Applications of Virus-Induced Gene Silencing in Cotton
Abstract
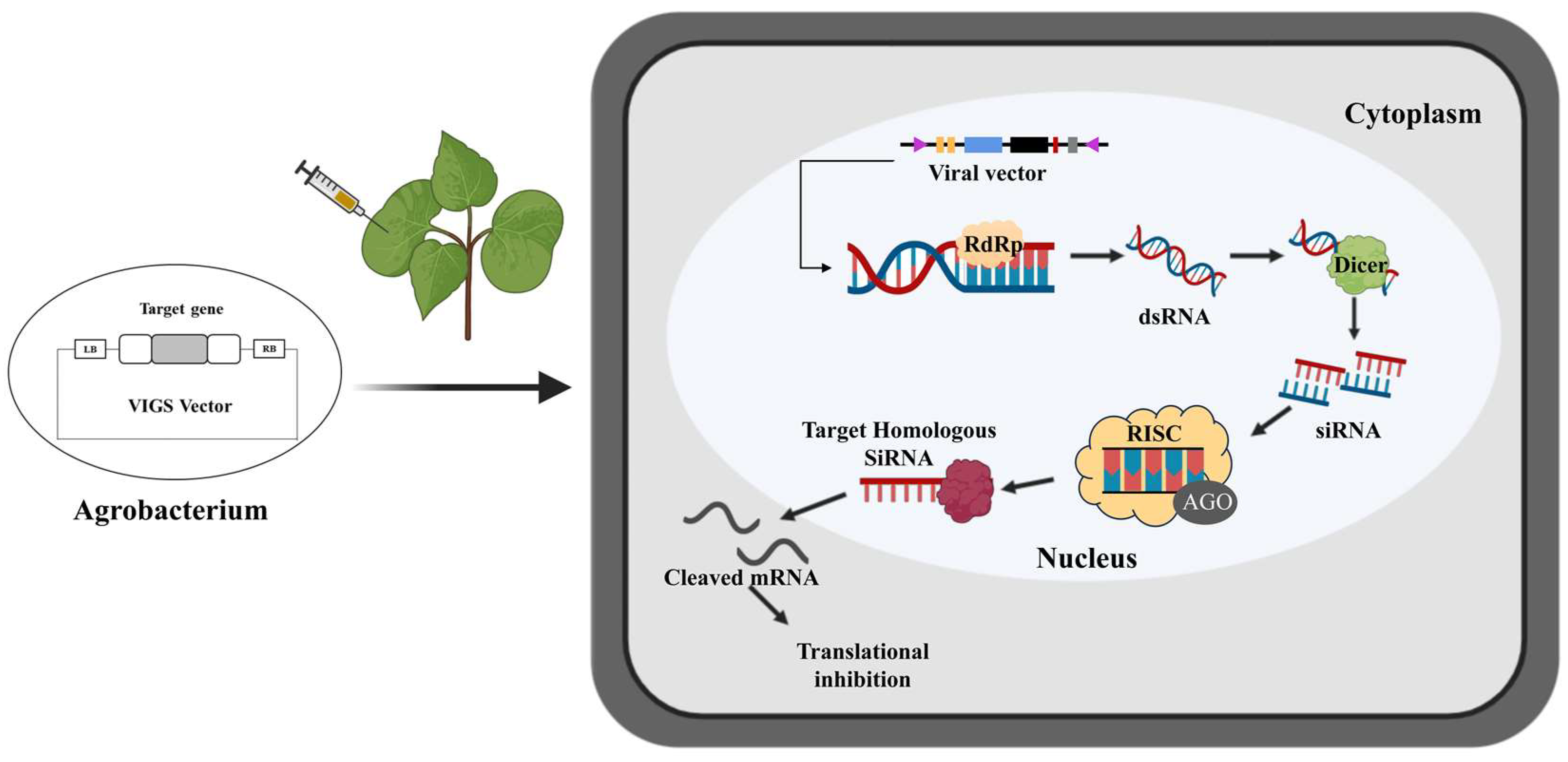
1. Introduction
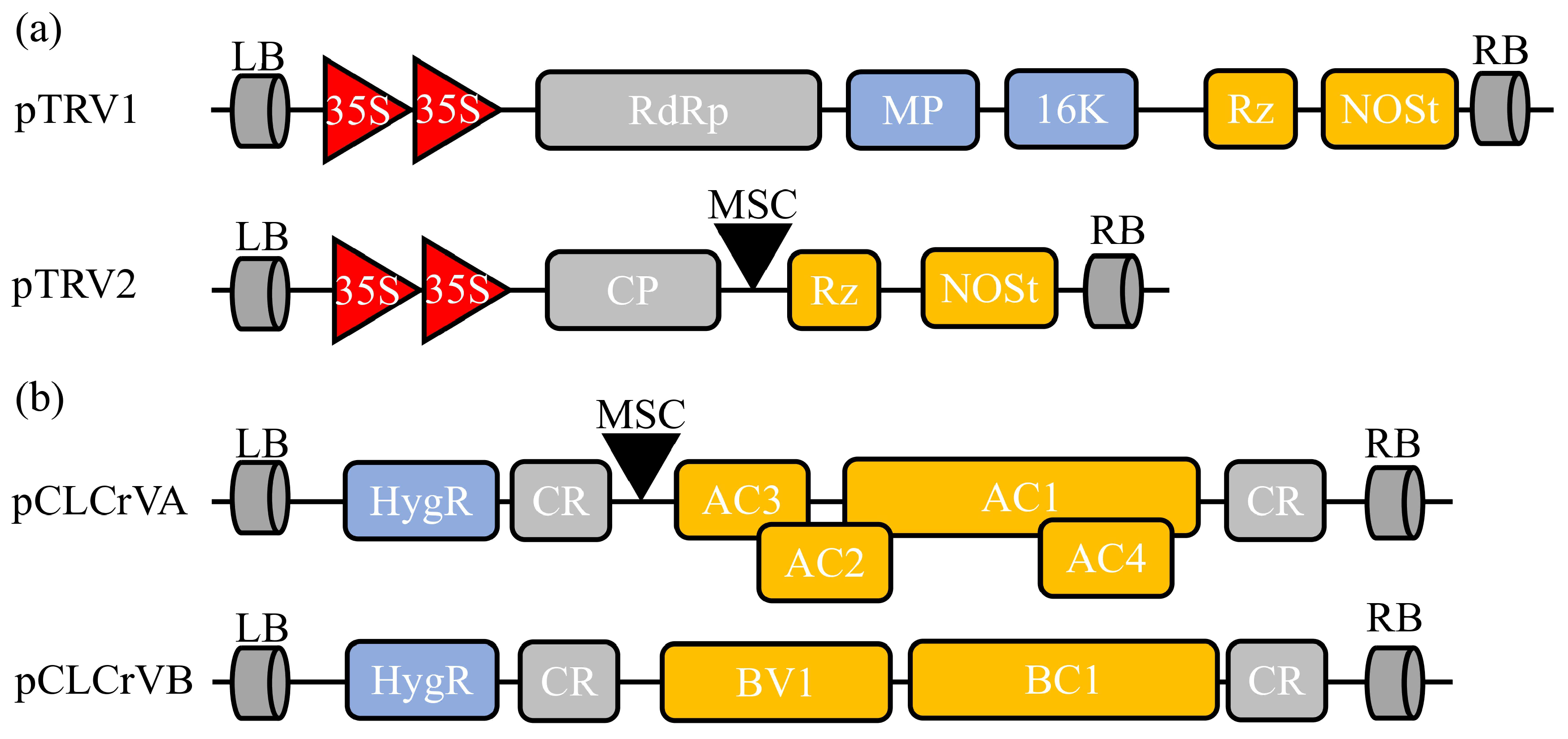
2. VIGS Vectors for Gene Functional Analysis in Cotton
3. Visible Markers for Endogenous Gene Silencing in Cotton
4. VIGS for Studying Abiotic Stress Response in Cotton
4.1. Drought Stress Tolerance
4.2. Salt Stress Tolerance
4.3. Cold and Heat Stress Tolerance
5. VIGS for Studying Biotic Stress Response in Cotton
5.1. Verticillium Stress Tolerance
5.2. Bemisia Tabaci (Whitefly) Stress Tolerance
6. VIGS for Studying Vegetative Development in Cotton
6.1. VIGS for Studying Seed Germination in Cotton
6.2. VIGS for Studying Root Development in Cotton
6.3. VIGS for Studying Stem Development in Cotton
6.4. VIGS for Studying Leaf Development in Cotton
7. VIGS for Studying Flowering in Cotton
8. VIGS for Studying Fiber Development in Cotton
| Gene | Category | Function | Reference |
|---|---|---|---|
| GhMAP3K15 | Response to drought stress | Positive regulator of drought stress | [44] |
| GhMKK4 | Positive regulator of drought stress | [44] | |
| GhMPK6 | Positive regulator of drought stress | [44] | |
| GhWRKY59 | Positive regulator of drought stress | [44] | |
| GhDREB2 | Positive regulator of drought stress | [44] | |
| GhVHA-A | Positive regulator of drought stress | [45] | |
| GhTULP30 | Positive regulator of drought stress | [46] | |
| GhECA1 | Positive regulator of drought stress | [47] | |
| GhCNGC4 | Positive regulator of drought stress | [47] | |
| GhLOX12 | Response to salt stress | Positive regulator of salt stress | [49] |
| GhLOX13 | Positive regulator of salt stress | [49] | |
| GhSOS1 | Positive regulator of salt stress | [50] | |
| GhDi19-3 | Negative regulator of salt stress | [51] | |
| GhDi19-4 | Negative regulator of salt stress | [51] | |
| GhBGLU24-A | Negative regulator of salt stress | [53] | |
| GhRUBL | Negative regulator of salt stress | [53] | |
| GbPATP | Response to temperature stress | Positive regulator of low-temperature stress | [54] |
| GhSAD1 | Positive regulator of low-temperature stress | [55] | |
| CAN1 | Negative regulator of low-temperature stress | [56] | |
| SnRK2.8 | Positive regulator of low-temperature stress | [56] | |
| GhMAP3K65 | Negative regulator of high temperature stress | [57] | |
| GhPMEI3 | Response to verticillium stress | Positive regulator of verticillium stress | [59] |
| GhGST | Positive regulator of verticillium stress | [60] | |
| GbCYP86A1-1 | Positive regulator of verticillium stress | [61] | |
| GhVLN2 | Negative regulator of verticillium stress | [62] | |
| GhlncNAT-ANX2 | Positive regulator of verticillium stress | [63] | |
| GhlncNAT-RLP7 | Positive regulator of verticillium stress | [63] | |
| GhMPK3 | Response to whitefly stress | Positive regulator of whitefly stress | [64] |
| GhGOLS2 | Seed germination | Positive regulator of seed germination | [65] |
| GhTRL1-A05 | Root development | Positive regulator of root length and surface area | [66] |
| GhPIN8-D04 | Positive regulator of root length and surface area | [66] | |
| DELLA | Stem development | Negative regulator of stem secondary cell wall formation | [67] |
| NAC | Positive regulator of stem secondary cell wall formation | [67] | |
| GoSTR | Negative regulator of stem trichome formation | [68] | |
| GhOKRA | Leave development | Regulator of leaf shape | [69,70] |
| GhLYI | Positive regulator of leaf senescence | [71] | |
| GhSAG20 | Positive regulator of leaf senescence | [71] | |
| GhACNAT | Flowering | Positive regulator of fertility | [72] |
| GhMYB80s | Positive regulator of male fertility | [73] | |
| GhCYP450 | Positive regulator of male fertility | [74] | |
| KATANIN | Fiber development | Positive regulator of fiber length | [75] |
| WRINKLED1 | Negative regulator of fiber length | [75] | |
| GhD15FAD | Positive regulator of fiber length | [76] | |
| GhPIS | Positive regulator of fiber length | [76] | |
| GhPIK | Positive regulator of fiber length | [76] | |
| GhMML3_A12 | Positive regulator of fiber initiation | [77] | |
| GhMML4_D12 | Positive regulator of fiber initiation | [78] | |
| GhARF7-1 | Positive regulator of fiber secondary cell wall formation | [79] | |
| GhARF7-2 | Positive regulator of fiber secondary cell wall formation | [79] |
9. Limitations of VIGS and How They Can Be Addressed in Cotton
10. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhai, Z.Y.; Zhang, K.X.; Fang, Y.; Yang, Y.J.; Cao, X.; Liu, L.; Tian, Y. Systematically and comprehensively understanding the regulation of cotton fiber initiation: A review. Plants 2023, 12, 3771. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Wendel, J.F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Llewellyn, D.; Showmaker, K.C.; Shu, S.; Udall, J.; et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012, 492, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Flagel, L.E.; Wendel, J.F.; Udall, J.A. Duplicate gene evolution, homoeologous recombination, and transcriptome characterization in allopolyploid cotton. BMC Genom. 2012, 13, 302. [Google Scholar] [CrossRef]
- Wen, X.; Chen, Z.; Yang, Z.; Wang, M.; Jin, S.; Wang, G.; Zhang, L.; Wang, L.; Li, J.; Saeed, S.; et al. A comprehensive overview of cotton genomics, biotechnology and molecular biological studies. Sci. China Life Sci. 2023, 66, 2214–2256. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Jones, D.C.; Liu, F.; Zhang, B. From sequencing to genome editing for cotton improvement. Trends Biotechnol. 2021, 39, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Dommes, A.B.; Gross, T.; Herbert, D.B.; Kivivirta, K.I.; Becker, A. Virus-induced gene silencing: Empowering genetics in non-model organisms. J. Exp. Bot. 2019, 70, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Hao, M.; Tian, B.; Cao, G.; Wei, F.; Xie, Z. A methodological advance of tobacco rattle virus-induced gene silencing for functional genomics in plants. Front. Plant Sci. 2021, 12, 671091. [Google Scholar] [CrossRef] [PubMed]
- Kammen, A.V. Virus-induced gene silencing in infected and transgenic plants. Trends Plant Sci. 1997, 2, 409–411. [Google Scholar] [CrossRef]
- 9 Jagram, N.; Dasgupta, I. Principles and practice of virus induced gene silencing for functional genomics in plants. Virus Genes 2023, 59, 173–187. [Google Scholar] [CrossRef]
- Choi, I.; Jeon, Y.; Pai, H.S. Brix protein APPAN plays a role in ribosomal RNA processing in Arabidopsis. Plant Sci. 2023, 333, 111721. [Google Scholar] [CrossRef]
- Chen, L.J.; Zou, W.S.; Wu, G.; Lin, H.H.; Xi, D.H. Tobacco alpha-expansion EXPA4 plays a role in Nicotiana benthamiana defence against Tobacco mosaic virus. Planta 2018, 247, 355–368. [Google Scholar] [CrossRef]
- Zhang, J.Q.; Shi, X.T.; Liu, W.D. Targeting wheat fusarium head blight with mycovirus-mediated VIGS. Trends Microbiol. 2023, 31, 1197–1198. [Google Scholar] [CrossRef] [PubMed]
- Serwatowska, J.; Lund, O.S.; Johansen, I.E. Transient posttranscriptional gene silencing in Medicago truncatula: Virus-induced gene silencing (VIGS). Methods Mol. Biol. 2018, 1822, 115–122. [Google Scholar] [PubMed]
- Xu, A.; Wei, L.; Ke, J.J.; Peng, C.F.; Li, P.Y.; Fan, C.Q.; Yu, X.; Li, B. ETI signaling nodes are involved in resistance of Hawaii 7996 to Ralstonia solanacearum-induced bacterial wilt disease in tomato. Plant Signal. Behav. 2023, 18, 2194747. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Sun, J.; Yao, J.; Wang, S.; Ding, M.; Zhang, H.; Qian, Z.; Zhao, N.; Sa, G.; Zhao, R.; et al. High rates of virus-induced gene silencing by tobacco rattle virus in Populus. Tree Physiol. 2015, 35, 1016–1029. [Google Scholar] [CrossRef]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A.; Ictv Report, C. ICTV virus taxonomy profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef]
- Aimone, C.D.; De Leon, L.; Dallas, M.M.; Ndunguru, J.; Ascencio-Ibanez, J.T.; Hanley-Bowdoin, L. A new type of satellite associated with cassava mosaic begomoviruses. J. Virol. 2021, 95, e0043221. [Google Scholar] [CrossRef]
- Lei, J.F.; Li, Y.; Dai, P.H.; Liu, C.; Zhao, Y.; You, Y.Z.; Qu, Y.Y.; Chen, Q.J.; Liu, X.D. Efficient virus-mediated genome editing in cotton using the CRISPR/Cas9 system. Front. Plant Sci. 2022, 13, 1032799. [Google Scholar] [CrossRef]
- Kumagai, M.H.; Donson, J.; Cioppa, G.D.; Harvey, D.; Hanley, H.; Grill, L.K. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 1995, 92, 1679–1683. [Google Scholar] [CrossRef]
- Singh, A.K.; Ghosh, D.; Chakraborty, S. Optimization of Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) in tomato. Methods Mol. Biol. 2022, 2408, 133–145. [Google Scholar]
- Jarugula, S.; Willie, K.; Stewart, L.R. Barley stripe mosaic virus (BSMV) as a virus-induced gene silencing vector in maize seedlings. Virus Genes 2018, 54, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Meziadi, C.; Lintz, J.; Naderpour, M.; Gautier, C.; Blanchet, S.; Noly, A.; Gratias-Weill, A.; Geffroy, V.; Pflieger, S. R-BPMV-mediated resistance to Bean pod mottle virus in Phaseolus vulgaris L. is heat-stable but elevated temperatures boost viral infection in susceptible genotypes. Viruses 2021, 13, 1239. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Gibbs, A.J.; Hajizadeh, M.; Perez, A.; Adams, I.P.; Fribourg, C.E.; Kreuze, J.; Fox, A.; Boonham, N.; Jones, R.A.C. The phylogeography of Potato virus X shows the fingerprints of its human vector. Viruses 2021, 13, 644. [Google Scholar] [CrossRef] [PubMed]
- Palukaitis, P. Satellite RNAs and satellite viruses. Mol. Plant Microbe Interact. 2016, 29, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Huang, C.; Li, F.; Zhou, X. A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 2014, 12, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Si, Z.; Wu, H.; Tian, Y.; Zhang, Z.; Zhang, T.; Hu, Y. Visible gland constantly traces virus-induced gene silencing in cotton. Front. Plant Sci. 2022, 13, 1020841. [Google Scholar] [CrossRef]
- Idris, A.M.; Brown, J.K. Cotton leaf crumple virus is a distinct western hemisphere begomovirus species with complex evolutionary relationships indicative of recombination and reassortment. Phytopathology 2004, 94, 1068–1074. [Google Scholar] [CrossRef]
- Tuttle, J.R.; Idris, A.M.; Brown, J.K.; Haigler, C.H.; Robertson, D. Geminivirus-mediated gene silencing from Cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol. 2008, 148, 41–50. [Google Scholar] [CrossRef]
- Fauquet, C.M.; Briddon, R.W.; Brown, J.K.; Moriones, E.; Stanley, J.; Zerbini, M.; Zhou, X. Geminivirus strain demarcation and nomenclature. Arch. Virol. 2008, 153, 783–821. [Google Scholar] [CrossRef]
- Fondong, V.N. Geminivirus protein structure and function. Mol. Plant Pathol. 2013, 14, 635–649. [Google Scholar] [CrossRef]
- Cheng, G.; Shu, X.; Wang, Z.; Wang, N.; Zhang, F. Establishing a virus-induced gene silencing system in Lycoris chinensis. Plants 2023, 12, 2458. [Google Scholar] [CrossRef] [PubMed]
- Bennypaul, H.S.; Mutti, J.S.; Rustgi, S.; Kumar, N.; Okubara, P.A.; Gill, K.S. Virus-induced gene silencing (VIGS) of genes expressed in root, leaf, and meiotic tissues of wheat. Funct. Integr. Genom. 2012, 12, 143–156. [Google Scholar] [CrossRef]
- Yin, X.; Zhang, Y.; Zhang, L.; Wang, B.; Zhao, Y.; Irfan, M.; Chen, L.; Feng, Y. Regulation of MYB transcription factors of anthocyanin synthesis in lily flowers. Front. Plant Sci. 2021, 12, 761668. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Mo, Q.; Li, J.; Zi, Z.; Xu, M.; Yue, S.; Zhao, H.; Zhu, H.; Wang, G. Establishment of virus-induced gene silencing (VIGS) system in Luffa acutangula using phytoene desaturase (PDS) and tendril synthesis related gene (TEN). Plant Methods 2023, 19, 94. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, F.; Zhao, J.; Xie, K.; Hong, Y.; Liu, Y. Virus-based microRNA expression for gene functional analysis in plants. Plant Physiol. 2010, 153, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Pei, H.; Zhang, S.; Chen, J.; Chen, W.; Yang, R.; Meng, Y.; You, J.; Gao, J.; Ma, N. TRV-GFP: A modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. J. Exp. Bot. 2014, 65, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Zhu, Y.; Li, Q.; Liu, J.; Tian, Y.; Liu, Y.; Wu, J. Development of Agrobacterium-mediated virus-induced gene silencing and performance evaluation of four marker genes in Gossypium barbadense. PLoS ONE 2013, 8, e73211. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, R.; Shafiq, M.; Mansoor, S.; Briddon, R.W.; Scheffler, B.E.; Scheffler, J.; Amin, I. Virus-induced gene silencing in cultivated cotton (Gossypium spp.) using Tobacco rattle virus. Mol. Biotechnol. 2015, 58, 65–72. [Google Scholar] [CrossRef]
- Khan, A.H.; Akram, A.; Saeed, M.; ur Rahman, M.; ur Rehman, A.; Mansoor, S.; Amin, I. Establishment of transcriptional gene silencing targeting the promoter regions of GFP, PDS, and PSY genes in cotton using virus-induced gene silencing. Mol. Biotechnol. 2022, 65, 1052–1061. [Google Scholar] [CrossRef]
- Ma, D.; Hu, Y.; Yang, C.; Liu, B.; Fang, L.; Wan, Q.; Liang, W.; Mei, G.; Wang, L.; Wang, H.; et al. Genetic basis for glandular trichome formation in cotton. Nat. Commun. 2016, 7, 10456. [Google Scholar] [CrossRef]
- Zulfiqar, S.; Farooq, M.A.; Zhao, T.; Wang, P.; Tabusam, J.; Wang, Y.; Xuan, S.; Zhao, J.; Chen, X.; Shen, S.; et al. Virus-induced gene silencing (VIGS): A powerful tool for crop improvement and its advancement towards epigenetics. Int. J. Mol. Sci. 2023, 24, 5608. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Saud, S.; Wang, L. Mechanism of cotton resistance to abiotic stress, and recent research advances in the osmoregulation related genes. Front. Plant Sci. 2022, 13, 972635. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, M.; Wang, P.; Cox, K.L.; Duan, L.; Dever, J.K.; Shan, L.; Li, Z.; He, P. Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of GhWRKY59. New Phytol. 2017, 215, 1462–1475. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Ni, Z.; Zhang, H.; Chen, Q.; Gao, W.; Cai, Y.; Li, M.; Sun, G.; Qu, Y.Y. The gene encoding subunit A of the vacuolar H+-ATPase from cotton plays an important role in conferring tolerance to water deficit. Front. Plant Sci. 2018, 9, 758. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, J.; Kuang, M.; Zhang, C.; Ma, Q.; Huang, L.; Wang, H.; Fan, S.; Peng, J. Improvement of plant tolerance to drought stress by cotton tubby-like protein 30 through stomatal movement regulation. J. Adv. Res. 2022, 42, 55–67. [Google Scholar] [CrossRef]
- Li, B.; Zhang, M.; Sun, W.; Yue, D.; Ma, Y.; Zhang, B.; Duan, L.; Wang, M.; Lindsey, K.; Nie, X.; et al. N6-methyladenosine RNA modification regulates cotton drought response in a Ca2+ and ABA-dependent manner. Plant Biotechnol. J. 2023, 21, 1270–1285. [Google Scholar] [CrossRef]
- Maryum, Z.; Luqman, T.; Nadeem, S.; Khan, S.; Wang, B.; Ditta, A.; Khan, M.K.R. An overview of salinity stress, mechanism of salinity tolerance and strategies for its management in cotton. Front. Plant Sci. 2022, 13, 907937. [Google Scholar] [CrossRef]
- Shaban, M.; Ahmed, M.M.; Sun, H.; Ullah, A.; Zhu, L. Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses. BMC Genom. 2018, 19, 599. [Google Scholar] [CrossRef]
- Che, B.; Cheng, C.; Fang, J.; Liu, Y.; Jiang, L.; Yu, B. The recretohalophyte tamarix TrSOS1 gene confers enhanced salt tolerance to transgenic hairy root composite cotton seedlings exhibiting virus-induced gene silencing of GhSOS1. Int. J. Mol. Sci. 2019, 20, 2930. [Google Scholar] [CrossRef]
- Zhao, L.; Li, Y.; Li, Y.; Chen, W.; Yao, J.; Fang, S.; Lv, Y.; Zhang, Y.; Zhu, S. Systematical characterization of the sotton Di19 gene family and the role of GhDi19-3 and GhDi19-4 as two negative regulators in response to salt stress. Antioxidants 2022, 11, 2225. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ma, L.; Jiang, W.; Yao, Y.; Tang, Y.; Pang, Y. Comprehensive identification and characterization of abiotic stress and hormone responsive glycosyl hydrolase family 1 genes in Medicago truncatula. Plant Physiol. Biochem. 2021, 158, 21–33. [Google Scholar] [CrossRef]
- Cui, C.; Wan, H.; Li, Z.; Ai, N.; Zhou, B. Long noncoding RNA TRABA suppresses beta-glucosidase-encoding BGLU24 to promote salt tolerance in cotton. Plant Physiol. 2023, 6, kiad530. [Google Scholar] [CrossRef]
- Liu, T.; Guo, S.; Lian, Z.; Chen, F.; Yang, Y.; Chen, T.; Ling, X.; Liu, A.; Wang, R.; Zhang, B. A P4-ATPase gene GbPATP of cotton confers chilling tolerance in plants. Plant Cell Physiol. 2015, 56, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Wang, L.; Yang, Y.; Liu, R.; Liu, S.; Chen, J.; Shen, Q.; Ma, H.; Li, Y.; Zhang, S.; et al. Genome-wide association study identifies variants of GhSAD1 conferring cold tolerance in cotton. J. Exp. Bot. 2022, 73, 2222–2237. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Long, X.; Gao, M.; Zhao, Y.; Guan, X. Global identification of natural antisense transcripts in Gossypium hirsutum and Gossypium barbadense under chilling stress. iScience 2023, 26, 107362. [Google Scholar] [CrossRef]
- Zhai, N.; Jia, H.; Liu, D.; Liu, S.; Ma, M.; Guo, X.; Li, H. GhMAP3K65, a cotton Raf-like MAP3K gene, enhances susceptibility to pathogen infection and heat stress by negatively modulating growth and development in transgenic Nicotiana benthamiana. Int. J. Mol. Sci. 2017, 18, 2462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.F.; Ding, Z.G.; Ma, Q.; Zhang, G.R.; Zhang, S.L.; Li, Z.K.; Wu, L.Q.; Zhang, G.Y.; Ma, Z.Y. Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genom. 2013, 14, 637. [Google Scholar] [CrossRef]
- Liu, N.; Sun, Y.; Pei, Y.; Zhang, X.; Wang, P.; Li, X.; Li, F.; Hou, Y. A pectin methylesterase inhibitor enhances resistance to Verticillium wilt. Plant Physiol. 2018, 176, 2202–2220. [Google Scholar] [CrossRef]
- Li, Z.K.; Chen, B.; Li, X.X.; Wang, J.P.; Zhang, Y.; Wang, X.F.; Yan, Y.Y.; Ke, H.F.; Yang, J.; Wu, J.H.; et al. A newly identified cluster of glutathione S-transferase genes provides Verticillium wilt resistance in cotton. Plant J. 2019, 98, 213–227. [Google Scholar] [CrossRef]
- Wang, G.; Xu, J.; Li, L.; Guo, Z.; Si, Q.; Zhu, G.; Wang, X.; Guo, W. GbCYP86A1-1 from Gossypium barbadense positively regulates defence against Verticillium dahliae by cell wall modification and activation of immune pathways. Plant Biotechnol. J. 2020, 18, 222–238. [Google Scholar] [CrossRef] [PubMed]
- Li, W.B.; Song, S.W.; Zhong, M.M.; Liu, L.G.; Su, L.; Han, L.B.; Xia, G.X.; Sun, Y.D.; Wang, H.Y. VILLIN2 regulates cotton defense against Verticillium dahliae by modulating actin cytoskeleton remodeling. Plant Physiol. 2023, 192, 666–679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, M.; Li, N.; Wang, H.; Qiu, P.; Pei, L.; Xu, Z.; Wang, T.; Gao, E.; Liu, J.; et al. Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol. J. 2018, 16, 1172–1185. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, L.; Hull, J.J.; Daniell, H.; Jin, S.; Zhang, X. Transcriptome analysis reveals a comprehensive insect resistance response mechanism in cotton to infestation by the phloem feeding insect Bemisia tabaci (whitefly). Plant Biotechnol. J. 2016, 14, 1956–1975. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Hao, M.; Tian, B.; Cao, G.; Chen, W.; Zhang, Q.; Zhang, Y.; Ling, H.; Li, J.; Xie, Z.; et al. A newly established virus-induced gene silencing method via seed imbibition for functional genomics at early germination stages in cotton. Ind. Crops Prod. 2021, 172, 114040. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, S.; Ge, C.; Shen, Q.; Zhang, S.; Ma, H.; Liu, R.; Zhao, X.; Liu, R.; Li, P.; et al. Genome-wide association study reveals that GhTRL1 and GhPIN8 affect cotton root development. Theor. Appl. Genet. 2022, 135, 3161–3176. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, W.; Ran, L.; Chen, Z.; Wang, C.; Dou, Y.; Qin, Y.; Suo, Q.; Li, Y.; Zeng, J.; et al. DELLA-NAC interactions mediate GA signaling to promote secondary cell wall formation in cotton stem. Front. Plant Sci. 2021, 12, 655127. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Zhang, J.; Shi, Y.; Dai, F.; Jiang, T.; Xuan, L.; He, Y.; Zhang, Z.; Deng, J.; Zhang, T.; et al. GoSTR, a negative modulator of stem trichome formation in cotton. Plant J. 2023, 116, 389–403. [Google Scholar] [CrossRef]
- Chang, L.; Fang, L.; Zhu, Y.; Wu, H.; Zhang, Z.; Liu, C.; Li, X.; Zhang, T. Insights into interspecific hybridization events in allotetraploid cotton formation from characterization of a gene-regulating leaf shape. Genetics 2016, 204, 799–806. [Google Scholar] [CrossRef]
- Andres, R.J.; Coneva, V.; Frank, M.H.; Tuttle, J.R.; Samayoa, L.F.; Han, S.W.; Kaur, B.; Zhu, L.; Fang, H.; Bowman, D.T.; et al. Modifications to a LATE MERISTEM IDENTITY1 gene are responsible for the major leaf shapes of upland cotton (Gossypium hirsutum L.). Proc. Natl. Acad. Sci. USA 2016, 114, E57–E66. [Google Scholar] [CrossRef]
- Zhang, Y.; Zang, Y.; Chen, J.; Feng, S.; Zhang, Z.; Hu, Y.; Zhang, T. A truncated ETHYLENE INSENSITIVE3-like protein, GhLYI, regulates senescence in cotton. Plant Physiol. 2023, 193, 1177–1196. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Shen, Y.; Hao, J.; Wu, J.; Ke, L.; Wu, C.; Huang, K.; Luo, B.; Xu, M.; Cheng, X.; et al. Acyl-CoA N-acyltransferase influences fertility by regulating lipid metabolism and jasmonic acid biogenesis in cotton. Sci. Rep. 2015, 5, 11790. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wei, H.; Hao, P.; Wu, A.; Ma, Q.; Zhang, J.; Wang, H.; Fu, X.; Ma, L.; Lu, J.; et al. GhGPAT12/25 are essential for the formation of anther cuticle and pollen exine in cotton (Gossypium hirsutum L.). Front. Plant Sci. 2021, 12, 667739. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Dai, F.; Si, Z.; Fang, L.; Zhang, T. Duplicate mutations of GhCYP450 lead to the production of ms5m6 male sterile line in cotton. Theor. Appl. Genet. 2023, 136, 2. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Ye, J.; Geng, Y.F.; Sun, Y.W.; Gao, S.Q.; Zhang, B.P.; Chen, W.; Chua, N.H. Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant Physiol. 2012, 160, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.J.; Xiao, G.H.; Liu, N.J.; Liu, D.; Chen, P.S.; Qin, Y.M.; Zhu, Y.X. Targeted lipidomics studies reveal that linolenic acid promotes cotton fiber elongation by activating phosphatidylinositol and phosphatidylinositol monophosphate biosynthesis. Mol. Plant 2015, 8, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Guan, X.; Yang, N.; Wu, H.; Pan, M.; Liu, B.; Fang, L.; Yang, S.; Hu, Y.; Ye, W.; et al. Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytol. 2016, 210, 1298–1310. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Tian, Y.; Wan, Q.; Fang, L.; Guan, X.; Chen, J.; Hu, Y.; Ye, W.; Zhang, H.; Guo, W.; et al. Genetics and evolution of MIXTA genes regulating cotton lint fiber development. New Phytol. 2017, 217, 883–895. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; He, S.P.; Xu, S.W.; Li, L.; Zheng, Y.; Li, X.B. The transcription factor ERF108 interacts with auxin response factors to mediate cotton fiber secondary cell wall biosynthesis. Plant Cell 2023, 35, 4133–4154. [Google Scholar] [CrossRef]
- Becker, A.; Lange, M. VIGS—Genomics goes functional. Trends Plant Sci. 2010, 15, 1–4. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Mysore, K.S. New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 2011, 16, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Wege, S.; Scholz, A.; Gleissberg, S.; Becker, A. Highly efficient virus-induced gene silencing (VIGS) in california poppy (Eschscholzia californica): An evaluation of VIGS as a strategy to obtain functional data from non-model plants. Ann. Bot. 2007, 100, 641–649. [Google Scholar] [CrossRef]
- Fu, D.Q.; Zhu, B.Z.; Zhu, H.L.; Zhang, H.X.; Xie, Y.H.; Jiang, W.B.; Zhao, X.D.; Luo, Y.B. Enhancement of virus-induced gene silencing in tomato by low temperature and low humidity. Mol. Cells 2006, 21, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Kotakis, C.; Vrettos, N.; Kotsis, D.; Tsagris, M.; Kotzabasis, K.; Kalantidis, K. Light intensity affects RNA silencing of a transgene in Nicotiana benthamiana plants. BMC Plant Biol. 2010, 10, 220. [Google Scholar] [CrossRef] [PubMed]
- Chellappan, P.; Vanitharani, R.; Ogbe, F.; Fauquet, C.M. Effect of temperature on geminivirus-induced RNA silencing in plants. Plant Physiol. 2005, 138, 1828–1841. [Google Scholar] [CrossRef]
- Dalakouras, A.; Papadopoulou, K.K. Epigenetic Modifications: An unexplored facet of exogenous RNA application in plants. Plants 2020, 9, 673. [Google Scholar] [CrossRef]
- Dalakouras, A.; Vlachostergios, D.; Manavella, P. Epigenetic approaches to crop breeding: Current status and perspectives. J. Exp. Bot. 2021, 72, 5356–5371. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Fang, Y.; Zhang, K.; Zhai, Z.; Yang, Y.; He, M.; Cao, X. Applications of Virus-Induced Gene Silencing in Cotton. Plants 2024, 13, 272. https://doi.org/10.3390/plants13020272
Tian Y, Fang Y, Zhang K, Zhai Z, Yang Y, He M, Cao X. Applications of Virus-Induced Gene Silencing in Cotton. Plants. 2024; 13(2):272. https://doi.org/10.3390/plants13020272
Chicago/Turabian StyleTian, Yue, Yao Fang, Kaixin Zhang, Zeyang Zhai, Yujie Yang, Meiyu He, and Xu Cao. 2024. "Applications of Virus-Induced Gene Silencing in Cotton" Plants 13, no. 2: 272. https://doi.org/10.3390/plants13020272
APA StyleTian, Y., Fang, Y., Zhang, K., Zhai, Z., Yang, Y., He, M., & Cao, X. (2024). Applications of Virus-Induced Gene Silencing in Cotton. Plants, 13(2), 272. https://doi.org/10.3390/plants13020272





