Comparative Physiological, Biochemical, and Leaf Proteome Responses of Contrasting Wheat Varieties to Drought Stress
Abstract
1. Introduction
2. Results
2.1. Growth and Physiological Responses of BW4074 and BW35695 Wheat Varieties towards Drought Stress
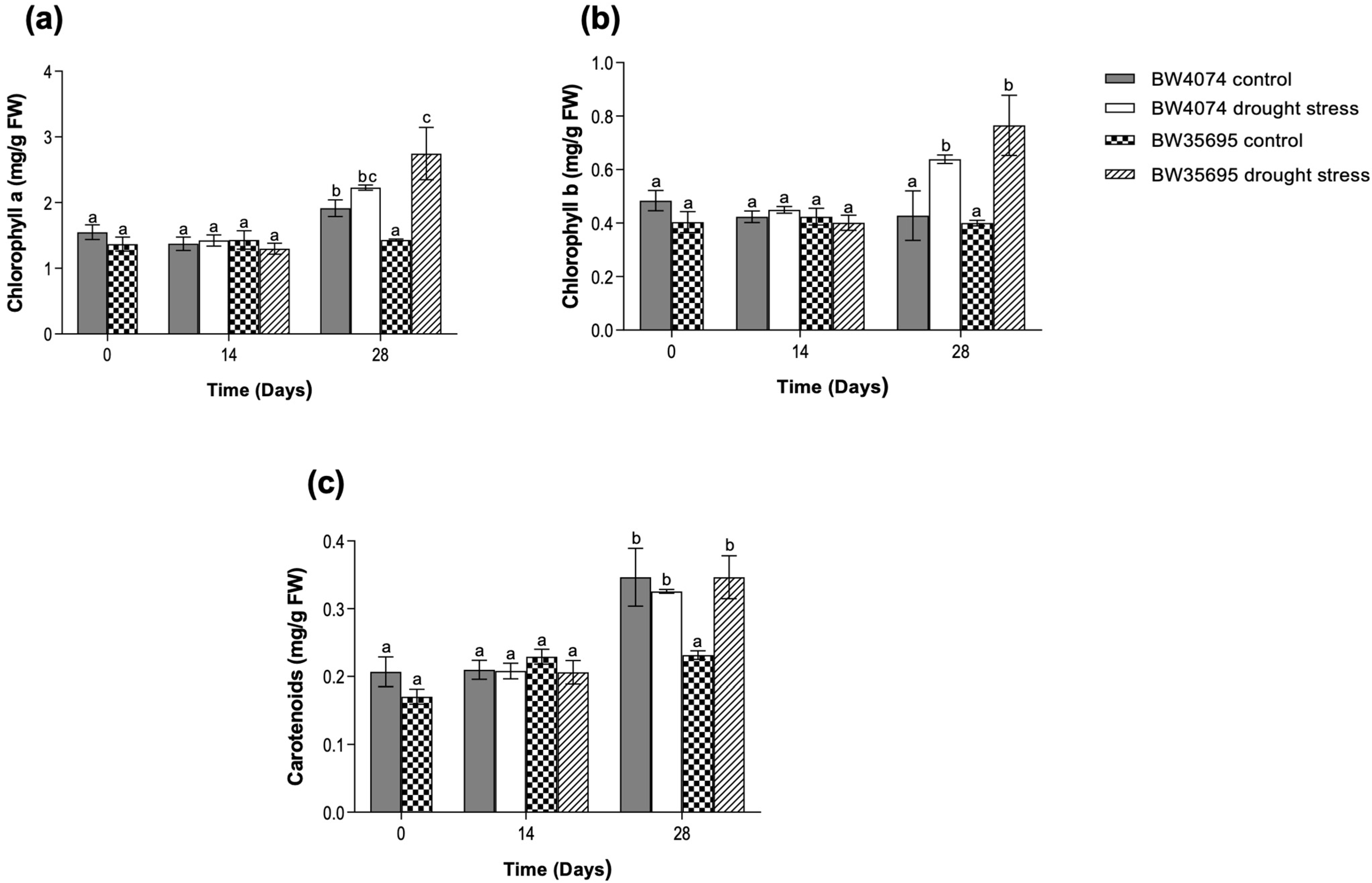
2.2. Drought Stress Increases the Levels of Photosynthetic Pigments Mainly in the Drought-Tolerant Wheat Variety
2.3. Drought Stress Induces Oxidative Stress in Wheat Tissues
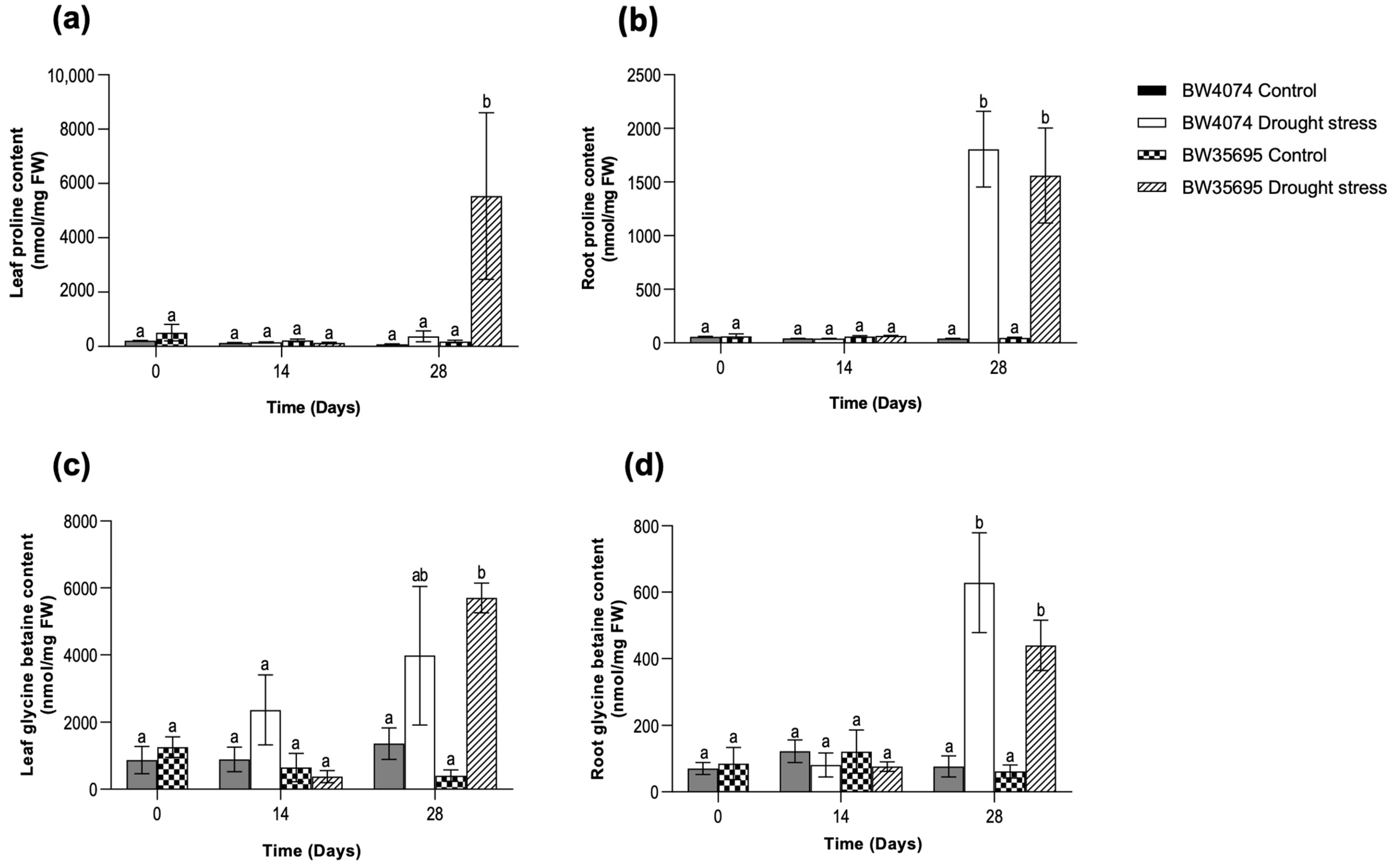
2.4. Drought-Induced Accumulation of Osmoprotectants in Wheat Leaf and Root Tissues
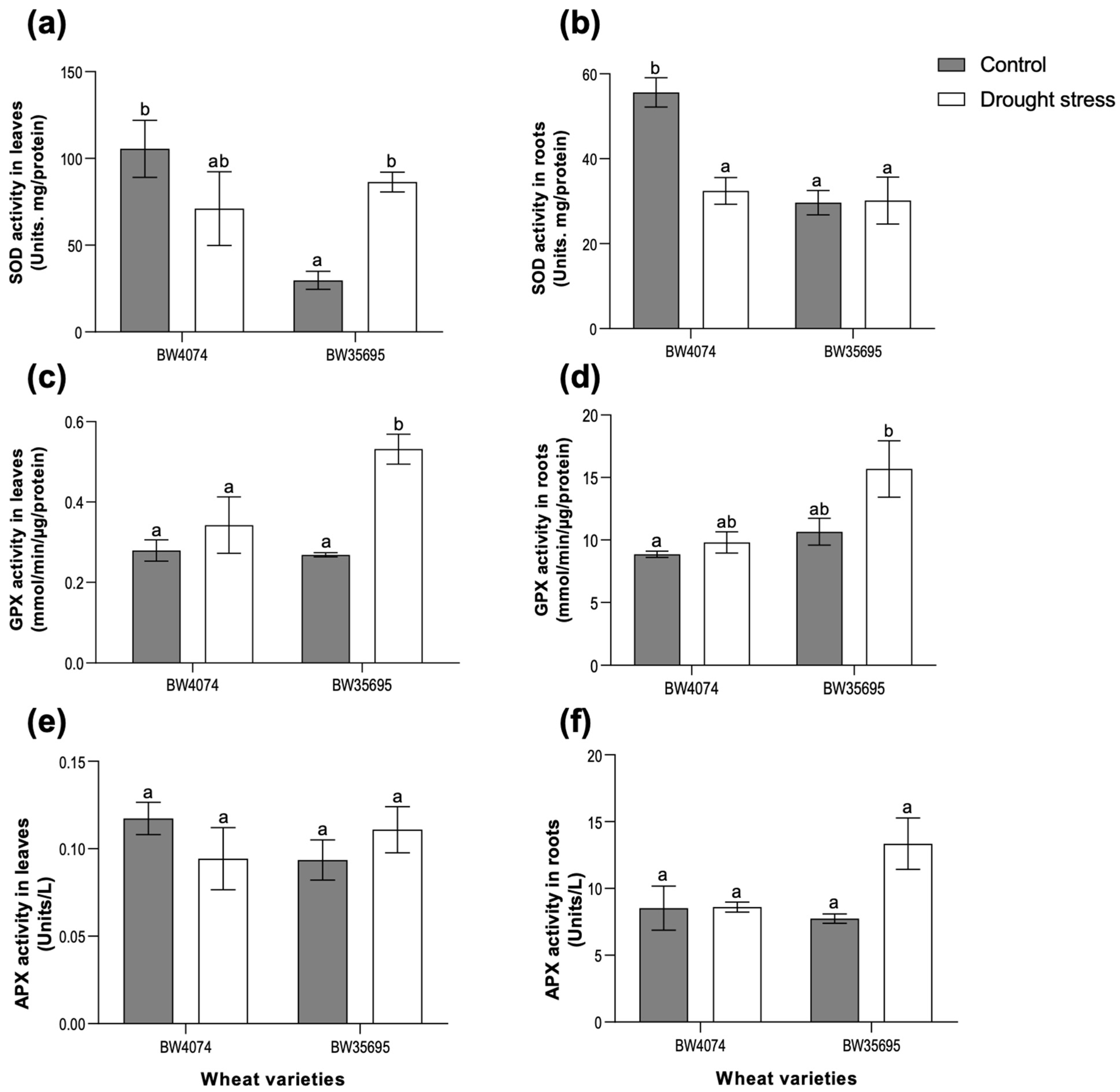
2.5. The Drought-Tolerant Wheat Variety BW35695 Possesses Greater Antioxidant Enzyme Activities Compared to Drought-Susceptible BW4074 under Drought Stress
2.6. The Drought-Responsive Leaf Proteome of Two Contrasting Wheat Varieties
2.6.1. Drought Stress Modulates the Accumulation of Total Soluble Leaf Proteins of Wheat
2.6.2. Drought-Responsive Wheat Leaf Proteins Are Implicated in Diverse Functional Roles
Drought Stress Largely Downregulates Energy-Related Proteins, Particularly Those Associated with Photosynthesis
Drought Stress Upregulates Proteins Involved in Defense Response, Protein Homeostasis, and Transcription
Other Functional Categories of Drought-Responsive Wheat Leaf Proteins
Common Drought-Responsive Proteins between BW4074 and BW35695
2.7. Pathway Enrichment Analysis and Protein–Protein Interactions
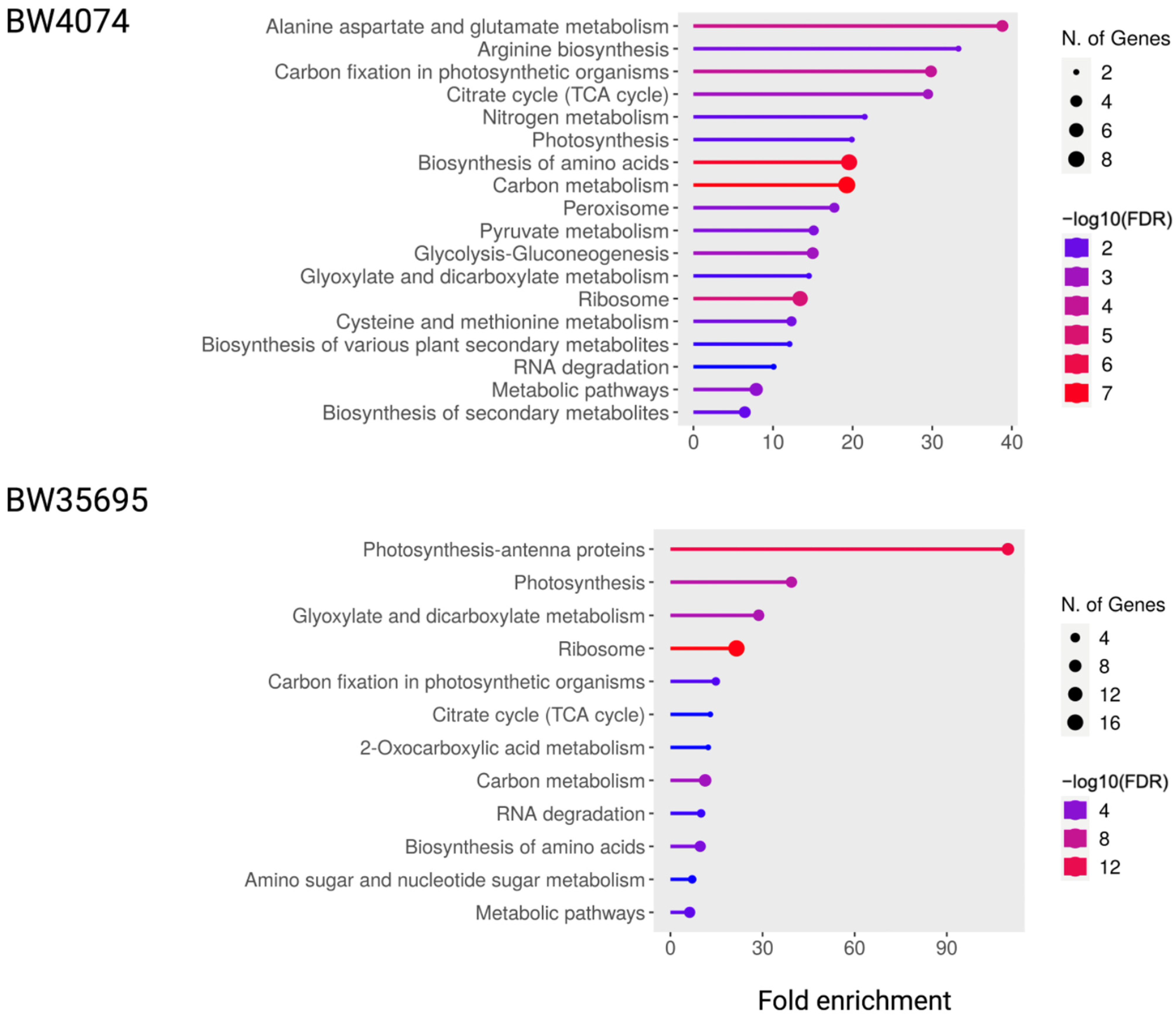
2.7.1. KEGG Pathway Enrichment Analysis
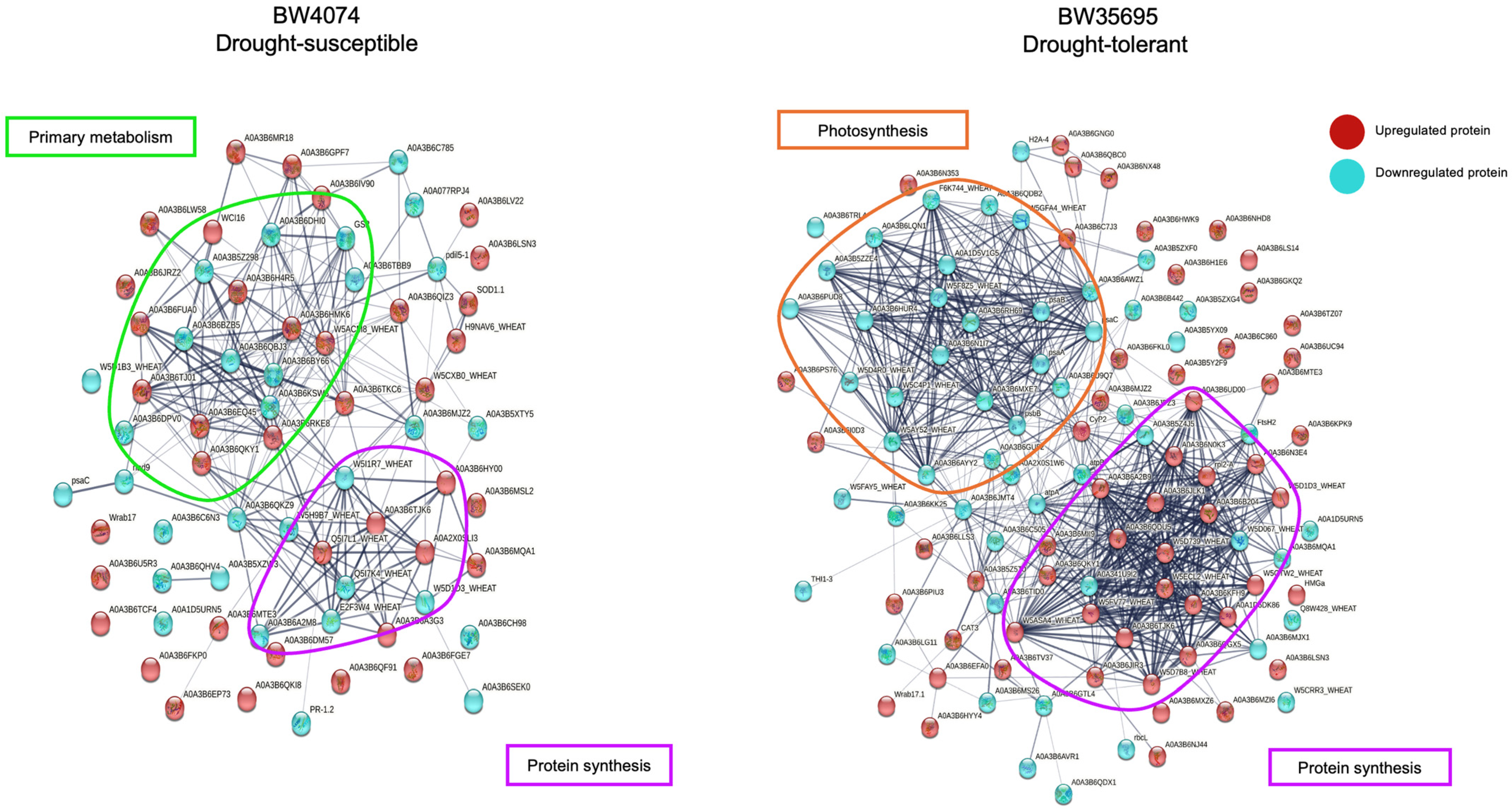
2.7.2. Protein–Protein Interaction Analysis
2.8. Gene Expression Analysis Using qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Drought Stress Treatments
4.2. Plant Growth Parameters, Relative Water Content, and Relative Shoot Water Loss Measurements
4.3. Biochemical Assays
4.3.1. Chlorophyll and Carotenoid Content
4.3.2. Lipid Peroxidation, Hydrogen Peroxide, and Superoxide Content
4.3.3. Proline and Glycine Betaine Content
4.3.4. Enzymatic Antioxidant Activity Assays
4.4. Leaf Protein Extraction, iTRAQ Labeling, LC-MS/MS, and Identification
4.4.1. Leaf Protein Extraction, iTRAQ Labeling, and Tandem Mass Spectrometry
4.4.2. Protein Identification and Quantification
4.4.3. Bioinformatics Analyses of Differentially Expressed Proteins
4.5. Gene Expression Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mirzabaev, A.; Kerr, R.B.; Hasegawa, T.; Pradhan, P.; Wreford, A.; Pahlen, M.C.T.V.; Gurney-Smith, H. Severe climate change risks to food security and nutrition. Clim. Risk Manag. 2023, 39, 100473. [Google Scholar] [CrossRef]
- Gan, T.Y.; Ito, M.; Hulsmann, S.; Qin, X.; Lu, X.X.; Liong, S.Y.; Rutschman, P.; Disse, M.; Koivusalo, H. Possible climate change/variability and human impacts, vulnerability of drought-prone regions, water resources and capacity building for Africa. Hydrol. Sci. J. 2016, 61, 1209–1226. [Google Scholar] [CrossRef]
- Lottering, S.; Mafongoya, P.; Lottering, R. Drought and its impacts on small-scale farmers in sub-Saharan Africa: A review. S. Afr. Geogr. J. 2021, 103, 319–341. [Google Scholar] [CrossRef]
- Mittler, R.; Blumwald, E. Genetic engineering for modern agriculture: Challenges and perspectives. Annu. Rev. Plant Biol. 2010, 61, 443–462. [Google Scholar] [CrossRef] [PubMed]
- Passioura, J.B. Phenotyping for drought tolerance in grain crops: When is it useful to breeders? Funct. Plant Biol. 2012, 39, 851–859. [Google Scholar] [CrossRef]
- Zheng, C.; Bochmann, H.; Liu, Z.; Kant, J.; Schrey, S.D.; Wojciechowski, T.; Postma, J.A. Plant root plasticity during drought and recovery: What do we know and where to go? Front. Plant Sci. 2023, 14, 1084355. [Google Scholar] [CrossRef]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef]
- Ghatak, A.; Chaturvedi, P.; Weckwerth, W. Cereal crop proteomics: Systemic analysis of crop drought stress responses towards marker-assisted selection breeding. Front. Plant Sci. 2017, 8, 757. [Google Scholar] [CrossRef]
- Ngara, R.; Goche, T.; Swanevelder, D.Z.H.; Chivasa, S. Sorghum’s whole-plant transcriptome and proteome responses to drought stress: A review. Life 2021, 11, 704. [Google Scholar] [CrossRef]
- Thanmalagan, R.R.; Jayaprakash, A.; Roy, A.; Arunachalam, A.; Lakshmi, P.T.V. A review on applications of plant network biology to understand the drought stress response in economically important cereal crops. Plant Gene 2022, 29, 100345. [Google Scholar] [CrossRef]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Qamer, Z.; Chaudhary, M.T.; Du, X.; Hinze, L.; Azhar, M.T. Review of oxidative stress and antioxidative defense mechanisms in Gossypium hirsutum L. in response to extreme abiotic conditions. J. Cotton Res. 2021, 4, 9. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Bubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Fang, Y.J.; Xiong, L.Z. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Takahashi, F.; Kuromori, T.; Sato, H.; Shinozaki, K. Regulatory gene networks in drought stress responses and resistance in plants. Adv. Exp. Med. Biol. 2018, 1081, 189–214. [Google Scholar]
- Rai, G.K.; Khanday, D.M.; Choudhary, S.M.; Kumar, P.; Kumari, S.; Martínez-Andújar, C.; Martinez-Melgarejo, P.A.; Rai, P.K.; Pérez-Alfocea, F. Unlocking nature’s stress buster: Abscisic acid’s crucial role in defending plants against abiotic stress. Plant Stress 2024, 11, 100359. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Liu, S.W.; Lv, Z.Y.; Liu, Y.H.; Li, L.; Zhang, L.D. Network analysis of ABA-dependent and ABA-independent drought responsive genes in Arabidopsis thaliana. Genet. Mol. Biol. 2018, 41, 624–637. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, J.; Singh, S.; Singh, V.P.; Prasad, S.M. Roles of osmoprotectants in improving salinity and drought tolerance in plants: A review. Rev. Environ. Sci. Biotechnol. 2015, 14, 407–426. [Google Scholar] [CrossRef]
- Turner, N.C. Turgor maintenance by osmotic adjustment: 40 years of progress. J. Exp. Bot. 2018, 69, 3223–3233. [Google Scholar] [CrossRef] [PubMed]
- Dumanovic, J.; Nepovimova, E.; Natic, M.; Kuca, K.; Jacevic, V. The significance of reactive oxygen species and antioxidant defense system in plants: A concise overview. Front. Plant Sci. 2021, 11, 552969. [Google Scholar] [CrossRef] [PubMed]
- Tardieu, F.; Simonneau, T.; Muller, B. The physiological basis of drought tolerance in crop plants: A scenario-dependent probabilistic approach. Annu. Rev. Plant Biol. 2018, 69, 733–759. [Google Scholar] [CrossRef] [PubMed]
- Grote, U.; Fasse, A.; Nguyen, T.T.; Erenstein, O. Food security and the dynamics of wheat and maize value chains in Africa and Asia. Front. Sustain. Food Syst. 2021, 4, 617009. [Google Scholar] [CrossRef]
- Sendhil, R.; Kiran Kumara, T.M.; Kandpal, A.; Kumari, B.; Mohapatra, S. Wheat production, trade, consumption, and stocks: Global trends and prospects. In Wheat Science: Nutritional and Anti-Nutritional Properties, Processing, Storage, Bioactivity, and Product Development, 1st ed.; Gupta, O.P., Kumar, S., Pandey, A., Khan, M.K., Singh, S.K., Singh, G.P., Eds.; CRC Press: Boca Raton, USA, 2023; p. 23. [Google Scholar]
- Ahmad, Z.; Waraich, E.A.; Akhtar, S.; Anjum, S.; Ahmad, T.; Mahboob, W.; Hafeez, O.B.; Tapera, T.; Labuschagne, M.; Rizwan, M. Physiological responses of wheat to drought stress and its mitigation approaches. Acta Physiol. Plant. 2018, 40, 80. [Google Scholar] [CrossRef]
- Farooq, M.; Hussain, M.; Siddique, K.H.M. Drought stress in wheat during flowering and grain-filling periods. Crit. Rev. Plant Sci. 2014, 33, 331–349. [Google Scholar] [CrossRef]
- Lopes, M.S.; El-Basyoni, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A.; et al. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef]
- Sansaloni, C.; Franco, J.; Santos, B.; Percival-Alwyn, L.; Singh, S.; Petroli, C.; Campos, J.; Dreher, K.; Payne, T.; Marshall, D.; et al. Diversity analysis of 80,000 wheat accessions reveals consequences and opportunities of selection footprints. Nat. Commun. 2020, 11, 4572. [Google Scholar] [CrossRef]
- Tadesse, W.; Bishaw, Z.; Assefa, S. Wheat production and breeding in Sub-Saharan Africa: Challenges and opportunities in the face of climate change. Int. J. Clim. Chang. Strateg. Manag. 2019, 11, 696–715. [Google Scholar] [CrossRef]
- De Sousa, T.; Ribeiro, M.; Sabença, C.; Igrejas, G. The 10,000-Year Success Story of Wheat! Foods 2021, 10, 2124. [Google Scholar] [CrossRef]
- Abid, M.; Ali, S.; Qi, L.K.; Zahoor, R.; Tian, Z.W.; Jiang, D.; Snider, J.L.; Dai, T.B. Physiological and biochemical changes during drought and recovery periods at tillering and jointing stages in wheat (Triticum aestivum L.). Sci Rep. 2018, 8, 4615. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.R.; Zhang, X.Y.; Han, Z.D.; Feng, H.X.; Wang, Y.Y.; Kang, J.; Han, X.J.; Wang, L.F.; Wang, C.Y.; Li, H.; et al. Analysis of physiological indicators associated with drought tolerance in wheat under drought and re-watering conditions. Antioxidants 2022, 11, 2266. [Google Scholar] [CrossRef]
- Amoah, J.N.; Ko, C.S.; Yoon, J.S.; Weon, S.Y. Effect of drought acclimation on oxidative stress and transcript expression in wheat (Triticum aestivum L.). J. Plant Interact. 2019, 14, 492–505. [Google Scholar] [CrossRef]
- Kirova, E.; Moskova, I.; Geneva, M.; Kocheva, K. Antioxidant potential of tolerant and susceptible wheat varieties under drought and recovery. Cereal Res. Commun. 2022, 50, 841–849. [Google Scholar] [CrossRef]
- Popova, A.V.; Mihailova, G.; Geneva, M.; Peeva, V.; Kirova, E.; Sichanova, M.; Dobrikova, A.; Georgieva, K. Different responses to water deficit of two common winter wheat varieties: Physiological and biochemical characteristics. Plants 2023, 12, 2239. [Google Scholar] [CrossRef]
- Faghani, E.; Gharechahi, J.; Komatsu, S.; Mirzaei, M.; Khavarinejad, R.A.; Najafi, F.; Farsad, L.K.; Salekdeh, G.H. Comparative physiology and proteomic analysis of two wheat genotypes contrasting in drought tolerance. J. Proteom. 2015, 114, 1–15. [Google Scholar] [CrossRef]
- Chaichi, M.; Sanjarian, F.; Razavi, K.; Gonzalez-Hernandez, J.L. Analysis of transcriptional responses in root tissue of bread wheat landrace (Triticum aestivum L.) reveals drought avoidance mechanisms under water scarcity. PLoS ONE 2019, 14, e0212671. [Google Scholar] [CrossRef]
- Lv, L.J.; Chen, X.Y.; Li, H.; Huang, J.A.; Liu, Y.P.; Zhao, A.J. Different adaptive patterns of wheat with different drought tolerance under drought stresses and rehydration revealed by integrated metabolomic and transcriptomic analysis. Front. Plant Sci. 2022, 13, 1008624. [Google Scholar] [CrossRef]
- Ford, K.L.; Cassin, A.; Bacic, A. Quantitative proteomic analysis of wheat cultivars with differing drought stress tolerance. Front. Plant Sci. 2011, 2, 44. [Google Scholar] [CrossRef]
- Michaletti, A.; Naghavi, M.R.; Toorchi, M.; Zolla, L.; Rinalducci, S. Metabolomics and proteomics reveal drought-stress responses of leaf tissues from spring-wheat. Sci. Rep. 2018, 8, 5710. [Google Scholar] [CrossRef]
- Kang, Z.Y.; Babar, M.A.; Khan, N.; Guo, J.; Khan, J.; Islam, S.; Shrestha, S.; Shahi, D. Comparative metabolomic profiling in the roots and leaves in contrasting genotypes reveals complex mechanisms involved in post-anthesis drought tolerance in wheat. PLoS ONE 2019, 14, e0213502. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.S.; Galbraith, D.W.; Dai, S.Y.; Griffin, P.; Stewart, C.N. Plant systems biology comes of age. Trends Plant Sci. 2008, 13, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A.; Kusano, M.; Redestig, H.; Arita, M.; Saito, K. Integrated omics approaches in plant systems biology. Curr. Opin. Chem. Biol. 2009, 13, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Sanchez, A.; Gonzalez-Melendi, P.; Santamaria, M.E.; Arbona, V.; Lopez-Gonzalvez, A.; Garcia, A.; Hensel, G.; Kumlehn, J.; Martinez, M.; Diaz, I. Repression of drought-induced cysteine-protease genes alters barley leaf structure and responses to abiotic and biotic stresses. J. Exp. Bot. 2019, 70, 2143–2155. [Google Scholar] [CrossRef]
- Wang, C.L.; Gao, B.; Chen, N.N.; Jiao, P.; Jiang, Z.Z.; Zhao, C.L.; Ma, Y.Y.; Guan, S.Y.; Liu, S.Y. A novel senescence-specific gene (ZmSAG39) negatively regulates darkness and drought responses in maize. Int. J. Mol. Sci. 2022, 23, 15984. [Google Scholar] [CrossRef]
- Shao, H.B.; Chu, L.Y.; Jaleel, C.A.; Manivannan, P.; Panneerselvam, R.; Shao, M.A. Understanding water deficit stress-induced changes in the basic metabolism of higher plants—Biotechnologically and sustainably improving agriculture and the ecoenvironment in arid regions of the globe. Crit. Rev. Biotechnol. 2009, 29, 131–151. [Google Scholar] [CrossRef]
- Bevan, M.; Bancroft, I.; Bent, E.; Love, K.; Goodman, H.; Dean, C.; Bergkamp, R.; Dirkse, W.; Van Staveren, M.; Stiekema, W.; et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 1998, 391, 485–488. [Google Scholar] [PubMed]
- Samarah, N.H. Effects of drought stress on growth and yield of barley. Agron. Sustain. Dev. 2005, 25, 145–149. [Google Scholar] [CrossRef]
- Bai, L.-P.; Sui, F.-G.; Ge, T.-D.; Sun, Z.-H.; Lu, Y.-Y.; Zhou, G.-S. Effect of soil drought stress on leaf water status, membrane permeability and enzymatic antioxidant system of maize. Pedosphere 2006, 16, 326–332. [Google Scholar] [CrossRef]
- Akter, N.; Brishty, T.A.; Karim, M.A.; Ahmed, M.J.U.; Islam, M.R. Leaf water status and biochemical adjustments as a mechanism of drought tolerance in two contrasting wheat (Triticum aestivum L.) varieties. Acta Physiol. Plant. 2023, 45, 50. [Google Scholar] [CrossRef]
- Goche, T.; Shargie, N.G.; Cummins, I.; Brown, A.P.; Chivasa, S.; Ngara, R. Comparative physiological and root proteome analyses of two sorghum varieties responding to water limitation. Sci. Rep. 2020, 10, 11835. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.W.; Song, D.; Qi, X.L.; Asad, M.; Wang, S.; Tong, X.H.; Jiang, Y.; Wang, S.D. Physiological responses and transcriptome analysis of soybean under gradual water deficit. Front. Plant Sci. 2023, 14, 1269884. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.B.; Zhao, Y.F.; Zhang, J.L.; Chao, M.N.; Xie, K.L.; Zhang, C.; Sun, F.L.; Liu, S.D.; Xi, Y.J. Proteomic analysis of the similarities and differences of soil drought and polyethylene glycol stress responses in wheat (Triticum aestivum L.). Plant Mol. Biol. 2019, 100, 391–410. [Google Scholar] [CrossRef]
- Melkozernov, A.N.; Blankenship, R.E. Photosynthetic Functions of Chlorophylls. In Chlorophylls and Bacteriochlorophylls: Biochemistry, Biophysics, Functions and Applications; Grimm, B., Porra, R.J., Rudiger, W., Scheer, H., Eds.; Springer: Dordrecht, The Netherlands, 2006; Volume 25, pp. 397–412. [Google Scholar]
- Taiz, L.; Zeiger, E. Plant Physiology, 4th ed.; Sinauer Associates Inc.: Sunderland, MA, USA, 2010. [Google Scholar]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef]
- Zeng, W.J.; Peng, Y.L.; Zhao, X.Q.; Wu, B.Y.; Chen, F.Q.; Ren, B.; Zhuang, Z.L.; Gao, Q.H.; Ding, Y.F. Comparative proteomics analysis of the seedling root response of drought-sensitive and drought-tolerant maize varieties to drought stress. Int. J. Mol. Sci. 2019, 20, 2793. [Google Scholar] [CrossRef]
- Pyngrope, S.; Bhoomika, K.; Dubey, R.S. Reactive oxygen species, ascorbate-glutathione pool, and enzymes of their metabolism in drought-sensitive and tolerant indica rice (Oryza sativa L.) seedlings subjected to progressing levels of water deficit. Protoplasma 2013, 250, 585–600. [Google Scholar] [CrossRef]
- Smith, K.T.; Workman, J.L. Chromatin proteins: Key responders to stress. PLoS Biol. 2012, 10, e1001371. [Google Scholar] [CrossRef]
- Yuan, L.Y.; Liu, X.C.; Luo, M.; Yang, S.G.; Wu, K.Q. Involvement of histone modifications in plant abiotic stress responses. J. Integr. Plant Biol. 2013, 55, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, S.; Liang, Y.J.; Qi, Y.H.; Chen, J.; Zhu, W.N.; Zhang, L.S. Label-free quantitative proteomic analysis of drought stress-responsive late embryogenesis abundant proteins in the seedling leaves of two wheat (Triticum aestivum L.) genotypes. J. Proteom. 2018, 172, 122–142. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Kaur, K.; Singh, M.; Kaur, G.; Singh, P. Plant cyclophilins: Multifaceted proteins with versatile roles. Front. Plant Sci. 2020, 11, 585212. [Google Scholar] [CrossRef]
- Li, Y.H.; Cui, J.Y.; Zhao, Q.; Yang, Y.Z.; Wei, L.; Yang, M.D.; Liang, F.; Ding, S.T.; Wang, T.C. Physiology and proteomics of two maize genotypes with different drought resistance. Biol. Plant. 2019, 63, 519–528. [Google Scholar] [CrossRef]
- Waititu, J.K.; Zhang, X.E.; Chen, T.C.; Zhang, C.Y.; Zhao, Y.; Wang, H. Transcriptome analysis of tolerant and susceptible maize genotypes reveals novel insights about the molecular mechanisms underlying drought responses in leaves. Int. J. Mol. Sci. 2021, 22, 6980. [Google Scholar] [CrossRef] [PubMed]
- Fracasso, A.; Trindade, L.M.; Amaducci, S. Drought stress tolerance strategies revealed by RNA-Seq in two sorghum genotypes with contrasting WUE. BMC Plant Biol. 2016, 16, 115. [Google Scholar] [CrossRef]
- Varoquaux, N.; Cole, B.; Gao, C.; Pierroz, G.; Baker, C.R.; Patel, D.; Madera, M.; Jeffers, T.; Hollingsworth, J.; Sievert, J.; et al. Transcriptomic analysis of field-droughted sorghum from seedling to maturity reveals biotic and metabolic responses. Proc. Natl. Acad. Sci. USA 2019, 116, 27124–27132. [Google Scholar] [CrossRef]
- Azzouz-Olden, F.; Hunt, A.G.; Dinkins, R. Transcriptome analysis of drought-tolerant sorghum genotype SC56 in response to water stress reveals an oxidative stress defense strategy. Mol. Biol. Rep. 2020, 47, 3291–3303. [Google Scholar] [CrossRef]
- Kosova, K.; Vitamvas, P.; Prasil, I.T.; Klima, M.; Renaut, J. Plant proteoforms under environmental stress: Functional proteins arising from a single gene. Front. Plant Sci. 2021, 12, 793113. [Google Scholar] [CrossRef]
- Nemati, M.; Piro, A.; Norouzi, M.; Vahed, M.M.; Nisticò, D.M.; Mazzuca, S. Comparative physiological and leaf proteomic analyses revealed the tolerant and sensitive traits to drought stress in two wheat parental lines and their F6 progenies. Environ. Exp. Bot. 2019, 158, 223–237. [Google Scholar] [CrossRef]
- Chang, L.L.; Wang, L.M.; Peng, C.Z.; Tong, Z.; Wang, D.; Ding, G.H.; Xiao, J.H.; Guo, A.P.; Wang, X.C. The chloroplast proteome response to drought stress in cassava leaves. Plant Physiol. Biochem. 2019, 142, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Vineeth, T.V.; Kumar, P.; Krishna, G.K. Bioregulators protected photosynthetic machinery by inducing expression of photorespiratory genes under water stress in chickpea. Photosynthetica 2016, 54, 234–242. [Google Scholar] [CrossRef]
- Barrs, H.; Weatherley, P. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust. J. Biol. Sci. 1962, 15, 413–428. [Google Scholar] [CrossRef]
- Rahman, S.; Shabeen, M.S.; Rahman, M.A.; Malik, T.A. Evaluation of excised leaf water loss and relative water content, as screening techniques for breeding drought resistant wheat. Pak. J. Biol. Sci. 2000, 3, 663–665. [Google Scholar]
- Hiscox, J.D.; Israelstam, G.F. A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 1979, 57, 1332–1334. [Google Scholar] [CrossRef]
- Wellburn, A.R. The spectral determination of chlorophyll-a and chlorophhyll-b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Gokul, A.; Carelse, M.F.; Niekerk, L.A.; Klein, A.; Ludidi, N.; Mendoza-Cozatl, D.; Keyster, M. Exogenous 3,3′-diindolylmethane improves vanadium stress tolerance in Brassica napus seedling shoots by modulating antioxidant enzyme activities. Biomolecules 2021, 11, 436. [Google Scholar] [CrossRef]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain treated bean plants—Protective role of exogenous polyamines. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Prinsen, H.; Schiebergen-Bronkhorst, B.G.M.; Roeleveld, M.W.; Jans, J.J.M.; de Sain-van der Velden, M.G.M.; Visser, G.; van Hasselt, P.M.; Verhoeven-Duif, N.M. Rapid quantification of underivatized amino acids in plasma by hydrophilic interaction liquid chromatography (HILIC) coupled with tandem mass-spectrometry. J. Inherit. Metab. Dis. 2016, 39, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Ngara, R. A Proteomic Analysis of Drought and Salt Stress Responsive Proteins of Different Sorghum Varieties. Ph.D. Thesis, University of the Western Cape, Cape Town, South Africa, 2009. [Google Scholar]
- Moloi, M.J.; van der Merwe, R. Drought tolerance responses in vegetable-type soybean involve a network of biochemical mechanisms at flowering and pod-filling stages. Plants 2021, 10, 1502. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Ngwenya, S.P.; Moloi, S.J.; Shargie, N.G.; Brown, A.P.; Chivasa, S.; Ngara, R. Regulation of proline accumulation and protein secretion in sorghum under combined osmotic and heat stress. Plants 2024, 13, 1874. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, R.Q.; Wang, H.G.; Li, D.X.; Wang, X.Y.; Zhang, Y.C.; Zhen, W.C.; Duane, H.J.; Yan, G.J.; Li, Y.M. Transcriptomics analyses reveal wheat responses to drought stress during reproductive stages under field conditions. Front. Plant Sci. 2017, 8, 592. [Google Scholar] [CrossRef]
- Kumar, J.; Gunapati, S.; Kianian, S.F.; Singh, S.P. Comparative analysis of transcriptome in two wheat genotypes with contrasting levels of drought tolerance. Protoplasma 2018, 255, 1487–1504. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]






| Wheat Variety | Positively Identified Proteins | Drought-Responsive Proteins | Protein Regulation | |
|---|---|---|---|---|
| Up | Down | |||
| BW4074 | 1062 | 69 | 41 | 28 |
| BW35695 | 882 | 110 | 58 | 52 |
| Accession a | Protein Name b | Ratio c | SD d | p-Value e | Family Name f |
|---|---|---|---|---|---|
| Primary Metabolism | |||||
| A0A3B6QKI8 | GDSL esterase/lipase | −1.86 | 0.22 | 1.29 × 10−2 | GDSL lipase/esterase-like, plant |
| A0A1D5URN5 | Fibronectin type III-LIKE domain-containing protein | −1.55 | 0.08 | 2.99 × 10−2 | Beta-D-xylosidase |
| A0A077RPJ4 | Tryptophan synthase | −1.67 | 0.19 | 4.51 × 10−2 | Tryptophan synthase, alpha chain |
| Q45NB2 | Glutamine synthetase | −1.68 | 0.26 | 3.75 × 10−2 | Glutamine synthetase |
| A0A3B6FGE7 | Enoyl reductase (ER) domain-containing protein | 1.89 | 0.41 | 2.32 × 10−2 | Medium-chain dehydrogenase/ reductase |
| A0A3B6DHI0 | Glutamate dehydrogenase | −2.08 | 0.19 | 4.52 × 10−2 | Glutamate dehydrogenase |
| W5ACM8 | Delta-1-pyrroline-5-carboxylate synthase | 1.52 | 0.24 | 3.17 × 10−2 | Delta-l-pyrroline-5-carboxylate synthetase |
| A0A3B6MSL2 | Aminocyclopropanecarboxylate oxidase | 1.58 | 0.36 | 2.09 × 10−2 | Plant 2-oxoglutarate-dependent oxidoreductases |
| Protein synthesis/folding/degradation | |||||
| W5I1R7 | 30S ribosomal protein S3, chloroplastic | 1.63 | 0.18 | 8.04 × 10−3 | Small ribosomal subunit protein uS3 |
| A0A3B6TJK6 | Small ribosomal subunit protein uS10 domain-containing protein | 1.64 | 0.40 | 2.86 × 10−2 | Small ribosomal subunit protein uS10 |
| W5H9B7 | Peptidyl-prolyl cis-trans isomerase | −1.57 | 0.21 | 3.52 × 10−2 | Cyclophilin-type peptidyl-prolyl cis-trans isomerase/CLD |
| A0A2 × 0SLI3 | 40S ribosomal protein S6 | 1.94 | 0.33 | 4.09 × 10−2 | Small ribosomal subunit protein eS6-like |
| A0A3B6QHV4 | Anion-transporting ATPase-like domain-containing protein | −1.68 | 0.25 | 2.63 × 10−2 | Arsenical pump ATPase, ArsA/GET3 |
| Defense/ROS detoxification | |||||
| A0A3B6HMK6 | Glutathione reductase | 1.94 | 0.66 | 4.69 × 10−2 | Glutathione reductase |
| S6AWC2 | Cold induced 16 | 1.68 | 0.42 | 4.43 × 10−2 | Nodulin-related protein1/2 |
| D8L9B5 | Putative PDI-like protein | −2.57 | 0.11 | 1.11 × 10−2 | Protein disulfide isomerase A6 |
| Energy | |||||
| A0A3B5Z298 | Phosphoglycerate kinase | −1.51 | 0.22 | 4.29 × 10−2 | Phosphoglycerate kinase family |
| A0A3B6FUA0 | Fructose-bisphosphate aldolase | 1.71 | 0.27 | 2.64 × 10−3 | Fructose-bisphosphate aldolase, class I |
| A0A3B6TBB9 | Ferredoxin--NADP reductase, chloroplastic | −2.02 | 0.31 | 1.77 × 10−2 | Ferredoxin-NADP reductase |
| A0A3B6KSW8 | Glyceraldehyde-3-phosphate dehydrogenase | −1.68 | 0.18 | 4.84 × 10−2 | Glyceraldehyde-3-phosphate dehydrogenase, type 1 |
| A0A3B6BY66 | Dihydrolipoyllysine-residue succinyltransferase | −1.61 | 0.18 | 3.67 × 10−2 | Dihydrolipoamide succinyltransferase |
| A0A3B6BZB5 | Transketolase | −1.88 | 0.04 | 1.89 × 10−2 | Transketolase, bacterial-like |
| Transcription | |||||
| A0A3B6LV22 | H15 domain-containing protein | 1.50 | 0.19 | 4.14 × 10−3 | Linker histone H1/H5 |
| A0A3B6A3G3 | Hyaluronan/mRNA-binding protein domain-containing protein | 1.99 | 0.59 | 2.54 × 10−2 | RNA binding protein HABP4/SERBP1-like |
| Secondary metabolism | |||||
| A0A3B6C785 | Zeta-carotene desaturase | −1.50 | 0.18 | 4.20 × 10−2 | Zeta-carotene desaturase |
| A0A3B6DPV0 | AB hydrolase-1 domain-containing protein | −1.78 | 0.07 | 3.66 × 10−2 | Epoxide hydrolase-like |
| Cell structure | |||||
| A0A3B6SEK0 | Cyanobacterial aminoacyl-tRNA synthetase CAAD domain-containing protein | −1.87 | 0.19 | 2.29 × 10−2 | Protein curvature thylakoid I |
| Unclear classification | |||||
| A0A3B6FKP0 | PH domain-containing protein | 1.71 | 0.34 | 7.96 × 10−3 | Ricin B-like lectin EULS3-like |
| A0A3B5XZW3 | Remorin C-terminal domain-containing protein | −1.65 | 0.02 | 1.98 × 10−2 | None predicted |
| Accession a | Protein Name b | Ratio c | SD d | p-Value e | Family Name f |
|---|---|---|---|---|---|
| Primary Metabolism | |||||
| A0A3B6UC94 | Acid phosphatase | 2.05 | 0.28 | 5.05 × 10−4 | Acid phosphatase, plant |
| A0A3B6FKL0 | Nucleoside phosphorylase domain-containing protein | 1.66 | 0.27 | 3.78 × 10−3 | Phosphorylase superfamily |
| A0A3B6B442 | Aspartate/glutamate/uridylate kinase domain-containing protein | −1.92 | 0.34 | 4.08 × 10−2 | Glutamate/acetylglutamate kinase |
| A0A3B6TUD9 | Thiamine thiazole synthase, chloroplastic | −1.74 | 0.13 | 3.42 × 10−2 | Thiamine thiazole synthase |
| A0A3B6NHD8 | O-methyltransferase ZRP4 | 1.56 | 0.41 | 3.89 × 10−2 | O-methyltransferase COMT-type |
| A0A3B5ZXG4 | Glycosyltransferase | −1.50 | 0.12 | 2.23 × 10−2 | UDP-glucuronosyl/UDP-glucosyltransferase |
| A0A3B6MS26 | Glucose-6-phosphate 1-epimerase | −2.37 | 0.15 | 1.86 × 10−3 | Glucose-6-phosphate 1-epimerase |
| A0A3B6KPK9 | Beta-glucosidase | 2.27 | 0.88 | 4.53 × 10−2 | Cellulase degradation glycosyl hydrolase 3 |
| Energy | |||||
| P24065 | Photosystem II CP47 reaction center protein | −2.02 | 0.12 | 1.01 × 10−2 | Photosystem II CP47 reaction centre protein |
| A0A3B6AYY2 | 23 kDa subunit of oxygen evolving system of photosystem II | 1.56 | 0.35 | 4.27 × 10−2 | PsbP |
| A0A3B6N1I7 | Photosystem I reaction center subunit II, chloroplastic | −1.57 | 0.19 | 1.60 × 10−2 | Photosystem I PsaD |
| A0A3B6LQN1 | Chlorophyll A-B binding protein, chloroplastic | −1.63 | 0.11 | 2.36 × 10−2 | Chlorophyll A-B binding protein |
| A0A3B6JMT4 | Glyceraldehyde-3-phosphate dehydrogenase | −2.20 | 0.42 | 4.92 × 10−2 | Glyceraldehyde-3-phosphate dehydrogenase, type 1 |
| W5C4P1 | Uncharacterized protein | −1.92 | 0.08 | 1.06 × 10−2 | Oxygen-evolving enhancer protein 3, plants |
| W5AY52 | Chlorophyll A-B binding protein, chloroplastic | −1.67 | 0.08 | 1.53 × 10−3 | Chlorophyll A-B binding protein |
| W5D4R0 | Photosystem II 22 kDa protein, chloroplastic | −1.63 | 0.13 | 5.52 × 10−3 | Chlorophyll A-B binding protein |
| A0A3B6QKY1 | Aconitate hydratase | 2.05 | 0.47 | 5.72 × 10−3 | Aconitase/Iron-responsive element-binding protein 2 |
| P58386 | Photosystem I P700 chlorophyll A apoprotein A2 | −2.39 | 0.09 | 2.93 × 10−3 | Photosystem I PsaB |
| A0A3B6QDB2 | Photosystem II protein D1 | −1.57 | 0.14 | 2.89 × 10−2 | Photosynthetic reaction centre, L/M |
| P58311 | Photosystem I P700 chlorophyll A apoprotein A1 | −2.39 | 0.04 | 9.95 × 10−3 | Photosystem I PsaA |
| A0A3B6HUR4 | Chlorophyll A-B binding protein, chloroplastic | −1.91 | 0.09 | 4.14 × 10−3 | Chlorophyll A-B binding protein |
| A0A3B6AWZ1 | Chlorophyll A-B binding protein, chloroplastic | −1.58 | 0.05 | 3.46 × 10−3 | Chlorophyll A-B binding protein |
| W5F8Z5 | Chlorophyll A-B binding protein, chloroplastic | −1.69 | 0.07 | 1.88 × 10−4 | Chlorophyll A-B binding protein |
| A0A3B5Z4J5 | ATP synthase subunit b, chloroplastic OS = Triticum aestivum | −1.55 | 0.09 | 2.09 × 10−2 | ATPase, FO complex, subunit b/b’ |
| P69415 | Photosystem I iron-sulfur center | −1.65 | 0.06 | 3.97 × 10−2 | Photosystem I protein PsaC |
| A0A3B6PUD8 | Photosystem II 10 kDa polypeptide, chloroplastic | −1.64 | 0.08 | 5.55 × 10−3 | Photosystem II PsbR |
| F6K744 | Chlorophyll A-B binding protein, chloroplastic | −2.27 | 0.06 | 1.71 × 10−3 | Chlorophyll A-B binding protein |
| A0A3B6U9Q7 | Cytochrome b/b6 C-terminal region profile domain-containing protein | −3.44 | 0.21 | 1.90 × 10−2 | Cytochrome b6/f complex, subunit IV |
| Protein synthesis/folding/degradation | |||||
| W5ASA4 | Uncharacterized protein | 1.61 | 0.41 | 4.51 × 10−2 | Small ribosomal subunit protein uS19 |
| A0A3B6JIR3 | Heat shock cognate 70kDa protein | 1.55 | 0.34 | 4.59 × 10−2 | Heat shock protein 70 family |
| A0A3B6B204 | Large ribosomal subunit protein uL23 N-terminal domain-containing protein | 1.59 | 0.16 | 5.55 × 10−4 | Large ribosomal subunit protein uL23 |
| W5D739 | KOW domain-containing protein | 1.64 | 0.24 | 4.49 × 10−3 | Large ribosomal subunit protein uL24 |
| A0A3B6MTE3 | Peptidylprolyl isomerase | 1.64 | 0.38 | 3.20 × 10−2 | Peptidyl-prolyl cis-trans isomerase FKBP18-like |
| A0A3B6N0K3 | 50S ribosomal protein L17, chloroplastic | 1.51 | 0.21 | 3.22 × 10−3 | Large ribosomal subunit protein bL17 |
| A0A3B6A2B9 | 60S ribosomal protein L37a | 2.54 | 0.42 | 9.27 × 10−3 | Large ribosomal subunit protein eL43 |
| A0A3B6QGX5 | 60S ribosomal protein L6 | 1.65 | 0.29 | 4.00 × 10−2 | Large ribosomal subunit protein eL6 |
| A0A3B6UD00 | 50S ribosomal protein L20 | 1.83 | 0.45 | 1.28 × 10−2 | Large ribosomal subunit protein bL20 |
| Transporters | |||||
| A0A3B6GKQ2 | Non-specific lipid-transfer protein | 1.71 | 0.40 | 1.31 × 10−2 | Plant non-specific lipid-transfer protein/Par allergen |
| A0A3B5YX09 | Chloroplast inner envelope protein | −1.50 | 0.10 | 2.53 × 10−2 | Protein TIC110, chloroplastic |
| A0A3B6I0D3 | STI1/HOP DP domain-containing protein | 1.74 | 0.39 | 1.84 × 10−2 | None predicted |
| Transcription | |||||
| A0A3B6LSN3 | MBD domain-containing protein | 1.52 | 0.18 | 7.18 × 10−3 | Methyl-CpG-binding domain-containing protein 10/11 |
| Q8LRU5 | HMG-I/Y protein HMGa | 1.60 | 0.40 | 3.88 × 10−2 | High-mobility group protein HMGA |
| A0A3B6MXZ6 | H15 domain-containing protein | 1.84 | 0.44 | 4.79 × 10−2 | Linker histone H1/H5 |
| A0A3B6GNG0 | Histone H2B | 1.53 | 0.24 | 5.14 × 10−3 | Histone H2B |
| Defense/ROS detoxification | |||||
| A0A3B6EFA0 | Uncharacterized protein | 2.66 | 0.73 | 4.24 × 10−3 | Nodulin-related protein ½ family |
| Q8W428 | Chitinase | −1.56 | 0.17 | 9.40 × 10−3 | Glycoside hydrolase, family 19 |
| A0A172WCB1 | Cold-responsive LEA/RAB-related COR protein | 3.23 | 1.19 | 1.01 × 10−2 | None predicted |
| A0A3B6TZ07 | GH18 domain-containing protein | 1.84 | 0.28 | 1.98 × 10−3 | Glycoside hydrolase 18 family chitinases |
| A0A3B6MJX1 | Pathogen-related protein | −2.00 | 0.13 | 1.68 × 10−2 | Pathogen-related defense protein |
| Secondary metabolism | |||||
| A0A3B6TV37 | Amine oxidase domain-containing protein | 1.67 | 0.33 | 2.10 × 10−2 | Flavin monoamine oxidase and related enzymes |
| A0A3B6QDX1 | Delta-aminolevulinic acid dehydratase | −1.59 | 0.17 | 8.35 × 10−3 | Delta-aminolevulinic acid dehydratase family |
| A0A3B5Y2F9 | Dienelactone hydrolase domain-containing protein | 1.60 | 0.22 | 2.30 × 10−3 | Dienelactone hydrolase family |
| Cell structure | |||||
| W5FAY5 | Actin | −1.50 | 0.15 | 3.53 × 10−2 | Actin family |
| Unclear classification | |||||
| A0A3B6GTL4 | DJ-1/PfpI domain-containing protein | −1.79 | 0.11 | 5.38 × 10−4 | Protein/nucleic acid deglycase DJ-1 |
| A0A3B6TRL4 | Thylakoid membrane protein slr0575 | −1.78 | 0.19 | 1.58 × 10−2 | Protein of unknown function DUF2854 |
| A0A3B6AVR1 | Uncharacterized protein | −1.63 | 0,06 | 4.05 × 10−2 | RidA family |
| A0A3B6N353 | Pentacotripeptide-repeat region of PRORP domain-containing protein | 2.10 | 0.41 | 3.42 × 10−3 | Tetratricopeptide-like helical domain superfamily |
| W5CRR3 | DUF538 domain-containing protein | −2.38 | 0.13 | 1.91 × 10−3 | Protein of unknown function DUF538 |
| A0A3B5ZXF0 | Protein kinase domain-containing protein | 1.92 | 0.44 | 9.42 × 10−3 | None predicted |
| Accession | Protein Name | Ratio a | BW4074 vs. BW35698 Ratio p-Value b | |
|---|---|---|---|---|
| BW4074 | BW35698 | |||
| Primary Metabolism | ||||
| A0A3B6MJZ2 | 5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase | 1.47 | 1.37 | 3.39 × 10−1 |
| A0A1D5URN5 | Fibronectin type III-like domain-containing protein | −1.55 | −1.15 | 3.30 × 10−3 * |
| Protein synthesis/folding/degradation | ||||
| A0A3B6TJK6 | Small ribosomal subunit protein uS10 domain-containing protein | 1.64 | 1.44 | 3.98 × 10−1 |
| A0A3B6MTE3 | Peptidylprolyl isomerase | 1.28 | 1.64 | 1.28 × 10−1 |
| W5D1D3 | 30S ribosomal protein S20, chloroplastic | −1.27 | 1.36 | 2.49 × 10−3 * |
| Energy | ||||
| A0A3B6QKY1 | Aconitate hydratase | 1.26 | 2.05 | 1.83 × 10−2 * |
| P69415 | Photosystem I iron-sulfur center | −1.46 | −1.65 | 4.52 × 10−1 |
| Transcription | ||||
| A0A3B6LSN3 | MBD domain-containing protein | 1.36 | 1.52 | 2.86 × 10−1 |
| Unclear classification | ||||
| A0A3B6MQA1 | RRM domain-containing protein | 1.20 | −1.24 | 9.37 × 10−4 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moloi, S.J.; Alqarni, A.O.; Brown, A.P.; Goche, T.; Shargie, N.G.; Moloi, M.J.; Gokul, A.; Chivasa, S.; Ngara, R. Comparative Physiological, Biochemical, and Leaf Proteome Responses of Contrasting Wheat Varieties to Drought Stress. Plants 2024, 13, 2797. https://doi.org/10.3390/plants13192797
Moloi SJ, Alqarni AO, Brown AP, Goche T, Shargie NG, Moloi MJ, Gokul A, Chivasa S, Ngara R. Comparative Physiological, Biochemical, and Leaf Proteome Responses of Contrasting Wheat Varieties to Drought Stress. Plants. 2024; 13(19):2797. https://doi.org/10.3390/plants13192797
Chicago/Turabian StyleMoloi, Sellwane J., Ali O. Alqarni, Adrian P. Brown, Tatenda Goche, Nemera G. Shargie, Makoena J. Moloi, Arun Gokul, Stephen Chivasa, and Rudo Ngara. 2024. "Comparative Physiological, Biochemical, and Leaf Proteome Responses of Contrasting Wheat Varieties to Drought Stress" Plants 13, no. 19: 2797. https://doi.org/10.3390/plants13192797
APA StyleMoloi, S. J., Alqarni, A. O., Brown, A. P., Goche, T., Shargie, N. G., Moloi, M. J., Gokul, A., Chivasa, S., & Ngara, R. (2024). Comparative Physiological, Biochemical, and Leaf Proteome Responses of Contrasting Wheat Varieties to Drought Stress. Plants, 13(19), 2797. https://doi.org/10.3390/plants13192797








