Genome-Wide Identification and Expression Analysis of GST Genes during Light-Induced Anthocyanin Biosynthesis in Mango (Mangifera indica L.)
Abstract
:1. Introduction
2. Results
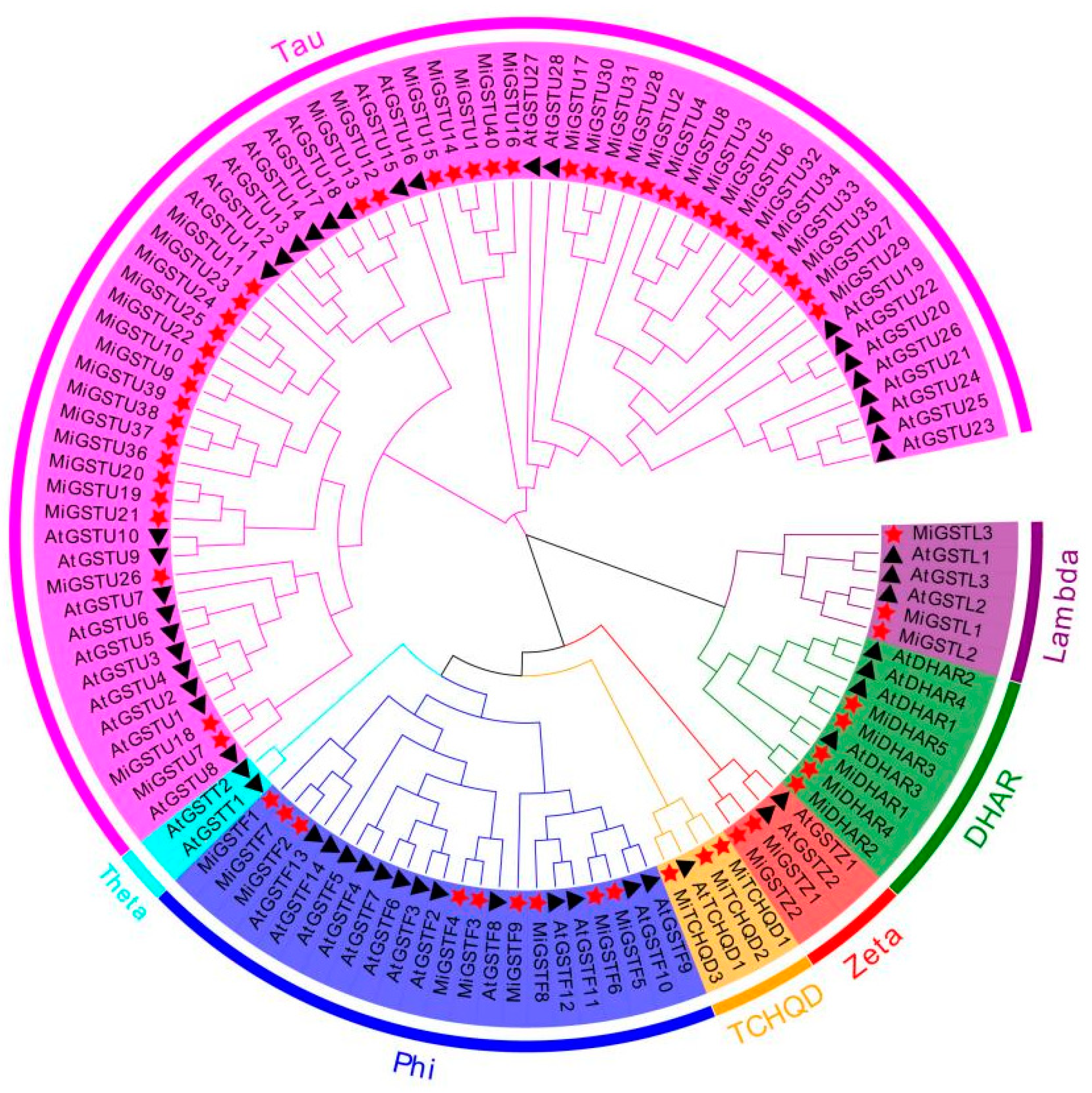
2.1. Identification, Phylogenetic Analysis, and Physicochemical Properties of GST Family in Mangifera indica
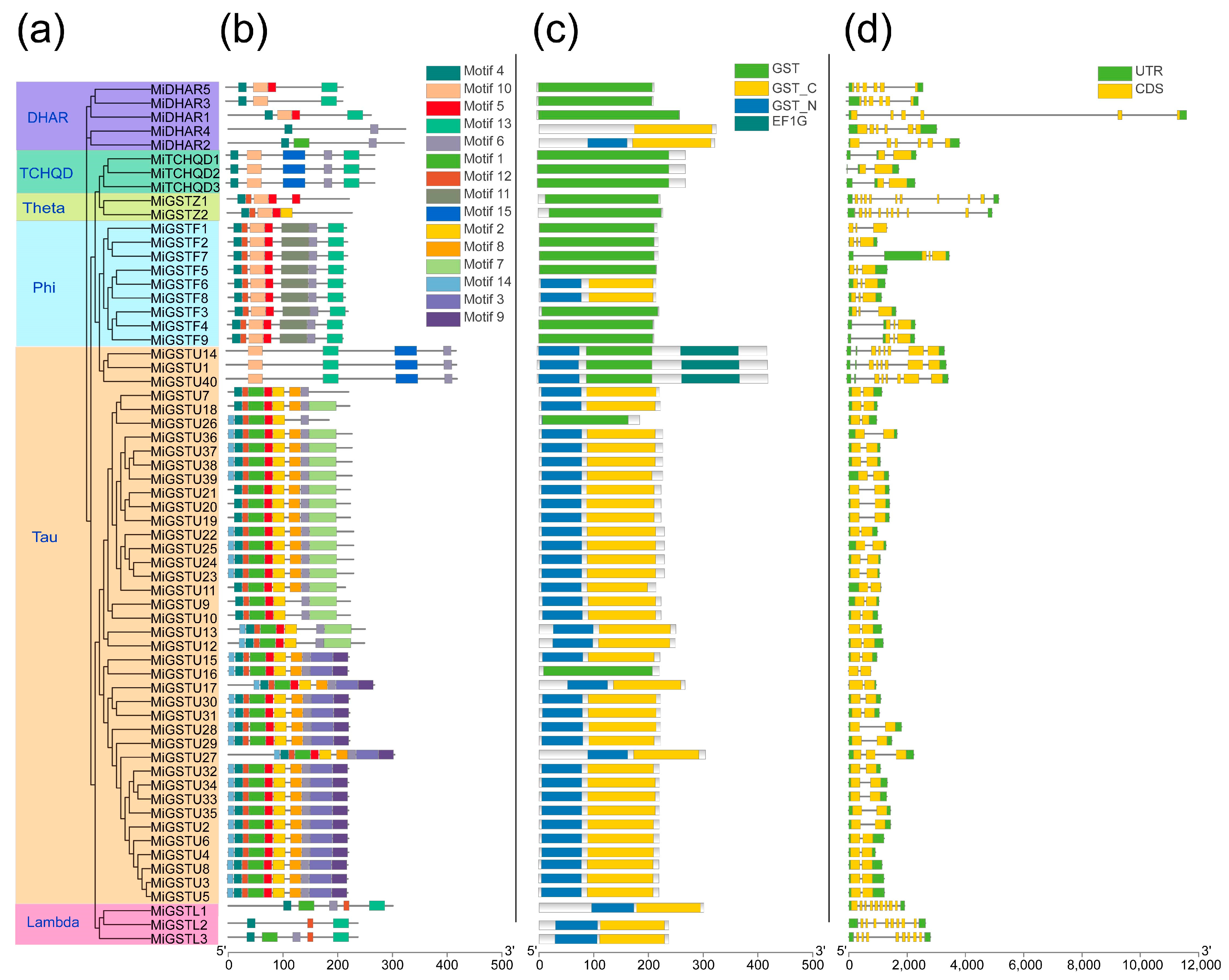
2.2. Conserved Motifs and Domains and Gene Structure Analyses of MiGST Proteins
2.3. Chromosomal Distribution and Syntenic Analysis of MiGST Genes
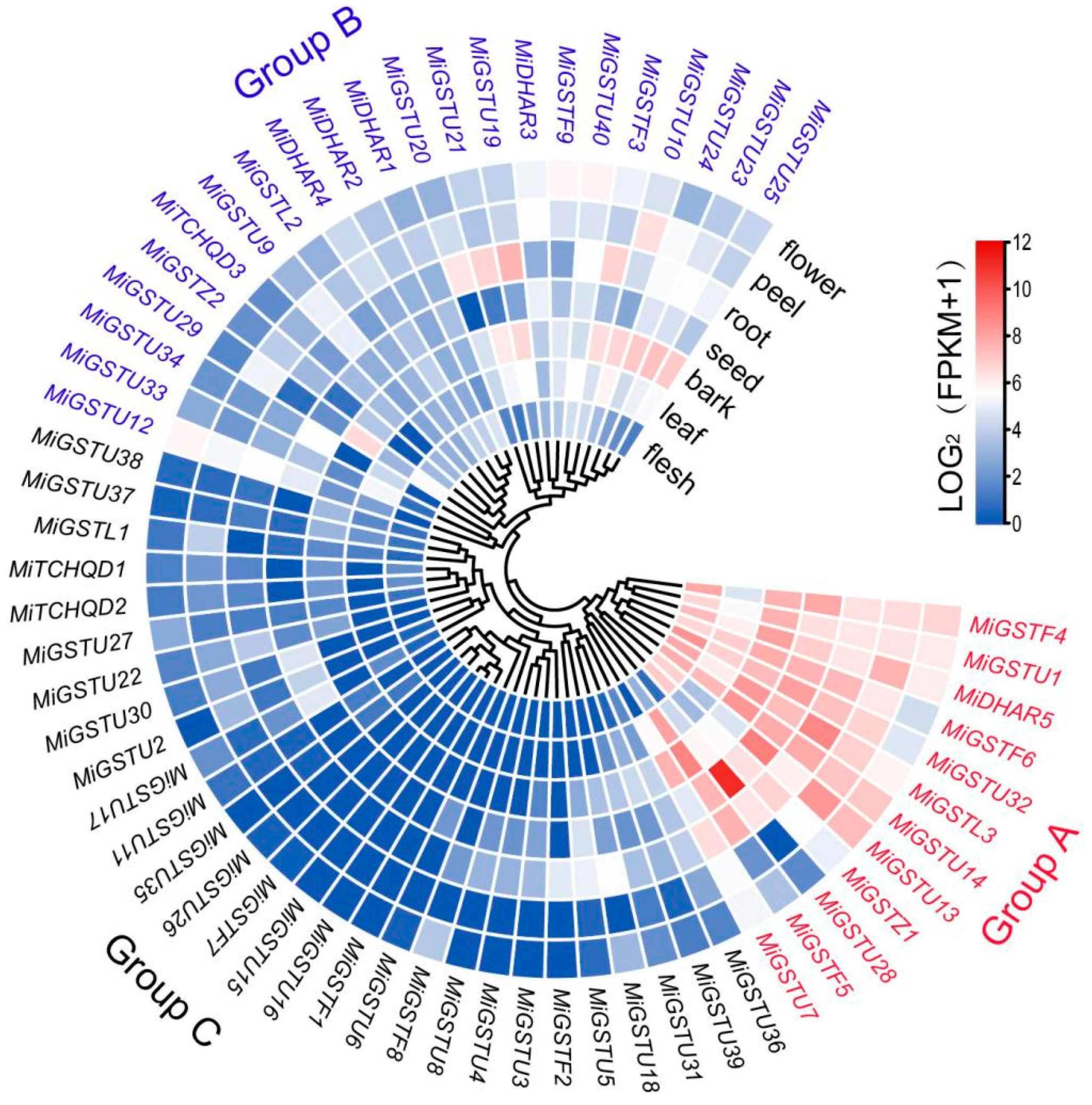
2.4. Tissue-Specific Expression Profiling of MiGST Genes
2.5. Expression of MiGST Genes under Bagging Treatment
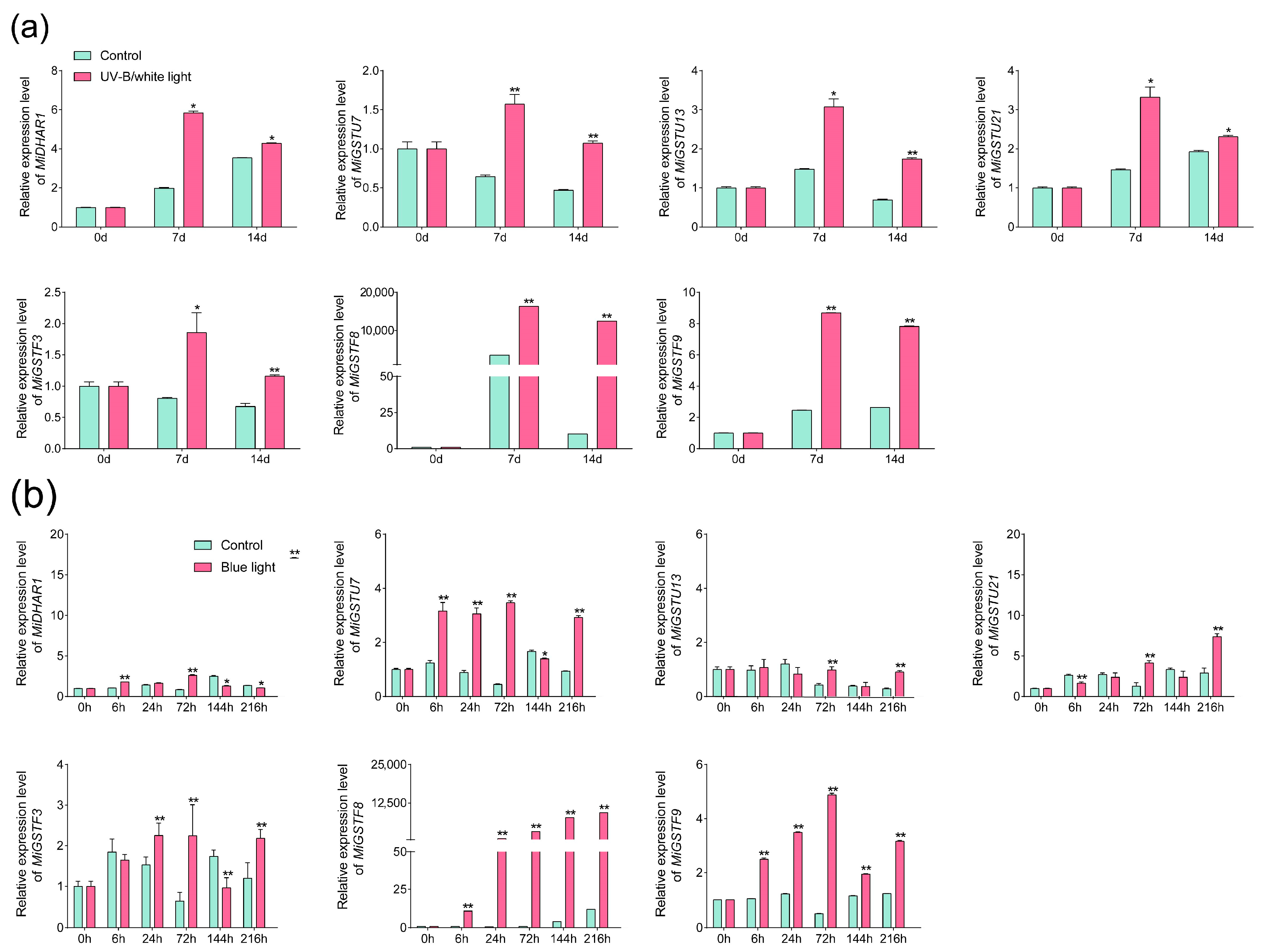
2.6. Anthocyanin Concentration and Expression of MiGST Genes under UV-B/White Light and Blue Light Treatments
2.7. The Expression Patterns of MiGSTs during Light-Induced Anthocyanin Accumulation Analyzed by qPCR
3. Discussion
4. Materials and Methods
4.1. Whole-Genome Identification of Mangifera indica GST
4.2. Phylogenetic, Gene Structure, and Conserved Motifs and Domains Analyses of MiGST
4.3. Chromosomal Distribution and Syntenic Analysis
4.4. Plant Material and Treatments
4.5. Acquisition and Analysis of RNA-Seq Data
4.6. Total Anthocyanin Extraction and Quantification
4.7. RNA Extraction and Gene Expression Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kanzaki, S.; Ichihi, A.; Tanaka, Y.; Fujishige, S.; Koeda, S.; Shimizu, K. The R2R3-MYB Transcription Factor MiMYB1 Regulates Light Dependent Red Coloration of ‘Irwin’ Mango Fruit Skin. Sci. Hortic. 2020, 272, 109567. [Google Scholar] [CrossRef]
- Medlicott, A.P.; Bhogal, M.; Reynolds, S.B. Changes in Peel Pigmentation during Ripening of Mango Fruit (Mangifera Indica Var. Tommy Atkins). Ann. Appl. Biol. 1986, 109, 651–656. [Google Scholar] [CrossRef]
- Shi, B.; Wu, H.; Zheng, B.; Qian, M.; Gao, A.; Zhou, K. Analysis of Light-Independent Anthocyanin Accumulation in Mango (Mangifera Indica L.). Horticulturae 2021, 7, 423. [Google Scholar] [CrossRef]
- Li, B.; Zhang, X.; Duan, R.; Han, C.; Yang, J.; Wang, L.; Wang, S.; Su, Y.; Wang, L.; Dong, Y.; et al. Genomic Analysis of the Glutathione S-Transferase Family in Pear (Pyrus Communis) and Functional Identification of PcGST57 in Anthocyanin Accumulation. Int. J. Mol. Sci. 2022, 23, 746. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.; Huang, D.; Xing, W.; Wu, B.; Wei, Q.; Xu, Y.; Zhan, R.; Ma, F.; Song, S.; et al. Research Progress on the MYB Transcription Factors in Tropical Fruit. Trop. Plants 2022, 1, 5. [Google Scholar] [CrossRef]
- Maier, A.; Hoecker, U. COP1/SPA Ubiquitin Ligase Complexes Repress Anthocyanin Accumulation under Low Light and High Light Conditions. Plant Signal. Behav. 2015, 10, e970440. [Google Scholar] [CrossRef]
- Ubi, B.E.; Honda, C.; Bessho, H.; Kondo, S.; Wada, M.; Kobayashi, S.; Moriguchi, T. Expression Analysis of Anthocyanin Biosynthetic Genes in Apple Skin: Effect of UV-B and Temperature. Plant Sci. 2006, 170, 571–578. [Google Scholar] [CrossRef]
- Qian, M.; Zhang, D.; Yue, X.; Wang, S.; Li, X.; Teng, Y. Analysis of Different Pigmentation Patterns in ‘Mantianhong’ (Pyrus Pyrifolia Nakai) and ‘Cascade’ (Pyrus Communis L.) under Bagging Treatment and Postharvest UV-B/Visible Irradiation Conditions. Sci. Hortic. 2013, 151, 75–82. [Google Scholar] [CrossRef]
- Tao, R.; Bai, S.; Ni, J.; Yang, Q.; Zhao, Y.; Teng, Y. The Blue Light Signal Transduction Pathway Is Involved in Anthocyanin Accumulation in ‘Red Zaosu’ Pear. Planta 2018, 248, 37–48. [Google Scholar] [CrossRef]
- Nguyen, C.T.T.; Lim, S.; Lee, J.G.; Lee, E.J. VcBBX, VcMYB21, and VcR2R3MYB Transcription Factors Are Involved in UV–B-Induced Anthocyanin Biosynthesis in the Peel of Harvested Blueberry Fruit. J. Agric. Food Chem. 2017, 65, 2066–2073. [Google Scholar] [CrossRef]
- Yang, C.; Wang, X.; Zhu, W.; Weng, Z.; Li, F.; Wu, H.; Zhou, K.; Strid, Å.; Qian, M. Postharvest White Light Combined with Different UV-B Doses Differently Promotes Anthocyanin Accumulation and Antioxidant Capacity in Mango Peel. LWT 2024, 203, 116385. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A Colorful Model for the Regulation and Evolution of Biochemical Pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent Advances in the Transcriptional Regulation of the Flavonoid Biosynthetic Pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of Flavonoids and Effects of Stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of Plant Pigments: Anthocyanins, Betalains and Carotenoids. Plant J. Cell Mol. Biol. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Gu, J.; Li, J.; Zhao, A.; Fu, Y.; Wang, T.; Li, T.; Li, X.; Sheng, Y.; Zhao, Y.; et al. Tyrosine Promotes Anthocyanin Biosynthesis in Pansy (Viola × Wittrockiana) by Inducing ABA Synthesis. Trop. Plants 2022, 1, 9. [Google Scholar] [CrossRef]
- Sabir, I.A.; Manzoor, M.A.; Shah, I.H.; Liu, X.; Jiu, S.; Wang, J.; Alam, P.; Abdullah, M.; Zhang, C. Identification and Comprehensive Genome-Wide Analysis of Glutathione S-Transferase Gene Family in Sweet Cherry (Prunus Avium) and Their Expression Profiling Reveals a Likely Role in Anthocyanin Accumulation. Front. Plant Sci. 2022, 13, 938800. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.; Dixon, D.P. Plant Glutathione Transferases. Methods Enzymol. 2005, 401, 169–186. [Google Scholar] [CrossRef]
- Wang, X.; Dong, J.; Hu, Y.; Huang, Q.; Lu, X.; Huang, Y.; Sheng, M.; Cao, L.; Xu, B.; Li, Y.; et al. Identification and Characterization of the Glutathione S-Transferase Gene Family in Blueberry (Vaccinium Corymbosum) and Their Potential Roles in Anthocyanin Intracellular Transportation. Plants Basel Switz. 2024, 13, 1316. [Google Scholar] [CrossRef]
- Loyall, L.; Uchida, K.; Braun, S.; Furuya, M.; Frohnmeyer, H. Glutathione and a UV Light–Induced Glutathione S-Transferase Are Involved in Signaling to Chalcone Synthase in Cell Cultures. Plant Cell 2000, 12, 1939–1950. [Google Scholar] [CrossRef]
- Vaish, S.; Gupta, D.; Mehrotra, R.; Mehrotra, S.; Basantani, M.K. Glutathione S-Transferase: A Versatile Protein Family. 3 Biotech 2020, 10, 321. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Ghanashyam, C.; Bhattacharjee, A. Comprehensive Expression Analysis Suggests Overlapping and Specific Roles of Rice Glutathione S-Transferase Genes during Development and Stress Responses. BMC Genom. 2010, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Sappl, P.G.; Carroll, A.J.; Clifton, R.; Lister, R.; Whelan, J.; Harvey Millar, A.; Singh, K.B. The Arabidopsis Glutathione Transferase Gene Family Displays Complex Stress Regulation and Co-Silencing Multiple Genes Results in Altered Metabolic Sensitivity to Oxidative Stress. Plant J. Cell Mol. Biol. 2009, 58, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Rahman, I.A.; Islam, T.; Ghosh, A. Genome-Wide Identification and Expression Analysis of Glutathione S-Transferase Gene Family in Tomato: Gaining an Insight to Their Physiological and Stress-Specific Roles. PLoS ONE 2017, 12, e0187504. [Google Scholar] [CrossRef]
- Fu, J.; Su, L.; Fan, J.; Yu, Q.; Huang, X.; Zhang, C.; Xian, B.; Yang, W.; Wang, S.; Chen, S.; et al. Systematic Analysis and Functional Verification of Citrus Glutathione S-Transferases Reveals That CsGSTF1 and CsGSTU18 Contribute Negatively to Citrus Bacterial Canker. Hortic. Plant J. 2023, S246801412300184X. [Google Scholar] [CrossRef]
- Fang, X.; An, Y.; Zheng, J.; Shangguan, L.; Wang, L. Genome-Wide Identification and Comparative Analysis of GST Gene Family in Apple (Malus Domestica) and Their Expressions under ALA Treatment. 3 Biotech 2020, 10, 307. [Google Scholar] [CrossRef] [PubMed]
- Marrs, K.A.; Alfenito, M.R.; Lloyd, A.M.; Walbot, V. A Glutathione S-Transferase Involved in Vacuolar Transfer Encoded by the Maize Gene Bronze-2. Nature 1995, 375, 397–400. [Google Scholar] [CrossRef]
- Hu, B.; Zhao, J.; Lai, B.; Qin, Y.; Wang, H.; Hu, G. LcGST4 Is an Anthocyanin-Related Glutathione S-Transferase Gene in Litchi Chinensis Sonn. Plant Cell Rep. 2016, 35, 831–843. [Google Scholar] [CrossRef]
- Pérez-Díaz, R.; Madrid-Espinoza, J.; Salinas-Cornejo, J.; González-Villanueva, E.; Ruiz-Lara, S. Differential Roles for VviGST1, VviGST3, and VviGST4 in Proanthocyanidin and Anthocyanin Transport in Vitis Vinífera. Front. Plant Sci. 2016, 7, 1166. [Google Scholar] [CrossRef]
- Jiang, S.; Chen, M.; He, N.; Chen, X.; Wang, N.; Sun, Q.; Zhang, T.; Xu, H.; Fang, H.; Wang, Y.; et al. MdGSTF6, Activated by MdMYB1, Plays an Essential Role in Anthocyanin Accumulation in Apple. Hortic. Res. 2019, 6, 40. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, W.; Zhu, Y.; Allan, A.C.; Lin-Wang, K.; Xu, C. PpGST1, an Anthocyanin-Related Glutathione S-Transferase Gene, Is Essential for Fruit Coloration in Peach. Plant Biotechnol. J. 2020, 18, 1284–1295. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, S.; Shikazono, N.; Tanaka, A. TRANSPARENT TESTA 19 Is Involved in the Accumulation of Both Anthocyanins and Proanthocyanidins in Arabidopsis. Plant J. 2004, 37, 104–114. [Google Scholar] [CrossRef]
- Shi, B.; Wu, H.; Zhu, W.; Zheng, B.; Wang, S.; Zhou, K.; Qian, M. Genome-Wide Identification and Expression Analysis of WRKY Genes during Anthocyanin Biosynthesis in the Mango (Mangifera Indica L.). Agriculture 2022, 12, 821. [Google Scholar] [CrossRef]
- Xue, L.; Huang, X.; Zhang, Z.; Lin, Q.; Zhong, Q.; Zhao, Y.; Gao, Z.; Xu, C. An Anthocyanin-Related Glutathione S-Transferase, MrGST1, Plays an Essential Role in Fruit Coloration in Chinese Bayberry (Morella Rubra). Front. Plant Sci. 2022, 13, 903333. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-W.; Wang, C.-K.; Huang, X.-Y.; Hu, D.-G. Genome-Wide Analysis of the Glutathione S-Transferase (GST) Genes and Functional Identification of MdGSTU12 Reveals the Involvement in the Regulation of Anthocyanin Accumulation in Apple. Genes 2021, 12, 1733. [Google Scholar] [CrossRef] [PubMed]
- Reiser, L.; Subramaniam, S.; Zhang, P.; Berardini, T. Using the Arabidopsis Information Resource (TAIR) to Find Information About Arabidopsis Genes. Curr. Protoc. 2022, 2, e574. [Google Scholar] [CrossRef] [PubMed]
- Mueller, L.A.; Goodman, C.D.; Silady, R.A.; Walbot, V. AN9, a Petunia Glutathione S -Transferase Required for Anthocyanin Sequestration, Is a Flavonoid-Binding Protein. Plant Physiol. 2000, 123, 1561–1570. [Google Scholar] [CrossRef]
- Luo, H.; Dai, C.; Li, Y.; Feng, J.; Liu, Z.; Kang, C. Reduced Anthocyanins in Petioles Codes for a GST Anthocyanin Transporter That Is Essential for the Foliage and Fruit Coloration in Strawberry. J. Exp. Bot. 2018, 69, 2595–2608. [Google Scholar] [CrossRef]
- Wang, P.; Li, Y.; Zhang, T.; Kang, Y.; Li, W.; Wang, J.; Yu, W.; Zhou, Y. Identification of the bZIP Gene Family and Investigation of Their Response to Drought Stress in Dendrobium Catenatum. Agronomy 2023, 13, 236. [Google Scholar] [CrossRef]
- Rensing, S.A. Gene Duplication as a Driver of Plant Morphogenetic Evolution. Curr. Opin. Plant Biol. 2014, 17, 43–48. [Google Scholar] [CrossRef]
- Zhao, Y.; Min, T.; Chen, M.; Wang, H.; Zhu, C.; Jin, R.; Allan, A.C.; Lin-Wang, K.; Xu, C. The Photomorphogenic Transcription Factor PpHY5 Regulates Anthocyanin Accumulation in Response to UVA and UVB Irradiation. Front. Plant Sci. 2021, 11, 603178. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons Control Fruit-Specific, Cold-Dependent Accumulation of Anthocyanins in Blood Oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.J.; Woo, J.-K.; Yi, K.U.; Park, Y.C.; Lee, H.-Y.; Kim, M.; Park, S.; Yun, S.-H.; Lee, Y.; Kim, H.-J.; et al. Development of molecular markers for genotyping of Ruby, a locus controlling anthocyanin pigment production in Citrus with its functional analysis. Sci. Hortic. 2021, 289, 110457. [Google Scholar] [CrossRef]
- Yuan, K.; Wang, C.; Wang, J.; Xin, L.; Zhou, G.; Li, L.; Shen, G. Analysis of the MdMYB1 gene sequence and development of new molecular markers related to apple skin color and fruit-bearing traits. Mol. Genet. Genom. 2014, 289, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- Azuma, A.; Kono, A.; Sato, A. Simple DNA marker system reveals genetic diversity of MYB genotypes that determine skin color in grape genetic resources. Tree Genet. Genomes 2020, 16, 29. [Google Scholar] [CrossRef]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef]
- Guo, T.; Wang, J.; Lu, X.; Wu, J.; Wang, L. The Development of Molecular Markers for Peach Skin Blush and Their Application in Peach Breeding Practice. Horticulturae 2023, 9, 887. [Google Scholar] [CrossRef]
- Lu, Z.; Cao, H.; Pan, L.; Niu, L.; Wei, B.; Cui, G.; Wang, L.; Yao, J.-L.; Zeng, W.; Wang, Z. Two loss-of-function alleles of the glutathione S-transferase (GST) gene cause anthocyanin deficiency in flower and fruit skin of peach (Prunus persica). Plant J. 2021, 107, 1320–1331. [Google Scholar] [CrossRef]
- Wang, P.; Luo, Y.; Huang, J.; Gao, S.; Zhu, G.; Dang, Z.; Gai, J.; Yang, M.; Zhu, M.; Zhang, H.; et al. The Genome Evolution and Domestication of Tropical Fruit Mango. Genome Biol. 2020, 21, 60. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, T.; Li, Y.; Zhao, X.; Liu, W.; Hu, Y.; Wang, J.; Zhou, Y. Comprehensive Analysis of Dendrobium Catenatum HSP20 Family Genes and Functional Characterization of DcHSP20–12 in Response to Temperature Stress. Int. J. Biol. Macromol. 2024, 258, 129001. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Liao, Y.; Zhang, M.; Pan, C.; Yang, Q.; Bai, S.; Teng, Y. Blue Light Simultaneously Induces Peel Anthocyanin Biosynthesis and Flesh Carotenoid/Sucrose Biosynthesis in Mango Fruit. J. Agric. Food Chem. 2022, 70, 16021–16035. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Shi, B.; Zhu, W.; Zheng, B.; Zhou, K.; Qian, M.; Wu, H. Genome-Wide Identification, Characterization and Expression Analysis of Mango (Mangifera Indica L.) Chalcone Synthase (CHS) Genes in Response to Light. Horticulturae 2022, 8, 968. [Google Scholar] [CrossRef]
- Qian, M.; Ni, J.; Niu, Q.; Bai, S.; Bao, L.; Li, J.; Sun, Y.; Zhang, D.; Teng, Y. Response of miR156-SPL Module during the Red Peel Coloration of Bagging-Treated Chinese Sand Pear (Pyrus Pyrifolia Nakai). Front. Physiol. 2017, 8, 550. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, S.; Yang, C.; Zheng, B.; Ni, J.; Zhou, K.; Qian, M.; Wu, H. Genome-Wide Identification and Expression Analysis of GST Genes during Light-Induced Anthocyanin Biosynthesis in Mango (Mangifera indica L.). Plants 2024, 13, 2726. https://doi.org/10.3390/plants13192726
Yuan S, Yang C, Zheng B, Ni J, Zhou K, Qian M, Wu H. Genome-Wide Identification and Expression Analysis of GST Genes during Light-Induced Anthocyanin Biosynthesis in Mango (Mangifera indica L.). Plants. 2024; 13(19):2726. https://doi.org/10.3390/plants13192726
Chicago/Turabian StyleYuan, Shiqing, Chengkun Yang, Bin Zheng, Junbei Ni, Kaibing Zhou, Minjie Qian, and Hongxia Wu. 2024. "Genome-Wide Identification and Expression Analysis of GST Genes during Light-Induced Anthocyanin Biosynthesis in Mango (Mangifera indica L.)" Plants 13, no. 19: 2726. https://doi.org/10.3390/plants13192726
APA StyleYuan, S., Yang, C., Zheng, B., Ni, J., Zhou, K., Qian, M., & Wu, H. (2024). Genome-Wide Identification and Expression Analysis of GST Genes during Light-Induced Anthocyanin Biosynthesis in Mango (Mangifera indica L.). Plants, 13(19), 2726. https://doi.org/10.3390/plants13192726







