Genome-Wide Characterization of IQD Family Proteins in Apple and Functional Analysis of the Microtubule-Regulating Abilities of MdIQD17 and MdIQD28 under Cold Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Analysis of MdIQDs in the Apple Genome
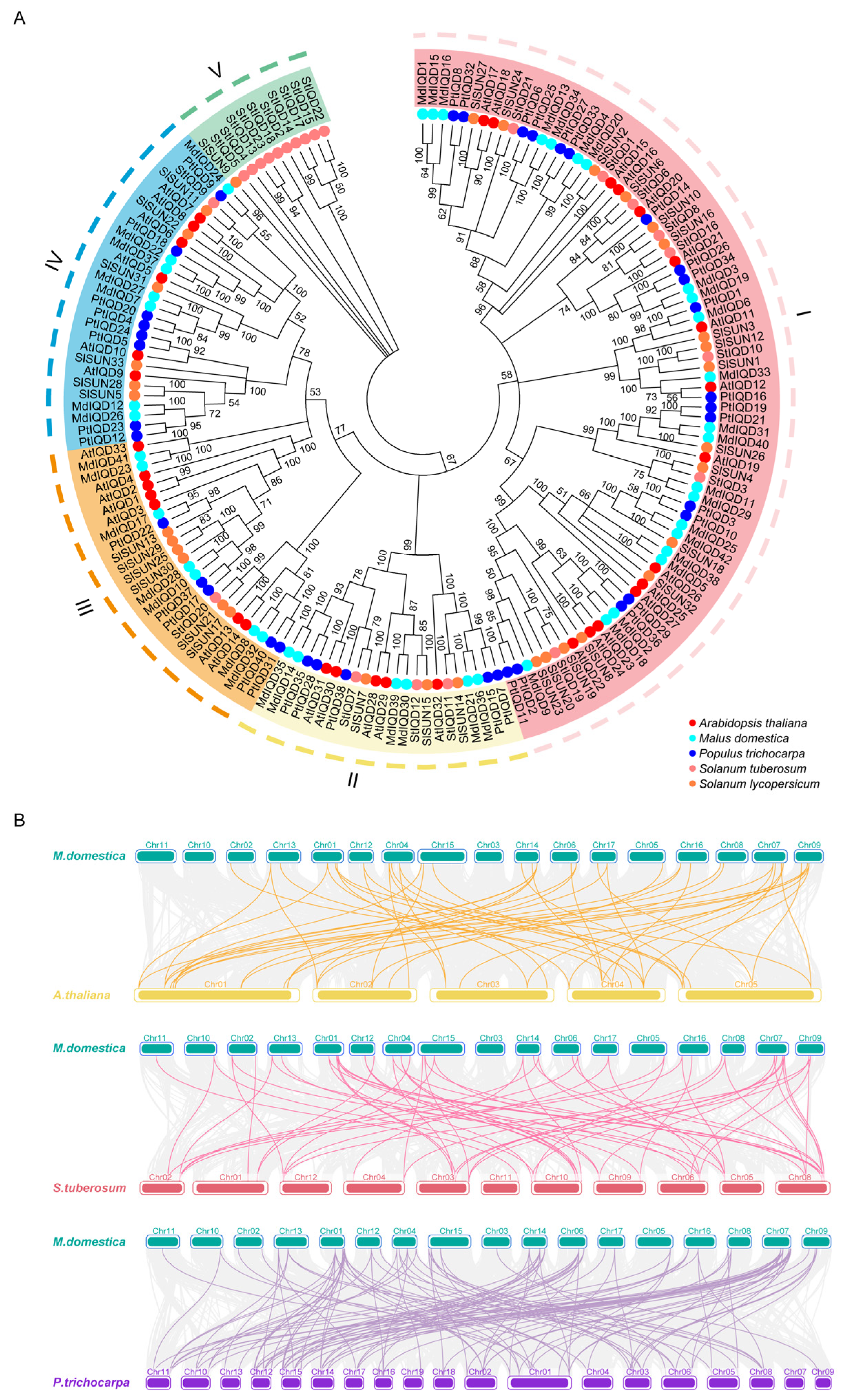
2.2. Evolutionary Analysis of MdIQDs
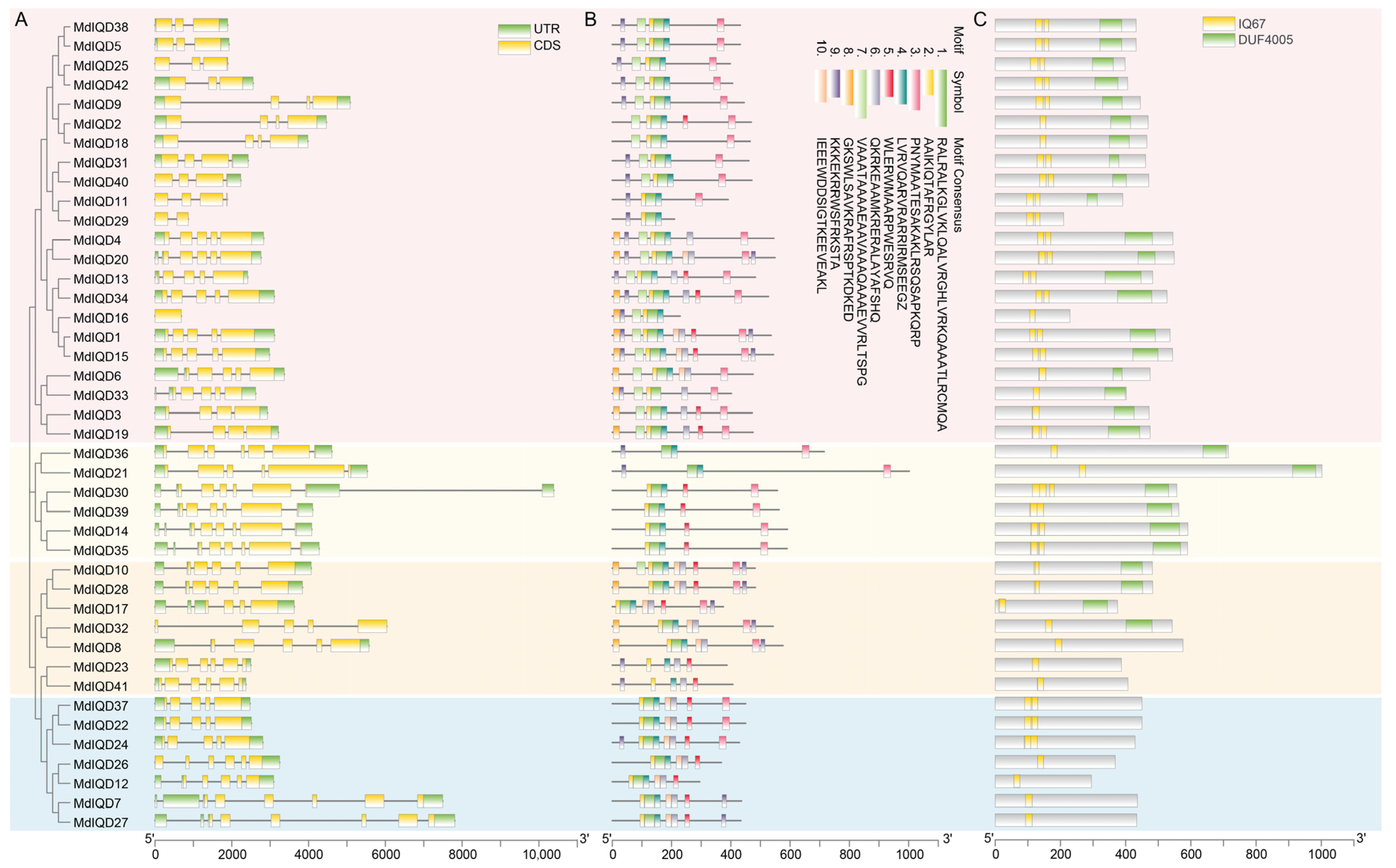
2.3. Structural Characterization Analysis of MdIQDs
2.4. Promoter Analysis and Expression Analysis under Cold Stress of MdIQDs
2.5. Subcellular Localization Analysis of MdIQD17 and MdIQD28
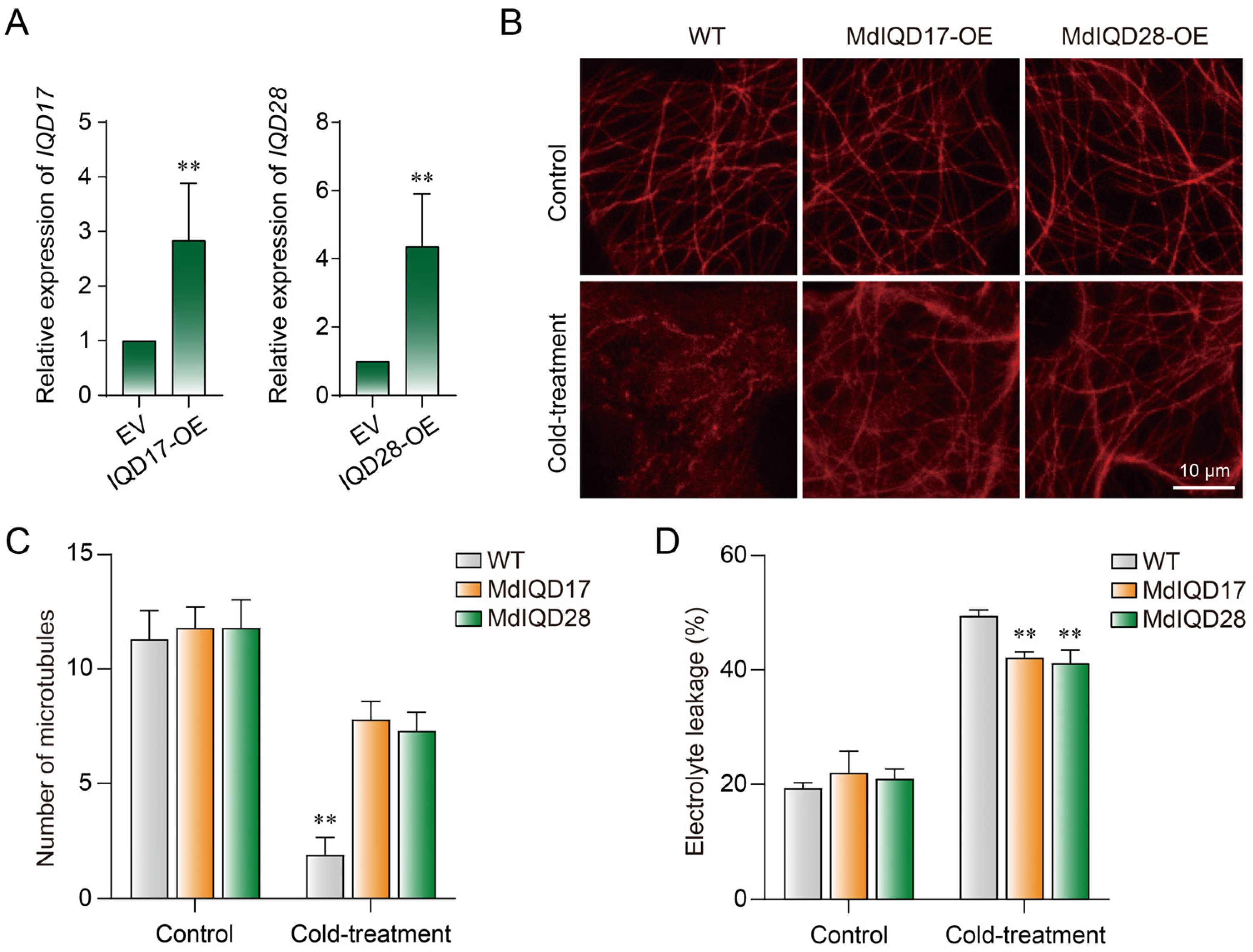
2.6. MdIQD17 and MdIQD28 Stabilize Microtubules under Cold Stress
3. Discussion
4. Materials and Methods
4.1. Identification of IQDs in Malus domestica
4.2. Phylogenetic Analysis
4.3. Characteristic Analysis of Gene and Protein Sequences
4.4. Species Homology and Chromosome Localization
4.5. Intrinsically Disordered Regions Analysis
4.6. Analysis of Malus domestica RNA-seq under Cold Stress
4.7. Plant Materials and Cold Treatments
4.8. Total RNA Extraction and cDNA Reverse Transcription Synthesis
4.9. Quantitative Real-Time PCR (qRT-PCR)
4.10. Subcellular Localization of MdIQD17 and MdIQD28 Proteins
4.11. Quantification of Cortical Microtubules
4.12. Electrolyte Leakage Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, X.; Mao, T. Understanding the functions and mechanisms of plant cytoskeleton in response to environmental signals. Curr. Opin. Plant Biol. 2019, 52, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Lian, N.; Wang, X.; Jing, Y.; Lin, J. Regulation of cytoskeleton-associated protein activities: Linking cellular signals to plant cytoskeletal function. J. Integr. Plant Biol. 2021, 63, 241–250. [Google Scholar] [CrossRef]
- Abdrakhamanova, A.; Wang, Q.Y.; Khokhlova, L.; Nick, P. Is microtubule disassembly a trigger for cold acclimation? Plant Cell Physiol. 2003, 44, 676–686. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Nick, P. Cold sensing in grapevine-which signals are upstream of the microtubular “thermometer”. Plant Cell Environ. 2017, 40, 2844–2857. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, C.; Hussey, P. Microtubule-associated proteins in plants—Why we need a map. Nat. Rev. Mol. Cell Biol. 2001, 2, 40–47. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Wang, Y.; Liu, H.; Hu, D.; Zhang, N.; Zhang, S.; Cao, H.; Cao, Q.; Zhang, Z.; et al. Arabidopsis PCaP2 Plays an Important Role in Chilling Tolerance and ABA Response by Activating CBF- and SnRK2-Mediated Transcriptional Regulatory Network. Front. Plant Sci. 2018, 9, 215. [Google Scholar] [CrossRef]
- Casal, J.J.; Balasubramanian, S. Thermomorphogenesis. Annu. Rev. Plant Biol. 2019, 70, 321–346. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Molecular regulation of plant responses to environmental temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef]
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef]
- Kidokoro, S.; Yoneda, K.; Takasaki, H.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Different Cold-Signaling Pathways Function in the Responses to Rapid and Gradual Decreases in Temperature. Plant Cell 2017, 29, 760–774. [Google Scholar] [CrossRef] [PubMed]
- Catalá, R.; Medina, J.; Salinas, J. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 16475–16480. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef]
- Hiraki, H.; Watanabe, M.; Uemura, M.; Kawamura, Y. Season specificity in the cold-induced calcium signal and the volatile chemicals in the atmosphere. Physiol. Plant. 2020, 168, 803–818. [Google Scholar] [CrossRef]
- Knight, H.; Trewavas, A.J.; Knight, M.R. Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 1996, 8, 489–503. [Google Scholar] [CrossRef]
- Medina, J.; Catalá, R.; Salinas, J. The CBFs: Three Arabidopsis transcription factors to cold acclimate. Plant Sci. 2011, 180, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, X.; Li, M.; Yang, H.; Fu, D.; Lv, J.; Ding, Y.; Gong, Z.; Shi, Y.; Yang, S. The direct targets of CBFs: In cold stress response and beyond. J. Integr. Plant Biol. 2021, 63, 1874–1887. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Örvar, B.L.; Sangwan, V.; Omann, F.; Dhindsa, R.S. Early steps in cold sensing by plant cells: The role of actin cytoskeleton and membrane fluidity. Plant J. 2000, 23, 785–794. [Google Scholar] [CrossRef]
- Abel, S.; Bürstenbinder, K.; Müller, J. The emerging function of IQD proteins as scaffolds in cellular signaling and trafficking. Plant Signal. Behav. 2013, 8, e24369. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Zhou, J.; Li, D. New insights into functions of IQ67-domain proteins. Front. Plant Sci. 2021, 11, 614851. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Dahiya, P.; Livanos, P.; Zergiebel, L.; Kölling, M.; Poeschl, Y.; Stamm, G.; Hermann, A.; Abel, S.; Müller, S.; et al. IQ67 DOMAIN proteins facilitate preprophase band formation and division-plane orientation. Nat. Plants 2021, 7, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, Y.; Wen, Y.; Wang, D.; Liu, H.; Li, Y.; Zhao, J.; An, L.; Yu, F.; Liu, X. The domain of unknown function 4005 (DUF4005) in an Arabidopsis IQD protein functions in microtubule binding. J. Biol. Chem. 2021, 297, 100849. [Google Scholar] [CrossRef]
- Bürstenbinder, K.; Savchenko, T.; Müller, J.; Adamson, A.W.; Stamm, G.; Kwong, R.; Zipp, B.J.; Dinesh, D.C.; Abel, S. Arabidopsis calmodulin-binding protein IQ67-domain 1 localizes to microtubules and interacts with kinesin light chain-related protein-1. J. Biol. Chem. 2013, 288, 1871–1882. [Google Scholar] [CrossRef]
- Dou, J.; Zhao, S.; Lu, X.; He, N.; Zhang, L.; Ali, A.; Kuang, H.; Liu, W. Genetic mapping reveals a candidate gene (ClFS1) for fruit shape in watermelon (Citrullus lanatus L.). Theor. Appl. Genet. 2018, 131, 947–958. [Google Scholar] [CrossRef]
- Ge, Q.; Wang, X.F.; Li, H.; Ren, Z.H.; Wang, L.N. Genome-wide identification and analysis of IQD/SUN gene family in cucumber. Genom. Appl. Biol. 2019, 38, 4110–4119. [Google Scholar]
- Mitra, D.; Klemm, S.; Kumari, P.; Quegwer, J.; Möller, B.; Poeschl, Y.; Pflug, P.; Stamm, G.; Abel, S.; Bürstenbinder, K. Microtubule-associated protein IQ67 DOMAIN5 regulates morphogenesis of leaf pavement cells in Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 529–543. [Google Scholar] [CrossRef]
- Yang, B.; Wendrich, J.R.; De Rybel, B.; Weijers, D.; Xue, H. Rice microtubule-associated protein IQ67-DOMAIN14 regulates grain shape by modulating microtubule cytoskeleton dynamics. Plant Biotechnol. J. 2020, 18, 1141–1152. [Google Scholar] [CrossRef]
- Yuan, J.; Liu, T.; Yu, Z.; Li, Y.; Ren, H.; Hou, X.; Li, Y. Genome-wide analysis of the Chinese cabbage IQD gene family and the response of BrIQD5 in drought resistance. Plant Mol. Biol. 2019, 99, 603–620. [Google Scholar] [CrossRef]
- Xiu, Y.; Kirungu, J.N.; Magwanga, R.O.; Xu, Y.; Pu, L.; Zhou, Z.; Hou, Y.; Cai, X.; Wang, K.; Liu, F. Knockdown of GhIQD31 and GhIQD32 increases drought and salt stress sensitivity in Gossypium hirsutum. Plant Physiol. Biochem. 2019, 144, 166–177. [Google Scholar] [CrossRef]
- An, J.-P.; Wang, X.-F.; Zhang, X.-W.; Xu, H.-F.; Bi, S.-Q.; You, C.-X.; Hao, Y.-J. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 2020, 18, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Mei, C.; Yang, J.; Mei, Q.; Jia, D.; Yan, P.; Feng, B.; Mamat, A.; Gong, X.; Guan, Q.; Mao, K.; et al. MdNAC104 positively regulates apple cold tolerance via CBF-dependent and CBF-independent pathways. Plant Biotechnol. J. 2023, 21, 2057–2073. [Google Scholar] [CrossRef]
- Sugiyama, Y.; Wakazaki, M.; Toyooka, K.; Fukuda, H.; Oda, Y. A Novel plasma membrane-anchored protein regulates Xylem cell-wall deposition through microtubule-dependent lateral inhibition of rho GTPase domains. Curr. Biol. 2017, 27, 2522–2528.e4. [Google Scholar] [CrossRef]
- Liang, H.; Zhang, Y.; Martinez, P.; Rasmussen, C.G.; Xu, T.; Yang, Z. The microtubule-associated protein IQ67 DOMAIN5 modulates microtubule dynamics and pavement cell shape. Plant Physiol. 2018, 177, 1555–1568. [Google Scholar] [CrossRef]
- Bao, Z.; Guo, Y.; Deng, Y.; Zang, J.; Zhang, J.; Deng, Y.; Ouyang, B.; Qu, X.; Bürstenbinder, K.; Wang, P. Microtubule-associated protein SlMAP70 interacts with IQ67-domain protein SlIQD21a to regulate fruit shape in tomato. Plant Cell 2023, 35, 4266–4283. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Zhang, C.; Zhao, Y.; Zhu, K.; Wang, Y.; Jiang, H.; Xiang, Y.; Cheng, B. Genome-wide analysis of the IQD gene family in maize. Mol. Genet. Genom. 2016, 291, 543–558. [Google Scholar] [CrossRef]
- Ma, H.; Feng, L.; Chen, Z.; Chen, X.; Zhao, H.; Xiang, Y. Genome-wide identification and expression analysis of the IQD gene family in Populus trichocarpa. Plant Sci. 2014, 229, 96–110. [Google Scholar] [CrossRef]
- Wu, M.; Li, Y.; Chen, D.; Liu, H.; Zhu, D.; Xiang, Y. Genome-wide identification and expression analysis of the IQD gene family in moso bamboo (Phyllostachys edulis). Sci. Rep. 2016, 6, 24520. [Google Scholar] [CrossRef]
- Wang, L.; Sadeghnezhad, E.; Riemann, M.; Nick, P. Microtubule dynamics modulate sensing during cold acclimation in grapevine suspension cells. Plant Sci. 2018, 280, 18–30. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; D’ehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Li, X.-L.; Meng, D.; Li, M.-J.; Zhou, J.; Yang, Y.-Z.; Zhou, B.-B.; Wei, Q.-P.; Zhang, J.-K. Transcription factors MhDREB2A/MhZAT10 play a role in drought and cold stress response crosstalk in apple. Plant Physiol. 2023, 192, 2203–2220. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Dong, B.; Song, Z.; Meng, D.; Fu, Y. Genome-wide identification and characterization of CIPK family and analysis responses to various stresses in apple (Malus domestica). Int. J. Mol. Sci. 2018, 19, 2131. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; He, K.; Peng, J.; Wang, X.; Mao, T. The E3 ligase MREL57 modulates microtubule stability and stomatal closure in response to ABA. Nat. Commun. 2021, 12, 2181. [Google Scholar] [CrossRef]
- Liu, X.; Qin, T.; Ma, Q.; Sun, J.; Liu, Z.; Yuan, M.; Mao, T. Light-regulated hypocotyl elongation involves proteasome-dependent degradation of the microtubule regulatory protein WDL3 in Arabidopsis. Plant Cell 2013, 25, 1740–1755. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.; Qin, T.; Zhang, Y.; Liu, X.; Sun, J.; Zhou, Y.; Zhu, L.; Zhang, Z.; Yuan, M.; et al. MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis. Plant Cell 2011, 23, 4411–4427. [Google Scholar] [CrossRef]
- An, J.-P.; Liu, Z.-Y.; Zhang, X.-W.; Wang, D.-R.; Zeng, F.; You, C.-X.; Han, Y. Brassinosteroid signaling regulator BIM1 integrates brassinolide and jasmonic acid signaling during cold tolerance in apple. Plant Physiol. 2023, 193, 1652–1674. [Google Scholar] [CrossRef]





| Protein | Number of Amino Acid | Molecular Weight (Da) | Theoretical pI | Instability Index (%) | Aliphatic Index (%) | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|
| MdIQD1 | 535 | 60,091 | 10.41 | 69.61 | 55.91 | −0.95 |
| MdIQD2 | 468 | 51,603 | 10.50 | 55.29 | 58.93 | −0.75 |
| MdIQD3 | 471 | 52,989 | 9.77 | 60.18 | 54.12 | −0.97 |
| MdIQD4 | 544 | 61,121 | 10.08 | 66.68 | 58.20 | −0.90 |
| MdIQD5 | 431 | 47,034 | 10.24 | 50.69 | 64.20 | −0.54 |
| MdIQD6 | 474 | 53,895 | 10.00 | 74.35 | 65.74 | −0.76 |
| MdIQD7 | 435 | 48,357 | 10.07 | 56.64 | 68.28 | −0.83 |
| MdIQD8 | 575 | 64,387 | 10.94 | 65.22 | 50.12 | −0.96 |
| MdIQD9 | 444 | 49,035 | 10.29 | 60.41 | 59.71 | −0.75 |
| MdIQD10 | 481 | 53,551 | 10.18 | 63.79 | 66.76 | −0.78 |
| MdIQD11 | 390 | 44,228 | 10.43 | 47.08 | 67.82 | −0.84 |
| MdIQD12 | 294 | 32,996 | 10.09 | 52.54 | 67.11 | −0.72 |
| MdIQD13 | 482 | 54,186 | 10.07 | 56.89 | 64.09 | −0.78 |
| MdIQD14 | 590 | 64,470 | 9.85 | 50.44 | 75.03 | −0.67 |
| MdIQD15 | 543 | 61,108 | 10.40 | 72.33 | 56.19 | −0.97 |
| MdIQD16 | 228 | 26,093 | 10.19 | 45.06 | 71.58 | −0.58 |
| MdIQD17 | 374 | 41,714 | 11.17 | 78.12 | 49.17 | −0.99 |
| MdIQD18 | 464 | 51,454 | 10.48 | 62.02 | 57.31 | −0.74 |
| MdIQD19 | 474 | 52,600 | 9.66 | 58.47 | 53.00 | −0.97 |
| MdIQD20 | 548 | 61,162 | 10.17 | 65.85 | 58.14 | −0.87 |
| MdIQD21 | 1001 | 109,765 | 4.91 | 67.91 | 59.13 | -0.97 |
| MdIQD22 | 449 | 50,382 | 10.27 | 49.23 | 55.90 | −0.84 |
| MdIQD23 | 386 | 43,424 | 9.33 | 60.49 | 49.25 | -0.94 |
| MdIQD24 | 428 | 48,167 | 10.19 | 50.31 | 60.40 | −0.82 |
| MdIQD25 | 397 | 44,785 | 9.93 | 65.38 | 57.86 | −0.75 |
| MdIQD26 | 367 | 41,220 | 10.34 | 50.60 | 79.05 | −0.46 |
| MdIQD27 | 433 | 47,871 | 10.07 | 56.05 | 68.15 | −0.85 |
| MdIQD28 | 482 | 53,797 | 10.09 | 63.76 | 63.80 | −0.84 |
| MdIQD29 | 209 | 23,277 | 11.32 | 38.06 | 95.31 | −0.19 |
| MdIQD30 | 556 | 60,935 | 9.92 | 54.73 | 75.52 | −0.68 |
| MdIQD31 | 460 | 50,918 | 9.82 | 57.21 | 54.89 | −0.79 |
| MdIQD32 | 542 | 61,072 | 10.41 | 64.54 | 53.87 | −0.94 |
| MdIQD33 | 401 | 45,295 | 10.14 | 51.33 | 71.37 | −0.71 |
| MdIQD34 | 526 | 58,926 | 9.93 | 64.54 | 61.16 | −0.83 |
| MdIQD35 | 589 | 64,370 | 9.85 | 49.50 | 72.85 | −0.72 |
| MdIQD36 | 714 | 77,924 | 5.57 | 65.74 | 64.19 | −0.88 |
| MdIQD37 | 449 | 50,343 | 10.22 | 51.41 | 57.84 | −0.85 |
| MdIQD38 | 431 | 47,428 | 10.27 | 52.09 | 61.25 | −0.67 |
| MdIQD39 | 562 | 61,994 | 9.76 | 50.42 | 72.31 | −0.73 |
| MdIQD40 | 470 | 52,206 | 9.88 | 59.97 | 56.64 | −0.79 |
| MdIQD41 | 406 | 45,574 | 9.39 | 67.43 | 55.47 | −0.87 |
| MdIQD42 | 405 | 45,263 | 10.26 | 59.47 | 58.42 | −0.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Wang, S.; Zhang, C.; Qi, M.; Liu, L.; Yang, L.; Lian, N. Genome-Wide Characterization of IQD Family Proteins in Apple and Functional Analysis of the Microtubule-Regulating Abilities of MdIQD17 and MdIQD28 under Cold Stress. Plants 2024, 13, 2532. https://doi.org/10.3390/plants13172532
Zhang Y, Wang S, Zhang C, Qi M, Liu L, Yang L, Lian N. Genome-Wide Characterization of IQD Family Proteins in Apple and Functional Analysis of the Microtubule-Regulating Abilities of MdIQD17 and MdIQD28 under Cold Stress. Plants. 2024; 13(17):2532. https://doi.org/10.3390/plants13172532
Chicago/Turabian StyleZhang, Yu, Shengjie Wang, Chaochao Zhang, Meng Qi, Luoqi Liu, Lipeng Yang, and Na Lian. 2024. "Genome-Wide Characterization of IQD Family Proteins in Apple and Functional Analysis of the Microtubule-Regulating Abilities of MdIQD17 and MdIQD28 under Cold Stress" Plants 13, no. 17: 2532. https://doi.org/10.3390/plants13172532
APA StyleZhang, Y., Wang, S., Zhang, C., Qi, M., Liu, L., Yang, L., & Lian, N. (2024). Genome-Wide Characterization of IQD Family Proteins in Apple and Functional Analysis of the Microtubule-Regulating Abilities of MdIQD17 and MdIQD28 under Cold Stress. Plants, 13(17), 2532. https://doi.org/10.3390/plants13172532







