Screening Germplasms and Detecting Quantitative Trait Loci for High Sucrose Content in Soybean
Abstract
1. Introduction
2. Results
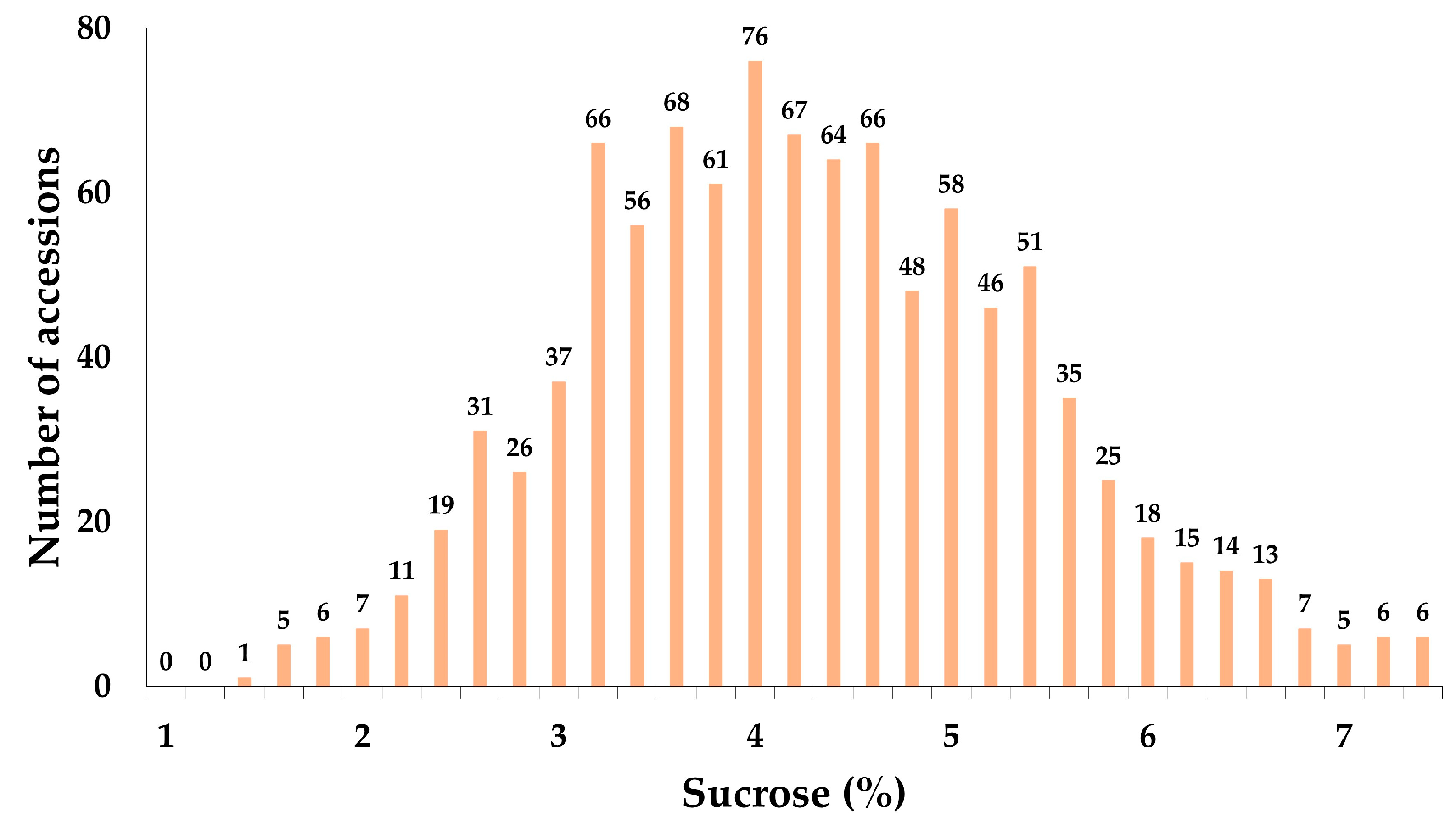
2.1. Analysis of the Sucrose Content of 1014 Soybean Accessions
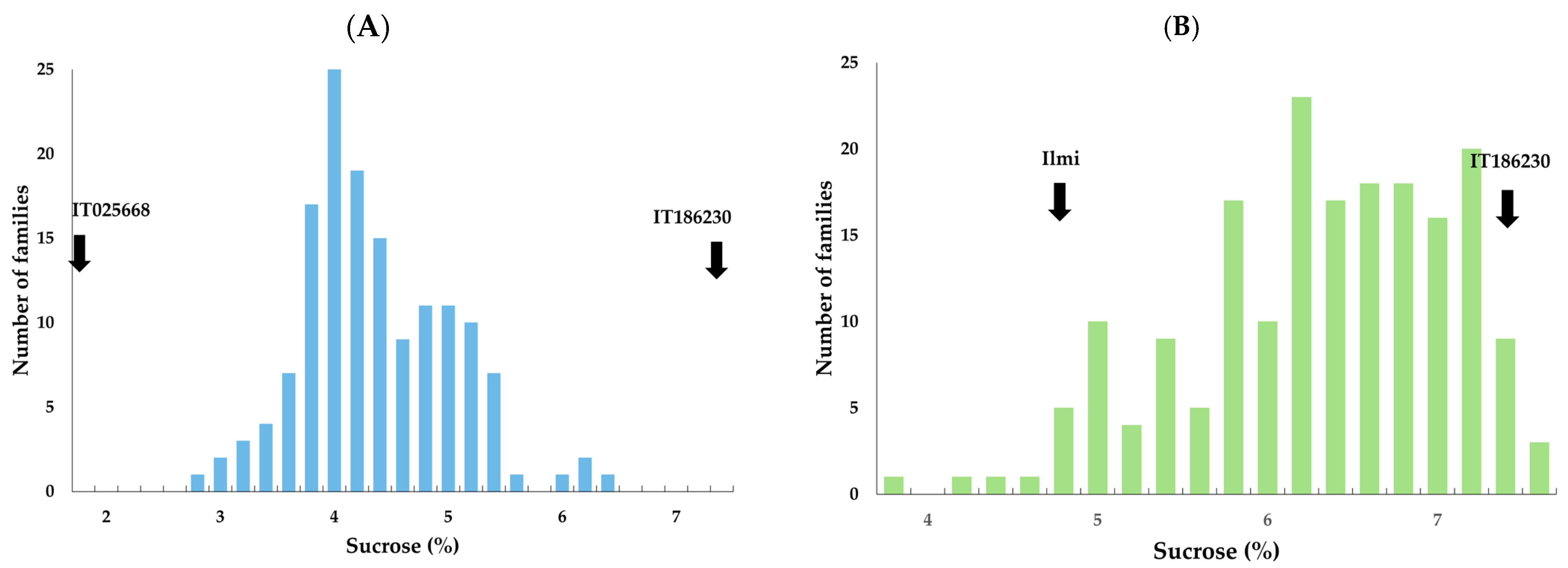
2.2. Sucrose Content Analysis of Two F2:3 Populations
2.3. Genotyping and Linkage Map Construction
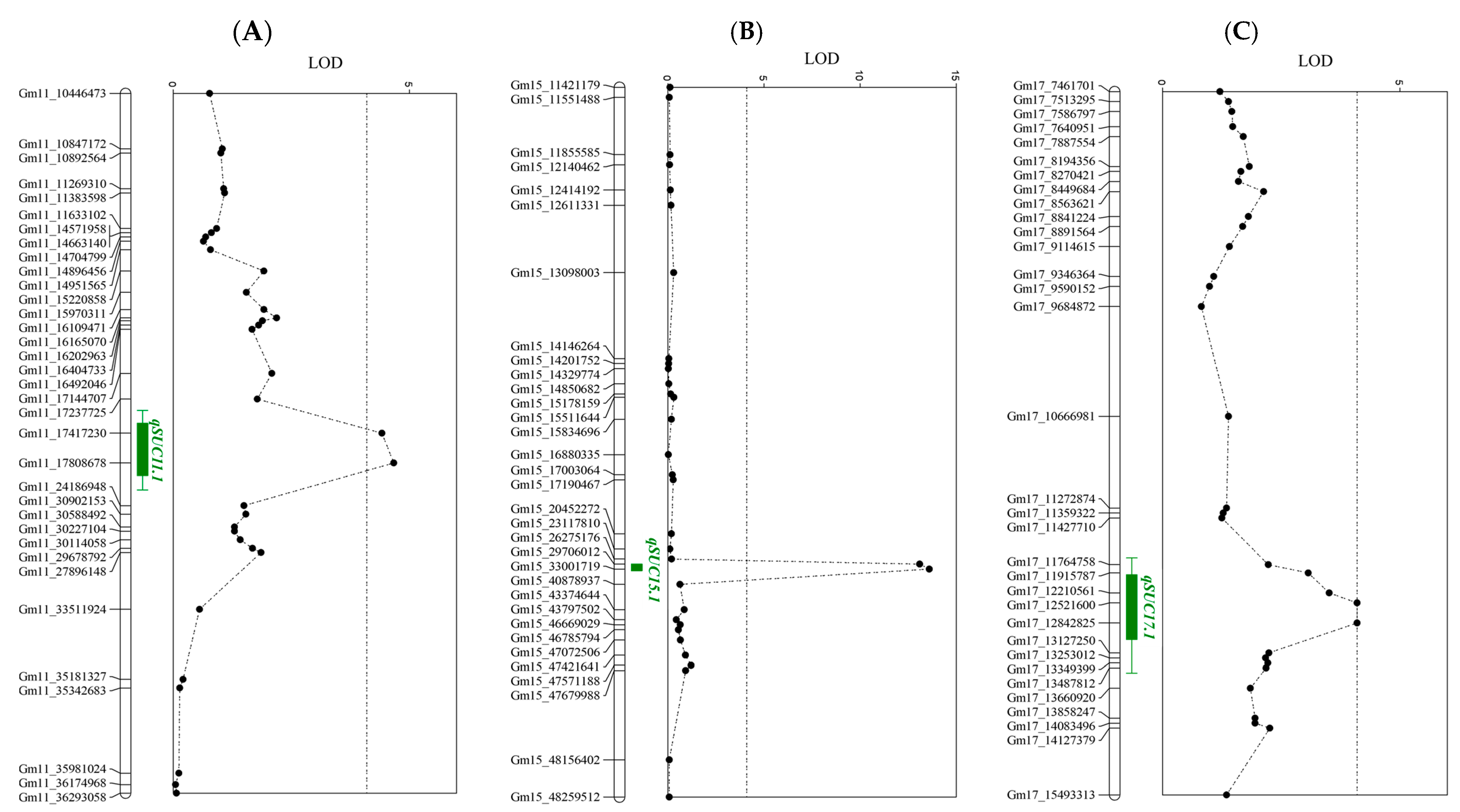
2.4. Quantitative Trait Locus (QTL) Analysis
2.5. Identification of Candidate Genes for Sucrose Content
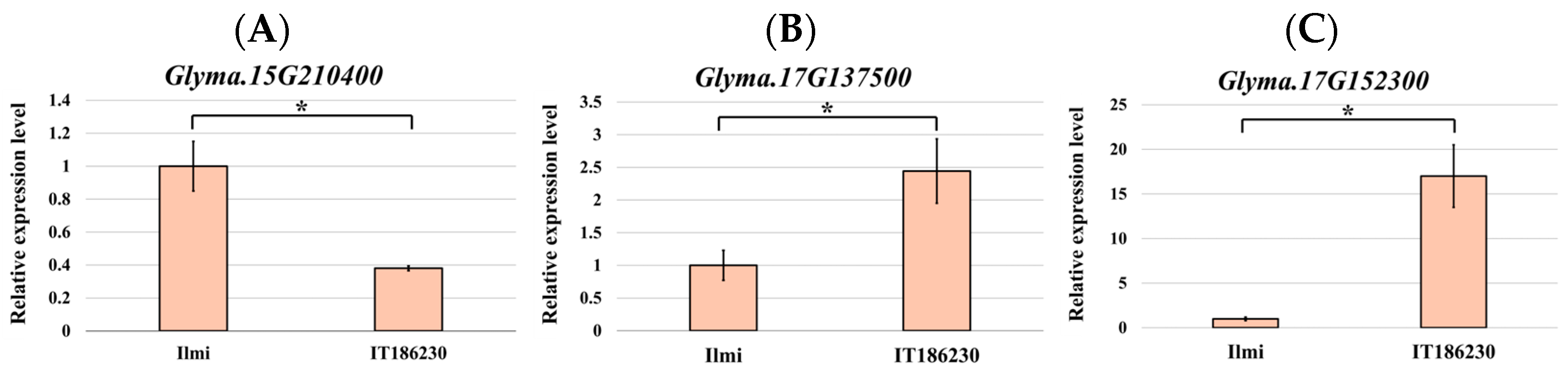
2.6. mRNA Expression Analysis of Sucrose-Related Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. GOD/INV Analysis of the Sucrose Content
4.3. Population Development
4.4. DNA Extraction and SNP Genotyping
4.5. Genetic Mapping and QTL Analysis
4.6. RT-qPCR Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ritchie, H.; Roser, M. Forests and Deforestation. 2021. Available online: https://ourworldindata.org/forests-and-deforestation (accessed on 6 October 2024).
- Cai, J.-S.; Feng, J.-Y.; Ni, Z.-J.; Ma, R.-H.; Thakur, K.; Wang, S.; Hu, F.; Zhang, J.-G.; Wei, Z.-J. An update on the nutritional, functional, sensory characteristics of soy products, and applications of new processing strategies. Trends Food Sci. Technol. 2021, 112, 676–689. [Google Scholar] [CrossRef]
- Voora, V.; Larrea, C.; Bermudez, S. Global Market Report: Soybeans. 2020. Available online: https://www.iisd.org/publications/report/global-market-report-soybeans (accessed on 6 October 2024).
- Liu, K.; Liu, K. Chemistry and nutritional value of soybean components. In Soybeans: Chemistry, Technology, and Utilization; Springer: Boston, MA, USA, 1997; pp. 25–113. [Google Scholar]
- Wang, Y.; Chen, P.; Zhang, B. Quantitative trait loci analysis of soluble sugar contents in soybean. Plant Breed. 2014, 133, 493–498. [Google Scholar] [CrossRef]
- Wilson, R.F. Seed composition. Soybeans Improv. Prod. Uses 2004, 16, 621–677. [Google Scholar]
- Huhn, M.R. Inheritance of Soluble Oligosaccharides in Soybean Seeds. Ph.D. Thesis, Virginia Tech, Blacksburg, VA, USA, 2003. [Google Scholar]
- Taira, H.; Tanaka, H.; Saito, M.; Saito, M. Effect of cultivar, seed size, and crop year on total and free sugar contents of domestic soybeans. Nippon Shokuhin Kogyo Gakkaishi 1990, 37, 203–213. [Google Scholar] [CrossRef]
- Xu, W.; Liu, H.; Li, S.; Zhang, W.; Wang, Q.; Zhang, H.; Liu, X.; Cui, X.; Chen, X.; Tang, W. GWAS and identification of candidate genes associated with seed soluble sugar content in vegetable soybean. Agronomy 2022, 12, 1470. [Google Scholar] [CrossRef]
- Valentine, M.F.; De Tar, J.R.; Mookkan, M.; Firman, J.D.; Zhang, Z.J. Silencing of soybean raffinose synthase gene reduced raffinose family oligosaccharides and increased true metabolizable energy of poultry feed. Front. Plant Sci. 2017, 8, 692. [Google Scholar] [CrossRef]
- Poysa, V.; Woodrow, L. Stability of soybean seed composition and its effect on soymilk and tofu yield and quality. Food Res. Int. 2002, 35, 337–345. [Google Scholar] [CrossRef]
- Taira, H. Quality of soybeans for processed foods in Japan. Jpn. Agric. Res. Q. 1990, 24, 224–230. [Google Scholar]
- Pradhananga, M. Effect of processing and soybean cultivar on natto quality using response surface methodology. Food Sci. Nutr. 2019, 7, 173–182. [Google Scholar] [CrossRef]
- Rada, V.; Bartoňová, J.; Vlková, E. Specific growth rate of bifidobacteria cultured on different sugars. Folia Microbiol. 2002, 47, 477–480. [Google Scholar] [CrossRef]
- Koch, K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 2004, 7, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.I.; Ribeiro, L.F.; Rezende, S.T.; Barros, E.G.; Moreira, M.A. Development of a method to quantify sucrose in soybean grains. Food Chem. 2012, 130, 1134–1136. [Google Scholar] [CrossRef][Green Version]
- Saldivar, X.; Wang, Y.-J.; Chen, P.; Hou, A. Changes in chemical composition during soybean seed development. Food Chem. 2011, 124, 1369–1375. [Google Scholar] [CrossRef]
- Alsajri, F.A.; Wijewardana, C.; Irby, J.T.; Bellaloui, N.; Krutz, L.J.; Golden, B.; Gao, W.; Reddy, K.R. Developing functional relationships between temperature and soybean yield and seed quality. Agron. J. 2020, 112, 194–204. [Google Scholar] [CrossRef]
- Xu, G.; Singh, S.; Barnaby, J.; Buyer, J.; Reddy, V.; Sicher, R. Effects of growth temperature and carbon dioxide enrichment on soybean seed components at different stages of development. Plant Physiol. Biochem. 2016, 108, 313–322. [Google Scholar] [CrossRef]
- Vu, J.C.; Gesch, R.W.; Pennanen, A.H.; Hartwell, L.A., Jr.; Boote, K.J.; Bowes, G. Soybean photosynthesis, Rubisco, and carbohydrate enzymes function at supraoptimal temperatures in elevated CO2. J. Plant Physiol. 2001, 158, 295–307. [Google Scholar] [CrossRef]
- Wang, M.; Ji, Q.; Lai, B.; Liu, Y.; Mei, K. Structure-function and engineering of plant UDP-glycosyltransferase. Comput. Struct. Biotechnol. J. 2023, 21, 5358–5371. [Google Scholar] [CrossRef]
- Guo, C.; Oosterhuis, D.M. Pinitol occurrence in soybean plants as affected by temperature and plant growth regulators. J. Exp. Bot. 1995, 46, 249–253. [Google Scholar] [CrossRef]
- Maughan, P.; Maroof, M.S.; Buss, G. Identification of quantitative trait loci controlling sucrose content in soybean (Glycine max). Mol. Breed. 2000, 6, 105–111. [Google Scholar] [CrossRef]
- Wolf, R.; Cavins, J.; Kleiman, R.; Black, L. Effect of temperature on soybean seed constituents: Oil, protein, moisture, fatty acids, amino acids and sugars. J. Am. Oil Chem. Soc. 1982, 59, 230–232. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Z.; Zhou, Q.; Wang, X.; Song, S.; Dong, S. Physiological response of soybean plants to water deficit. Front. Plant Sci. 2022, 12, 809692. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Jensen, C.R.; Andersen, M.N. Drought stress effect on carbohydrate concentration in soybean leaves and pods during early reproductive development: Its implication in altering pod set. Field Crops Res. 2004, 86, 1–13. [Google Scholar] [CrossRef]
- Pinheiro, C.; Rodrigues, A.P.; de Carvalho, I.S.; Chaves, M.M.; Ricardo, C.P. Sugar metabolism in developing lupin seeds is affected by a short-term water deficit. J. Exp. Bot. 2005, 56, 2705–2712. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Yang, R.; Yang, X.; Sun, S.; Mentreddy, S.R.; Jiang, B.; Wu, T.; Tian, S.; Sapey, E.; Wu, C. Spatial differences in soybean bioactive components across China and their influence by weather factors. Crop J. 2018, 6, 659–668. [Google Scholar] [CrossRef]
- Blackman, S.A.; Obendorf, R.L.; Leopold, A.C. Maturation Proteins and Sugars in Desiccation Tolerance of Developing Soybean Seeds 1. Plant Physiol. 1992, 100, 225–230. [Google Scholar] [CrossRef]
- Kuo, T.M.; VanMiddlesworth, J.F.; Wolf, W.J. Content of raffinose oligosaccharides and sucrose in various plant seeds. J. Agric. Food Chem. 1988, 36, 32–36. [Google Scholar] [CrossRef]
- Lowell, C.A.; Kuo, T.M. Oligosaccharide metabolism and accumulation in developing soybean seeds. Crop Sci. 1989, 29, 459–465. [Google Scholar] [CrossRef]
- Kim, G.; Park, A.; Kim, W.J.; Moon, C.Y.; Kang, B.H.; Kim, S.-H.; Choi, Y.-M.; Ha, B.-K. Development of a Simple Enzymatic Method for Screening Sucrose Content in Legume Seeds. Plant Breed. Biotechnol. 2021, 9, 250–258. [Google Scholar] [CrossRef]
- Kwok, P.-Y. Single Nucleotide Polymorphisms: Methods and Protocols; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2008; Volume 212. [Google Scholar]
- Yoon, M.; Song, Q.; Choi, I.; Specht, J.E.; Hyten, D.; Cregan, P. BARCSoySNP23: A panel of 23 selected SNPs for soybean cultivar identification. Theor. Appl. Genet. 2007, 114, 885–899. [Google Scholar] [CrossRef]
- Kim, H.-K.; Kang, S.-T.; Cho, J.-H.; Choung, M.-G.; Suh, D.-Y. Quantitative trait loci associated with oligosaccharide and sucrose contents in soybean (Glycine max L.). J. Plant Biol. 2005, 48, 106–112. [Google Scholar] [CrossRef]
- Kim, H.K.; Kang, S.T.; Oh, K.W. Mapping of putative quantitative trait loci controlling the total oligosaccharide and sucrose content of Glycine max seeds. J. Plant Res. 2006, 119, 533–538. [Google Scholar] [CrossRef]
- Knizia, D.; Bellaloui, N.; Yuan, J.; Lakhssasi, N.; Anil, E.; Vuong, T.; Embaby, M.; Nguyen, H.T.; Mengistu, A.; Meksem, K. Quantitative Trait Loci and Candidate Genes That Control Seed Sugars Contents in the Soybean ‘Forrest’by ‘Williams 82’Recombinant Inbred Line Population. Plants 2023, 12, 3498. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, H.; Yu, Q.; Gu, H.; Li, Y.; Tu, B.; Zhang, H.; Zhang, Q.; Liu, X. Identification of quantitative trait loci and candidate genes for seed sucrose and soluble sugar concentrations in soybean. Crop Sci. 2023, 63, 2976–2992. [Google Scholar] [CrossRef]
- Du, Y.; Zhao, Q.; Chen, L.; Yao, X.; Zhang, H.; Wu, J.; Xie, F. Effect of drought stress during soybean R2–R6 growth stages on sucrose metabolism in leaf and seed. Int. J. Mol. Sci. 2020, 21, 618. [Google Scholar] [CrossRef] [PubMed]
- Jamison, D.R.; Chen, P.; Hettiarachchy, N.S.; Miller, D.M.; Shakiba, E. Identification of Quantitative Trait Loci (QTL) for Sucrose and Protein Content in Soybean Seed. Plants 2024, 13, 650. [Google Scholar] [CrossRef]
- Sonnewald, U. SWEETS—The missing sugar efflux carriers. Front. Plant Sci. 2011, 2, 10785. [Google Scholar] [CrossRef]
- Chen, L.Q. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014, 201, 1150–1155. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Feng, W.; Ishiguro, Y.; Hotta, K.; Watanabe, H.; Suga, H.; Kageyama, K. Simple detection of Pythium irregulare using loop-mediated isothermal amplification assay. FEMS Microbiol. Lett. 2015, 362, fnv174. [Google Scholar] [CrossRef]
- Chen, H.Y.; Huh, J.H.; Yu, Y.C.; Ho, L.H.; Chen, L.Q.; Tholl, D.; Frommer, W.B.; Guo, W.J. The Arabidopsis vacuolar sugar transporter SWEET 2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. 2015, 83, 1046–1058. [Google Scholar] [CrossRef]
- Wei, X.; Liu, F.; Chen, C.; Ma, F.; Li, M. The Malus domestica sugar transporter gene family: Identifications based on genome and expression profiling related to the accumulation of fruit sugars. Front. Plant Sci. 2014, 5, 569. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, B.M.; Errafi, S.; Bucher, R.; Dobrev, P.; Geisler, M.; Bigler, L.; Zažímalová, E.; Ringli, C. 7-Rhamnosylated flavonols modulate homeostasis of the plant hormone auxin and affect plant development. J. Biol. Chem. 2016, 291, 5385–5395. [Google Scholar] [CrossRef]
- Baksi, R.; Singh, D.P.; Borse, S.P.; Rana, R.; Sharma, V.; Nivsarkar, M. In vitro and in vivo anticancer efficacy potential of Quercetin loaded polymeric nanoparticles. Biomed. Pharmacother. 2018, 106, 1513–1526. [Google Scholar] [CrossRef]
- Lin, J.S.; Huang, X.X.; Li, Q.; Cao, Y.; Bao, Y.; Meng, X.F.; Li, Y.J.; Fu, C.; Hou, B.K. UDP-glycosyltransferase 72B1 catalyzes the glucose conjugation of monolignols and is essential for the normal cell wall lignification in Arabidopsis thaliana. Plant J. 2016, 88, 26–42. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ji, K.; Liang, B.; Du, Y.; Jiang, L.; Wang, J.; Kai, W.; Zhang, Y.; Zhai, X.; Chen, P. Suppressing ABA uridine diphosphate glucosyltransferase (Sl UGT 75C1) alters fruit ripening and the stress response in tomato. Plant J. 2017, 91, 574–589. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yan, J.-P.; Li, D.-K.; Luo, Q.; Yan, Q.; Liu, Z.-B.; Ye, L.-M.; Wang, J.-M.; Li, X.-F.; Yang, Y. UDP-glucosyltransferase71c5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol. 2015, 167, 1659–1670. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Dong, G.-r.; Ma, Y.-q.; Zhao, S.-m.; Liu, X.; Li, X.-k.; Li, Y.-j.; Hou, B.-k. Rice glycosyltransferase gene UGT85E1 is involved in drought stress tolerance through enhancing abscisic acid response. Front. Plant Sci. 2021, 12, 790195. [Google Scholar] [CrossRef]
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef]
- Keegstra, K.; Raikhel, N. Plant glycosyltransferases. Curr. Opin. Plant Biol. 2001, 4, 219–224. [Google Scholar] [CrossRef]
- Yu, X.; Fu, X.; Yang, Q.; Jin, H.; Zhu, L.; Yuan, F. Genome-wide variation analysis of four vegetable soybean cultivars based on re-sequencing. Plants 2021, 11, 28. [Google Scholar] [CrossRef]
- Flügge, U.I.; Häusler, R.E.; Ludewig, F.; Fischer, K. Functional genomics of phosphate antiport systems of plastids. Physiol. Plant. 2003, 118, 475–482. [Google Scholar] [CrossRef]
- Fliege, R.; Flügge, U.-I.; Werdan, K.; Heldt, H.W. Specific transport of inorganic phosphate, 3-phosphoglycerate and triosephosphates across the inner membrane of the envelope in spinach chloroplasts. Biochim. Biophys. Acta 1978, 502, 232–247. [Google Scholar] [CrossRef] [PubMed]
- Zeeman, S.C.; Tiessen, A.; Pilling, E.; Kato, K.L.; Donald, A.M.; Smith, A.M. Starch synthesis in Arabidopsis. Granule synthesis, composition, and structure. Plant Physiol. 2002, 129, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Keim, P.; Olson, T.; Shoemaker, R. A rapid protocol for isolating soybean DNA. Soybean Genet. Newsl. 1988, 15, 150–152. [Google Scholar]
- Churchill, G.A.; Doerge, R. Empirical threshold values for quantitative trait mapping. Genetics 1994, 138, 963–971. [Google Scholar] [CrossRef]
- Falconer, D.S. Introduction to Quantitative Genetics; Pearson Education: Noida, India, 1996. [Google Scholar]

| Genotype | Origin | Sucrose Content (%) | ||
|---|---|---|---|---|
| 2020 | 2021 | |||
| High sucrose | IT186230 | KOR | 6.44 ± 0.09 | 7.77 ± 0.03 |
| IT195321 | COL | 6.34 ± 0.06 | 6.60 ± 0.07 | |
| IT263138 | KOR | 7.37 ± 0.18 | 7.61 ± 0.01 | |
| IT263276 | KOR | 6.97 ± 0.04 | 7.43 ± 0.09 | |
| IT263286 | KOR | 7.27 ± 0.25 | 7.65 ± 0.00 | |
| IT276521 | KOR | 6.60 ± 0.37 | 7.49 ± 0.06 | |
| Low sucrose | IT025668 | KOR | 1.23 ± 0.00 | 1.71 ± 0.03 |
| IT274054 | CSK † | 1.85 ± 0.11 | 2.18 ± 0.05 | |
| Control | Ilmi | KOR | - | 7.39 ± 0.03 |
| Populations | Parent | F2:3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P1 † | P2 | N | Min. | Max. | Mean. | CV | Skew. | Kurt. | h2 | |
| IT186230 × IT025668 | 7.25 | 1.73 | 147 | 2.66 | 6.22 | 4.27 | 15.33 | 0.51 | 0.32 | 0.96 |
| Ilmi × IT186230 | 4.73 | 7.25 | 188 | 3.79 | 7.59 | 6.21 | 12.39 | −0.60 | −0.14 | 0.95 |
| QTL | Chr. | Interval | Physical Interval (Mb) | LOD | Peak or Flanking Marker | IT186230 Allele Effect (%) | IT025668 Allele Effect (%) | R2 | Additive Effect |
|---|---|---|---|---|---|---|---|---|---|
| qSUC6.1 | 6 | Gm06_790146–Gm06_2393421 | 0.7–2.3 | 5.12 | Gm06_1655912 | 4.3 | 3.5 | 13.4 | 0.39 |
| QTL | Chr. | Interval | Physical Interval (Mb) | LOD | Peak or Flanking Marker | Ilmi Allele Effect (%) | IT186230 Allele Effect (%) | R2 | Additive Effect |
|---|---|---|---|---|---|---|---|---|---|
| qSUC11.1 | 11 | Gm11_17237725–Gm11_24186948 | 17.2–24.1 | 5.31 | Gm11_17808678 | 5.9 | 6.6 | 9.6 | –0.32 |
| qSUC15.1 | 15 | Gm15_29706012–Gm15_40878937 | 29.7–40.8 | 13.6 | Gm15_33001719 | 6.8 | 5.7 | 27.6 | 0.56 |
| qSUC17.1 | 17 | Gm17_12210561–Gm17_13127250 | 12.2–13.1 | 4.1 | Gm17_12842825 | 6.1 | 6.4 | 7.3 | –0.18 |
| QTL | Gene ID | Physical Position (bp) | Annotation | Protein ID |
|---|---|---|---|---|
| qSUC6.1 | ||||
| Glyma.06G011700 | Gm06:873613..878088 | Glucose-1-phosphate adenylyl transferase family protein | ||
| Glyma.06G015000 | Gm06:1127434..1132895 | Tonoplast monosaccharide transporter 2 | ||
| Glyma.06G015900 | Gm06:1187405..1190649 | Glyceraldehyde-3-phosphate dehydrogenase B subunit | ||
| Glyma.06G018000 | Gm06:1359828..1368182 | 1,4-alpha-glucan-branching enzyme/starch-branching enzyme II | ||
| Glyma.06G028800 | Gm06:2237126..2239413 | Fructose-2,6-bisphosphatase | ||
| Glyma.06G030400 | Gm06:2390673..2396324 | Glucose-1-phosphate adenylyltransferase large subunit 2 | ||
| Glyma.06G032500 | Gm06:2515199..2519958 | Glucose-6-phosphate isomerase | ||
| qSUC11.1 | ||||
| Glyma.11G148900 | Gm11:11489777..11495616 | Glycerol-3-phosphate dehydrogenase | ||
| Glyma.11G150600 | Gm11:11808846..11810660 | Sugar-1-phosphate guanyl transferase | ||
| qSUC15.1 | ||||
| Glyma.15G210100 | Gm15:32175249..32210061 | Trehalose-6-phosphate synthase | ||
| Glyma.15g210200 | Gm15:32217111..32218567 | Protein kinase superfamily protein | ||
| Glyma.15G210400 | Gm15:32262290..32264065 | RAG1-activating protein 1 | SWEET41 | |
| Glyma.15G211800 | Gm15:32835023..32838031 | RAG1-activating protein 1 | SWEET42 | |
| Glyma.15G212600 | Gm15:33402757..33405141 | Glutathione S-transferase THETA 2 | ||
| Glyma.15G221300 | Gm15:39951868..39954192 | UDP-glucosyl transferase 73B3 | ||
| qSUC17.1 | ||||
| Glyma.17G137200 | Gm17:11073919..11077576 | Nucleotide-sugar transporter family protein | ||
| Glyma.17G137500 | Gm17:11097469..11100931 | UDP-glycosyltransferase superfamily protein | ||
| Glyma.17G138700 | Gm17:11248118..11252772 | Trehalose-phosphate synthase | ||
| Glyma.17G152300 | Gm17:12696012..12697117 | Triose-phosphate transporter family | ||
| Glyma.17G152400 | Gm17:12704557..12706988 | UDP-galactose transporter |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, S.-H.; Shin, S.-Y.; Kang, B.H.; Chowdhury, S.; Lee, W.-H.; Kim, W.J.; Lee, J.-D.; Lee, S.; Choi, Y.-M.; Ha, B.-K. Screening Germplasms and Detecting Quantitative Trait Loci for High Sucrose Content in Soybean. Plants 2024, 13, 2815. https://doi.org/10.3390/plants13192815
Kang S-H, Shin S-Y, Kang BH, Chowdhury S, Lee W-H, Kim WJ, Lee J-D, Lee S, Choi Y-M, Ha B-K. Screening Germplasms and Detecting Quantitative Trait Loci for High Sucrose Content in Soybean. Plants. 2024; 13(19):2815. https://doi.org/10.3390/plants13192815
Chicago/Turabian StyleKang, Se-Hee, Seo-Young Shin, Byeong Hee Kang, Sreeparna Chowdhury, Won-Ho Lee, Woon Ji Kim, Jeong-Dong Lee, Sungwoo Lee, Yu-Mi Choi, and Bo-Keun Ha. 2024. "Screening Germplasms and Detecting Quantitative Trait Loci for High Sucrose Content in Soybean" Plants 13, no. 19: 2815. https://doi.org/10.3390/plants13192815
APA StyleKang, S.-H., Shin, S.-Y., Kang, B. H., Chowdhury, S., Lee, W.-H., Kim, W. J., Lee, J.-D., Lee, S., Choi, Y.-M., & Ha, B.-K. (2024). Screening Germplasms and Detecting Quantitative Trait Loci for High Sucrose Content in Soybean. Plants, 13(19), 2815. https://doi.org/10.3390/plants13192815








