 2016, 419, 64–77. [Google Scholar] [CrossRef]
- Nianiou-Obeidat, I.; Madesis, P.; Kissoudis, C.; Voulgari, G.; Chronopoulou, E.; Tsaftaris, A.; Labrou, N.E. Plant glutathione transferase-mediated stress tolerance: Functions and biotechnological applications. Plant Cell Rep. 2017, 36, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Natalia, C.Q.; Harro, J.B.; Keurentjes, J.J. Genetic analysis of metabolome-phenotype interactions: From model to crop species. Trends Genet. 2013, 29, 41–50. [Google Scholar]
- Saito, K. Phytochemical genomics-a new trend. Curr. Opin. Plant Biol. 2013, 16, 373–380. [Google Scholar] [CrossRef]
- Wu, H.Z.; Zhao, W.R.; Zhou, J.J.; Xie, X.Q.; Zhong, X.Y.; Liu, Y.J.; Shi, L.L. Extraction, analysis of antioxidant activities and structural characteristics of flavonoids in fruits of Diospyros Lotus L. Food Sci. Technol. 2024, 201, 116248. [Google Scholar] [CrossRef]
- Heim, K.E.; Tagliaferro, A.R.; Bobilya, D.J. Flavonoid antioxidants: Chemistry, metabolism and structure-activity relationships. J. Nutr. Biochem. 2002, 13, 572–584. [Google Scholar] [CrossRef]
- Bassard, J.E.; Ullmann, P.; Bernier, F.; Werck-Reichhart, D. Phenolamides: Bridging polyamines to the phenolic metabolism. Phytochemistry 2010, 71, 1808–1824. [Google Scholar] [CrossRef]
- Zheng, Y.C.; Wang, P.J.; Chen, X.J.; Yue, C.; Guo, Y.C.; Yang, J.F.; Sun, Y.; Ye, N.X. Integrated transcriptomics and metabolomics provide novel insight into changes in specialized metabolites in an albino tea cultivar (Camellia Sinensis (L.) O. Kuntz). Plant Physiol. Biochem. 2021, 160, 27–36. [Google Scholar] [CrossRef]
- Tasiu, I. Stress and defense responses in plant secondary metabolites production. Biol. Research. 2019, 52, 1–25. [Google Scholar]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Gordon, V.L.; Marianne, E.B.; Michelle, C.M.; Thomas, J.B.; Bradley, S.M.; Joseph, P.N. Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases. Chem. Biol. 2006, 13, 1327–1338. [Google Scholar]
- Zhang, C.; Zang, Y.; Liu, P.; Zheng, Z.J.; Ouyang, J. Characterization, functional analysis and application of 4-coumarate: CoA ligase genes from Populus trichocarpa. J. Biotechnol. 2019, 302, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Burbulis, I.E.; Brenda, W.S. Interactions among enzymes of the Arabidopsis flavonoid bioynthetic pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 12929–12934. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, Y.J.; Kim, B.G.; Lim, Y.H.; Ahn, J.H. Flavanone 3β-hydroxylases from rice: Key enzymes for favonol and anthocyanin biosynthesis. Mol. Cells 2008, 25, 312–316. [Google Scholar] [CrossRef]
- Zeng, Q.; Deng, X.L.; Zhou, S.F.; Wang, C.; Yang, L.; Tang, Y.; Liu, S.H. The Effect and Mechanism of Gibberellin Alleviate the Phytotoxicity of S-Metolachlor on Rice Seedlings. Agrochemicals 2019, 58, 519–522. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zhang, C.H.; Ge, Y. Response of Glutathione and Glutathione S-transferase in Rice Seedlings Exposed to Cadmium Stress. Rice Sci. 2008, 15, 73–76. [Google Scholar] [CrossRef]
- Alexander, V.P.; Christine, C.W. A microtiter plate assay for superoxide dismutase using a water-soluble tetrazolium salt (WST-1). Clin. Chim. Acta 2000, 293, 157–166. [Google Scholar]




| DAPs | CKvsM2 | CKvsM3 | M2vsM3 | ||||
|---|---|---|---|---|---|---|---|
| Fold Change | p-Value | Fold Change | p-Value | Fold Change | p Value | Protein Description | |
| B8AFQ6 | 1.5456 | 6.1 × 10−5 | 0.9837 | 0.15610 | 1.5713 | 4.3 × 10−5 | phenylalanine ammonia-lyase [Oryza sativa Japonica Group] |
| A2X1J7 | 1.9066 | 0.00044 | 0.9110 | 0.02346 | 2.0928 | 1.5 × 10−6 | probable 4-coumarate–CoA ligase 3 [Oryza sativa Japonica Group] |
| A2YFS4 | 1.5471 | 0.00138 | 0.9791 | 0.56018 | 1.5801 | 0.00053 | probable 4-coumarate–CoA ligase 4 [Oryza sativa Japonica Group] |
| Q38G74 | 1.4559 | 0.00286 | 0.8860 | 0.00106 | 1.6433 | 3.7 × 10−5 | Cinnamyl-alcohol dehydrogenase OS = Oryza sativa subsp. indica OX = 39946 GN = GH2 PE = 3 SV = 1 |
| A2WN51 | 2.3890 | 0.00012 | 0.9935 | 0.75944 | 2.4047 | 0.00012 | Peroxidase OS = Oryza sativa subsp. indica OX = 39946 GN = OsI_01277 PE = 3 SV = 1 |
| A2WPA0 | 1.2030 | 0.01086 | 0.7691 | 0.00020 | 1.5641 | 0.00026 | peroxidase 1 [Oryza sativa Japonica Group] |
| A2WZD6 | 1.2390 | 0.00050 | 0.7602 | 2.1 × 10−5 | 1.6300 | 1.5 × 10−5 | Peroxidase OS = Oryza sativa subsp. indica OX = 39946 GN = OsI_05314 PE = 3 SV = 1 |
| A2Y667 | 1.7234 | 0.00010 | 1.0376 | 0.07656 | 1.6609 | 0.00012 | Peroxidase OS = Oryza sativa subsp. indica OX = 39946 GN = OsI_20490 PE = 3 SV = 1 |
| B8ARU3 | 1.0433 | 0.19318 | 0.6404 | 4.2 × 10−6 | 1.6290 | 9.6 × 10−5 | Peroxidase OS = Oryza sativa subsp. indica OX = 39946 GN = OsI_18017 PE = 3 SV = 1 |
| B8ARU5 | 0.5657 | 2.4 × 10−5 | 0.3445 | 3.6 × 10−5 | 1.6420 | 0.00072 | Peroxidase OS = Oryza sativa subsp. indica OX = 39946 GN = OsI_18021 PE = 3 SV = 1 |
| B8B653 | 1.9473 | 0.00090 | 0.6799 | 0.00024 | 2.8643 | 0.00016 | Peroxidase OS = Oryza sativa subsp. indica OX = 39946 GN = OsI_27330 PE = 3 SV = 1 |
| A2YIB0 | / | / | 1.542 | 0.00894 | 1.874 | 0.00084 | Peroxidase OS = Oryza sativa subsp. indica OX = 39946 GN = OsI_24946 PE = 3 SV = 1 |
| A2Z9A1 | 2.1725 | 0.00026 | 1.1701 | 0.05896 | 1.8567 | 0.00028 | cytochrome P450 84A1 [Oryza sativa Japonica Group] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Yang, H.; Liu, Z.; Bai, L.; Wang, L.; Zhou, S. Multiomics Analysis of the Mechanism by Which Gibberellin Alleviates S-Metolachlor Toxicity in Rice Seedlings. Plants 2024, 13, 2517. https://doi.org/10.3390/plants13172517
Wang C, Yang H, Liu Z, Bai L, Wang L, Zhou S. Multiomics Analysis of the Mechanism by Which Gibberellin Alleviates S-Metolachlor Toxicity in Rice Seedlings. Plants. 2024; 13(17):2517. https://doi.org/10.3390/plants13172517
Chicago/Turabian StyleWang, Cong, Haona Yang, Zhixuan Liu, Lianyang Bai, Lifeng Wang, and Shangfeng Zhou. 2024. "Multiomics Analysis of the Mechanism by Which Gibberellin Alleviates S-Metolachlor Toxicity in Rice Seedlings" Plants 13, no. 17: 2517. https://doi.org/10.3390/plants13172517






