Molecular Phylogenetics and Historical Biogeography of Subtribe Ecliptinae (Asteraceae, Heliantheae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Taxon Sampling and Molecular Protocols
2.2. Phylogenetic Analyses
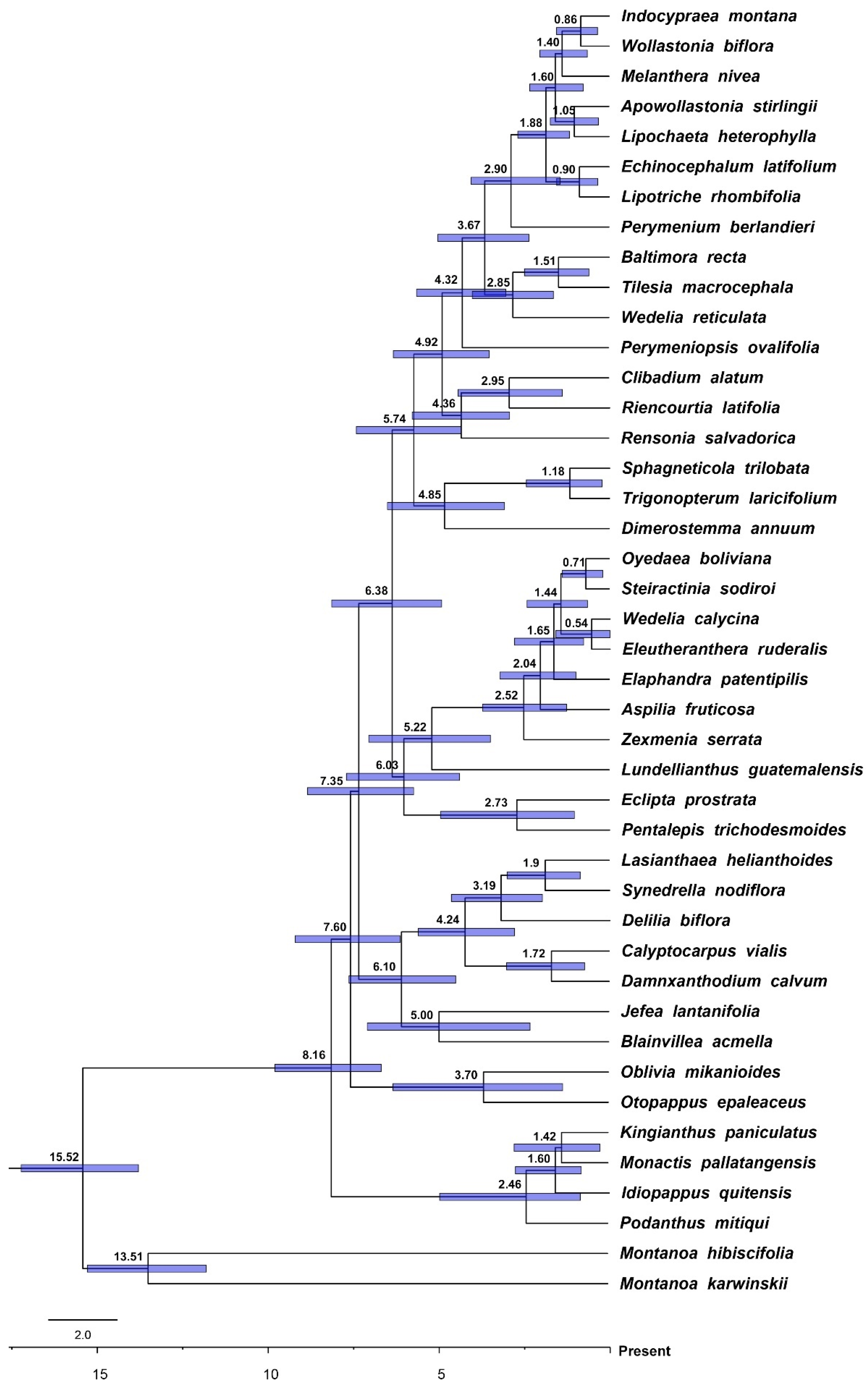
2.3. Calibration
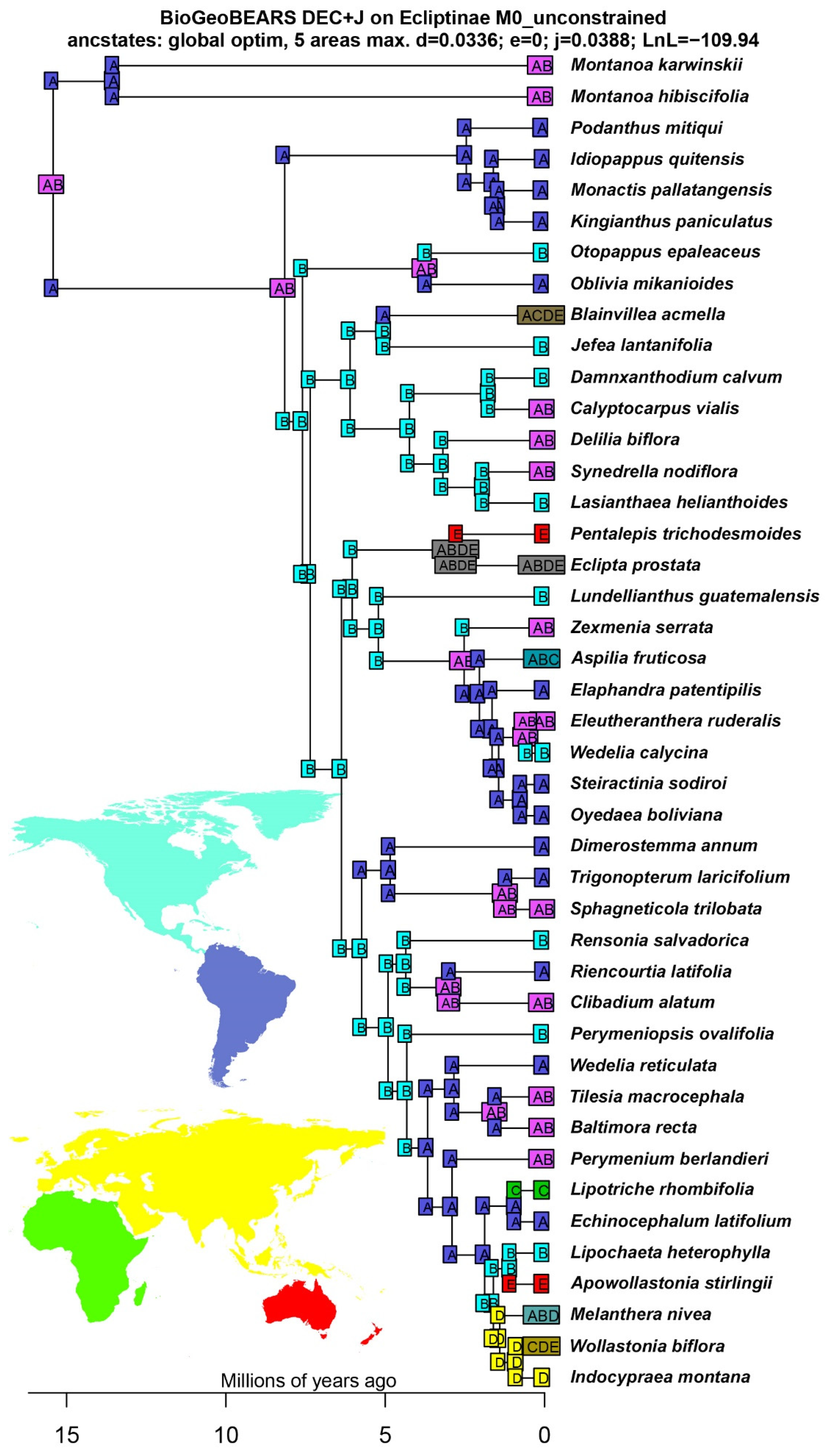
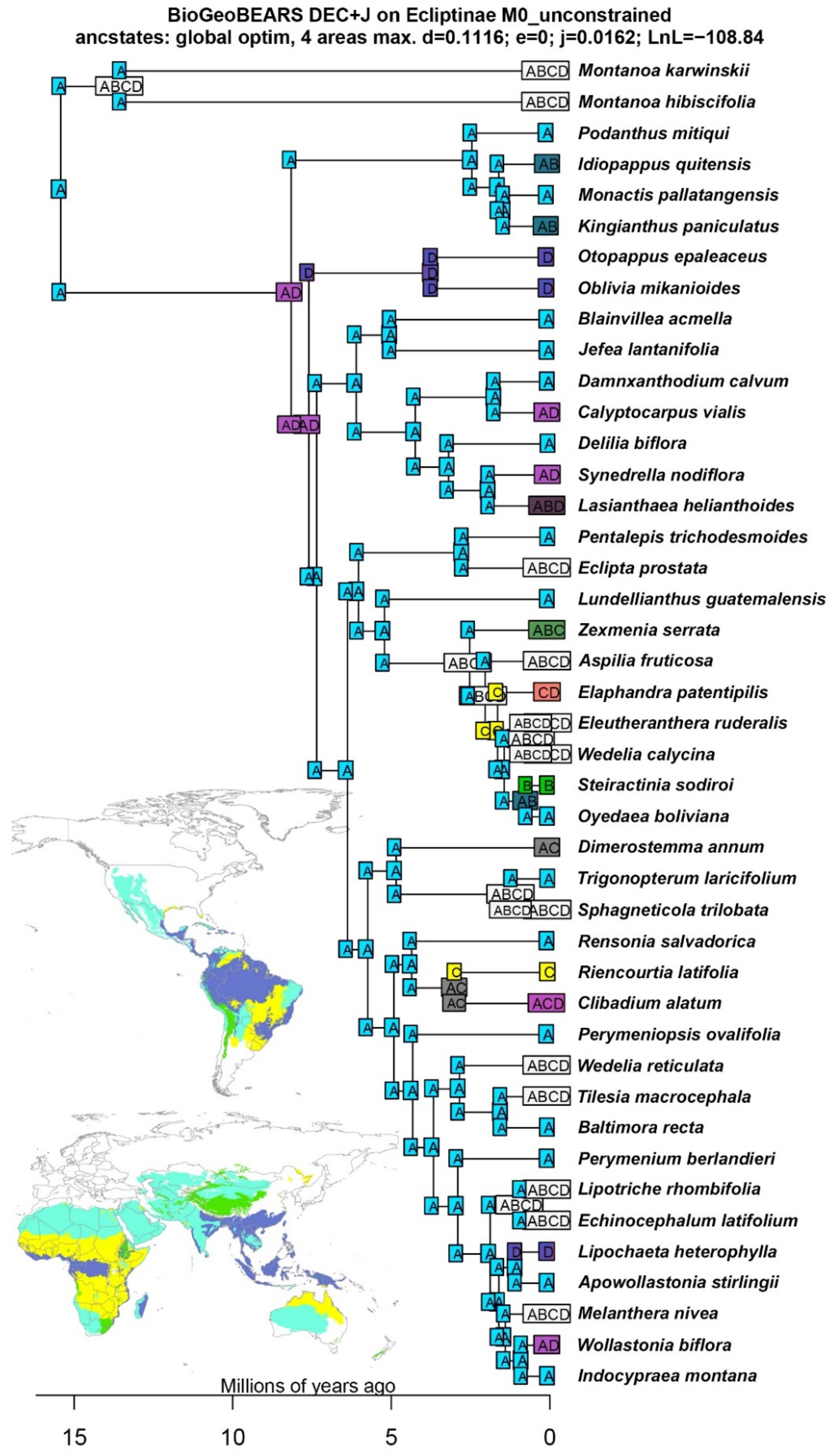
2.4. Ancestral Range Reconstruction
3. Results
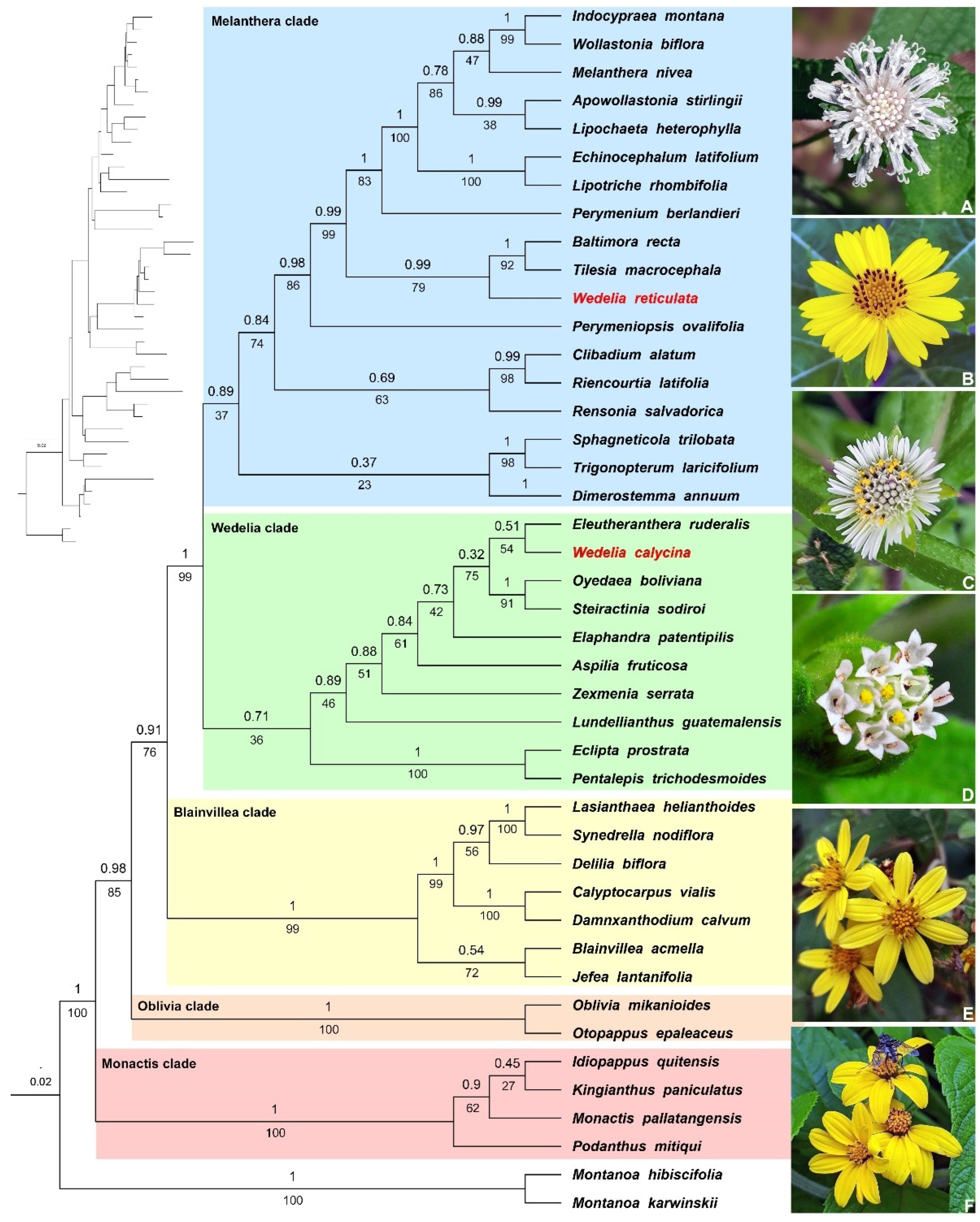
3.1. Phylogenetics of Ecliptinae
3.2. Divergence Times and Ancestral Ranges of Ecliptinae
4. Discussion
4.1. Phylogenetics and Systematics of Ecliptinae
4.2. Historical Biogeography of Ecliptinae
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baldwin, B.G. Heliantheae alliance. In Systematics, Evolution, and Biogeography of Compositae; Funk, V.A., Susanna, A., Stuessy, T.F., Bayer, R.J., Eds.; International Association for Plant Taxonomy: Bratislava, Slovakia, 2009; pp. 689–711. [Google Scholar]
- Panero, J.L.; Funk, V.A. Toward a phylogenetic subfamilial classification for the Compositae (Asteraceae). Proc. Biol. Soc. 2002, 115, 909–922. [Google Scholar]
- Panero, J.L. Compositae: Tribe Heliantheae. In Families and Genera of Vascular Plants, vol. VIII, Flowering Plants, Eudicots, Asterales; Kadereit, J.W., Jeffrey, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 440–477. [Google Scholar]
- Funk, V.A.; Susanna, A.; Stuessy, T.F.; Bayer, R.J. Systematics, Evolution, and Biogeography of Compositae; International Association for Plant Taxonomy: Bratislava, Slovakia, 2009; pp. 1–1000. [Google Scholar]
- Susanna, A.; Baldwin, B.G.; Bayer, R.J.; Bonifacino, J.M.; Garcia-Jacas, N.; Keeley, S.C.; Mandel, J.R.; Ortiz, S.; Robinson, H.; Stuessy, T.F. The classification of the Compositae: A tribute to Vicki Ann Funk (1947–2019). Taxon 2020, 69, 807–814. [Google Scholar] [CrossRef]
- Panero, J.L.; Jansen, R.K.; Clevinger, J.A. Phylogenetic relationships of subtribe Ecliptinae (Asteraceae: Heliantheae) based on chloroplast DNA restriction site data. Am. J. Bot. 1999, 86, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Schilling, E.E. Phylogeny of Helianthus and related genera. OCL 2001, 8, 22–25. [Google Scholar] [CrossRef]
- Urbatsch, L.E.; Baldwin, B.G.; Donoghue, M.J. Phylogeny of the coneflowers and relatives (Heliantheae: Asteraceae) based on nuclear rDNA Internal Transcribed Spacer (ITS) sequences and chloroplast DNA restriction site data. Syst. Bot. 2000, 25, 539–565. [Google Scholar] [CrossRef]
- Baldwin, B.G.; Wessa, B.L.; Panero, J.L. Nuclear rDNA evidence for major lineages of Helenioid Heliantheae (Compositae). Syst. Bot. 2002, 27, 161–198. [Google Scholar]
- Moraes, M.D.; Panero, J.L.; Semir, J. Relações filogenéticas na subtribo Ecliptinae (Asteraceae: Heliantheae). Rev. Bras. Biocienc. 2007, 5, 705–707. [Google Scholar]
- POWO. Plants of the World Online. Available online: http://powo.science.kew.org/ (accessed on 7 August 2024).
- Moraes, M.D.; Panero, J.L. A phylogeny of Dimerostemma (Asteraceae, Heliantheae, Ecliptinae) based on the ITS and ETS. Phytotaxa 2016, 245, 289–296. [Google Scholar] [CrossRef]
- Ren, C. Phylogenetic position and independent generic status of Indocypraea (Asteraceae-Heliantheae-Ecliptinae): Evidence from chloroplast DNA sequences. Phytotaxa 2016, 277, 146–158. [Google Scholar] [CrossRef]
- Edwards, R.D.; Cantley, J.T.; Chau, M.M.; Keeley, S.C.; Funk, V.A. Biogeography and relationships within the Melanthera alliance: A pantropical lineage (Compositae: Heliantheae: Ecliptinae). Taxon 2018, 67, 552–564. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.S. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Paithankar, K.R.; Prasad, K.S.N. Precipitation of DNA by polyethylene glycol and ethanol. Nucleic Acids Res. 1991, 19, 1346. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organisation and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. Mr Bayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J. Tracer v1.6. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 7 August 2024).
- Stamakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Sereno, P.C. Logical basis for morphological characters in phylogenetics. Cladistics 2007, 23, 565–587. [Google Scholar] [CrossRef]
- De Pinna, M.C.C. Concepts and tests of homology in the cladistic paradigm. Cladistics 1991, 7, 367–394. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.61. Available online: http://www.mesquiteproject.org (accessed on 7 August 2024).
- Nixon, K.C. Winclada (Beta) ver. 0.9. Published by the Author, Ithaca, NY. Available online: http://www.cladistics.com (accessed on 7 August 2024).
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Mandel, J.R.; Dikow, R.B.; Siniscalchi, C.M.; Thapa, R.; Watson, L.E.; Funk, V.A. A fully resolved backbone phylogeny reveals numerous dispersals and explosive diversifications throughout the history of Asteraceae. Proc. Natl. Acad. Sci. USA 2019, 116, 14083–14088. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree Version 1.3.1: Tree Figure Drawing Tool. Computer Program and Documentation Distributed by the Author. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 7 August 2024).
- GBIF. Global Biodiversity Information Facility. Available online: http://gbif.org (accessed on 7 August 2024).
- Matzke, N.J. BioGeoBEARS: Biogeography with Bayesian (and Likelihood) Evolutionary Analysis in R Scripts; CRAN: The Comprehensive R Archive Network: Berkeley, CA, USA, 2013; Available online: http://CRAN.R-project.org/package=BioGeoBEARS (accessed on 7 August 2024).
- Harold, R.E. A revision of the tribal and subtribal limits of the Heliantheae (Asteraceae). Smithson. Contrib. Bot. 1981, 51, 1–102. [Google Scholar]
- Alves, M.; Roque, N. Flora of Bahia: Asteraceae—Tribe Heliantheae. Sitientibus Ser. Cienc. Biol. 2016, 16, 1–63. [Google Scholar] [CrossRef]
- Magenta, M.A.G.; Loeuille, B.; Pirani, J.B. Phytogeography of Aldama (Asteraceae, Heliantheae) in South America. Rodriguesia 2017, 68, 463–480. [Google Scholar] [CrossRef]
- Alves, M.B.B. Filogenia Molecular, Evolução e Redelimitação do Complexo Aspilia-Wedelia (Asteraceae). Ph.D. Thesis, Universidade Estadual de Feira de Santana, Bahia, Brazil, 2019; 98 p. [Google Scholar]
- Moraes, M.D.; Semir, J. A revision of Brazilian Dimerostemma (Asteraceae, Heliantheae, Ecliptinae), with a new species and taxonomic adjustments. Brittonia 2009, 61, 341–365. [Google Scholar] [CrossRef]
- Mandel, J.R.; Dikow, R.B.; Funk, V.A. Using phylogenomics to resolve mega-families: An example from Compositae. J. Syst. Evol. 2015, 53, 391–402. [Google Scholar] [CrossRef]
- Moreira, G.L.; Panero, J.L.; Inglis, P.W.; Zappi, D.C.; Cavalcanti, T.B. A time-calibrated phylogeny of Verbesina (Heliantheae–Asteraceae) based on nuclear ribosomal ITS and ETS sequences. Edinb. J. Bot. 2023, 80, 1–22. [Google Scholar] [CrossRef]
- Soejima, A.; Tanabe, A.S.; Takayama, I.; Kawahara, T.; Watanabe, K.; Nakazawa, M.; Mishima, M.; Yahara, T. Phylogeny and biogeography of the genus Stevia (Asteraceae: Eupatorieae): An example of diversification in the Asteraceae in the new world. J. Plant Res. 2017, 130, 953–972. [Google Scholar] [CrossRef]
- Willis, C.G.; Franzone, B.F.; Xi, Z.; Davis, C.C. The establishment of Central American migratory corridors and the biogeographic origins of seasonally dry tropical forests in Mexico. Front. Genet. 2014, 5, 433. [Google Scholar] [CrossRef]
- Jaramillo, C.A. Evolution of the Isthmus of Panama: Biological, Palaeoceanographic and Palaeoclimatological Implications. In Mountains, Climate and Biodiversity; Hoorn, C., Perrigo, A., Antonelli, A., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2018; pp. 323–338. [Google Scholar]
- Simon, M.F.; Grether, R.; Queiroz, L.P.; Skema, C.; Pennington, R.T.; Hughes, C.E. Recent assembly of the Cerrado, a neotropical plant diversity hotspot, by in situ evolution of adaptations to fire. Proc. Natl. Acad. Sci. USA 2009, 106, 20359–20364. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, R.F.; Francener, A.; Mamede, M.C.H.; van den Berg, C. Molecular phylogeny and historical biogeography of Byrsonima (Malpighiaceae) corroborates the mid-Miocene origins of Neotropical Savannas. Diversity 2024, 16, 488. [Google Scholar] [CrossRef]
- Davis, C.C.; Bell, C.D.; Mathews, S.; Donoghue, M.J. Laurasian migration explains Gondwanan disjunctions: Evidence from Malpighiaceae. Proc. Natl. Acad. Sci. USA 2002, 99, 6833–6837. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, R.F.; van den Berg, C. Biogeography of Stigmaphyllon (Malpighiaceae) and a meta-analysis of vascular plant lineages diversified in the Brazilian Atlantic Rainforests point to the Late Eocene origins of this megadiverse biome. Plants 2020, 9, 1569. [Google Scholar] [CrossRef]
- Lohman, D.J.; de Bruyn, M.; Page, T.; von Rintelen, K.; Hall, R.; Ng, P.K.L.; Shih, H.T.; Carvalho, G.R.; von Rintelen, T. Biogeography of the Indo-Australian Archipelago. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 205–226. [Google Scholar] [CrossRef]
- Antonelli, A.; Sanmartín, I. Why are there so many plant species in the Neotropics? Taxon 2011, 60, 403–414. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Almeida, R.F.; Alves, M.; van den Berg, C.; Pellegrini, M.O.O.; Gostel, M.R.; Roque, N. Molecular Phylogenetics and Historical Biogeography of Subtribe Ecliptinae (Asteraceae, Heliantheae). Plants 2024, 13, 2817. https://doi.org/10.3390/plants13192817
de Almeida RF, Alves M, van den Berg C, Pellegrini MOO, Gostel MR, Roque N. Molecular Phylogenetics and Historical Biogeography of Subtribe Ecliptinae (Asteraceae, Heliantheae). Plants. 2024; 13(19):2817. https://doi.org/10.3390/plants13192817
Chicago/Turabian Stylede Almeida, Rafael Felipe, Maria Alves, Cássio van den Berg, Marco O. O. Pellegrini, Morgan R. Gostel, and Nádia Roque. 2024. "Molecular Phylogenetics and Historical Biogeography of Subtribe Ecliptinae (Asteraceae, Heliantheae)" Plants 13, no. 19: 2817. https://doi.org/10.3390/plants13192817







