Transcriptomic Analysis Reveals the Mechanism of Color Formation in the Peel of an Evergreen Pomegranate Cultivar ‘Danruo No.1’ During Fruit Development
Abstract
1. Introduction
2. Results
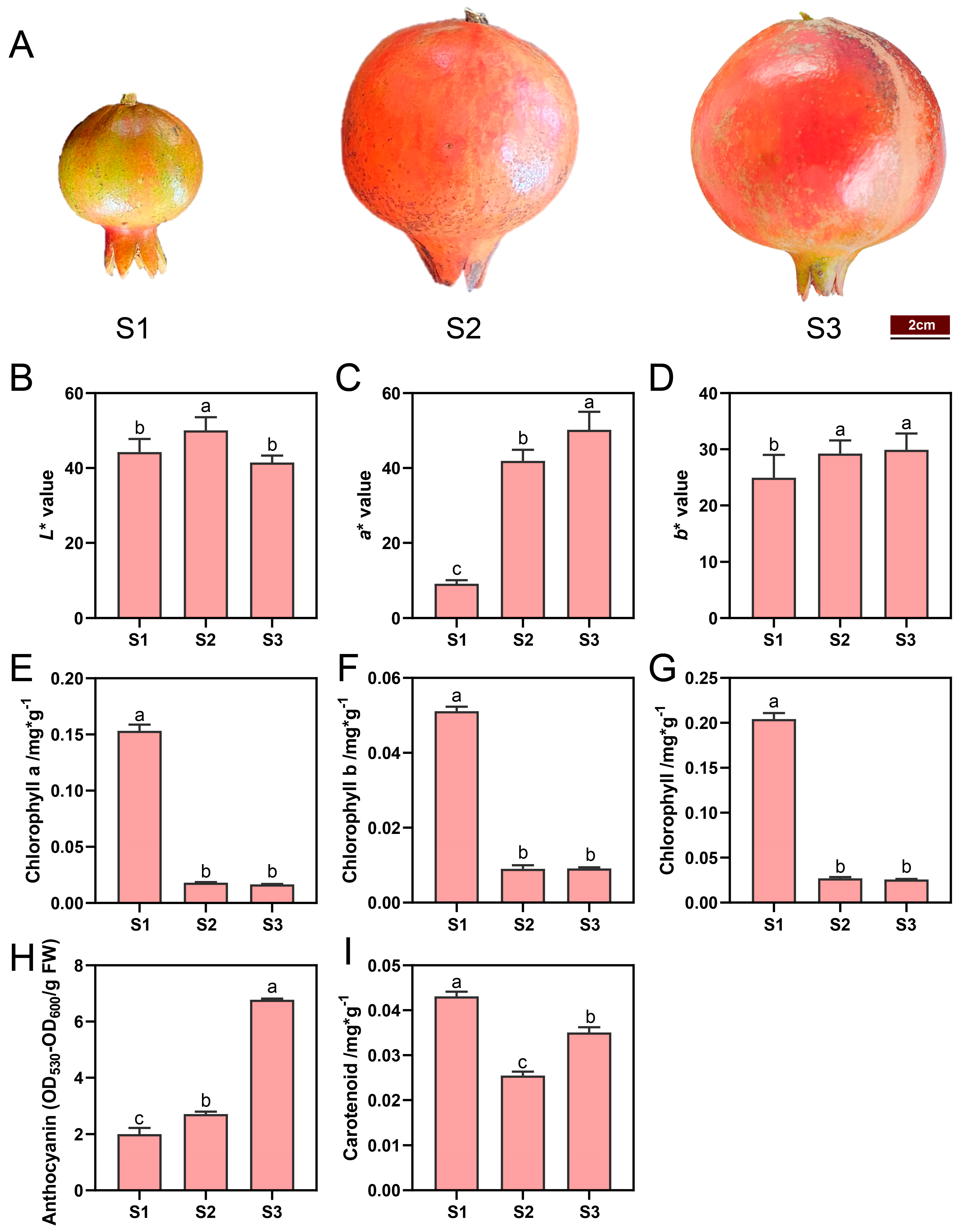
2.1. Coloration and Pigments Contents in ‘Danruo No.1’ Pomegranate Peel During Fruit Development
2.2. RNA-seq Data Overview
2.3. Differentially Expressed Genes (DEGs) Identification, and Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analyses
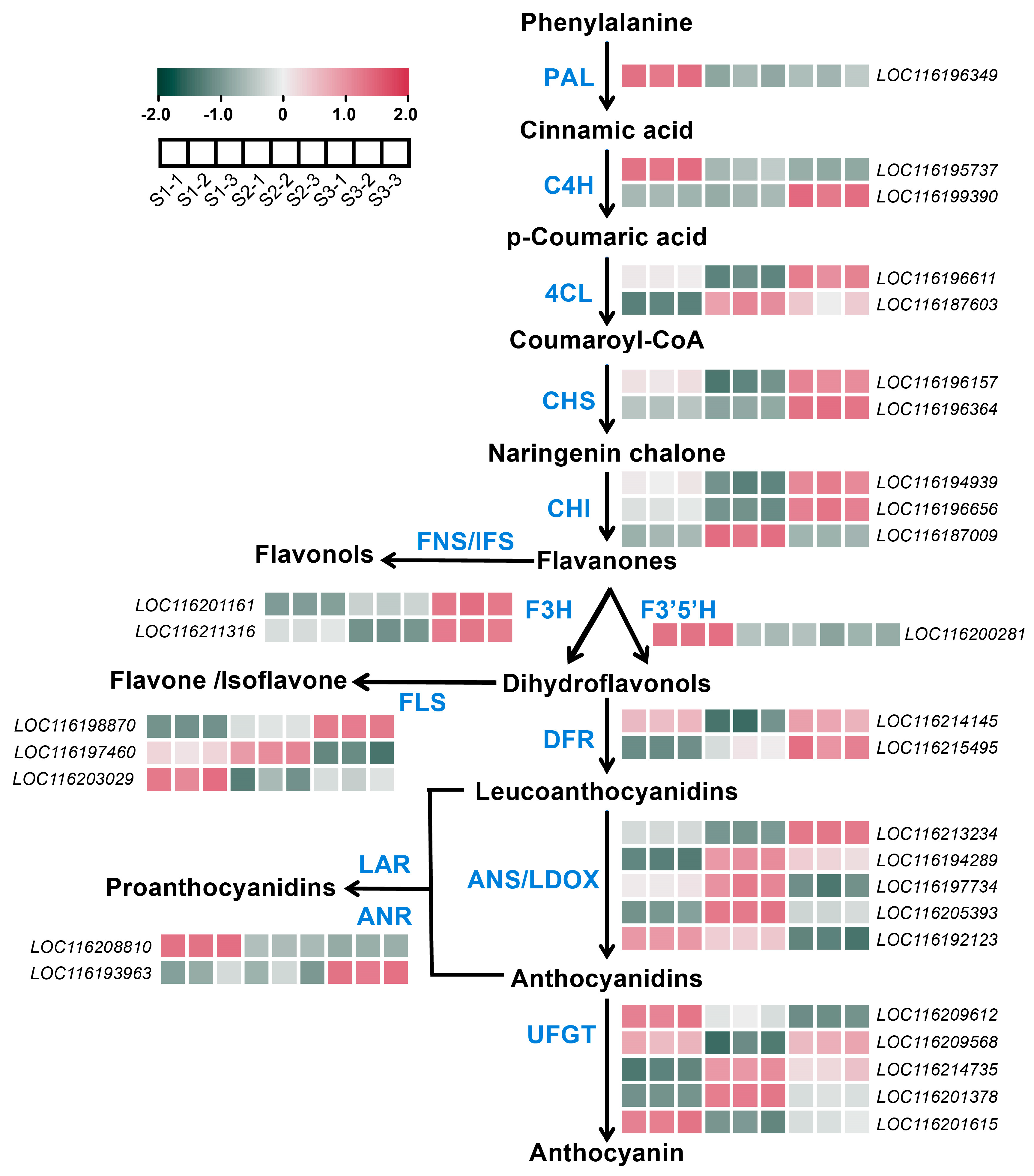
2.4. DEGs Associated with Anthocyanin Biosynthesis in Pomegranate Peel During Fruit Development
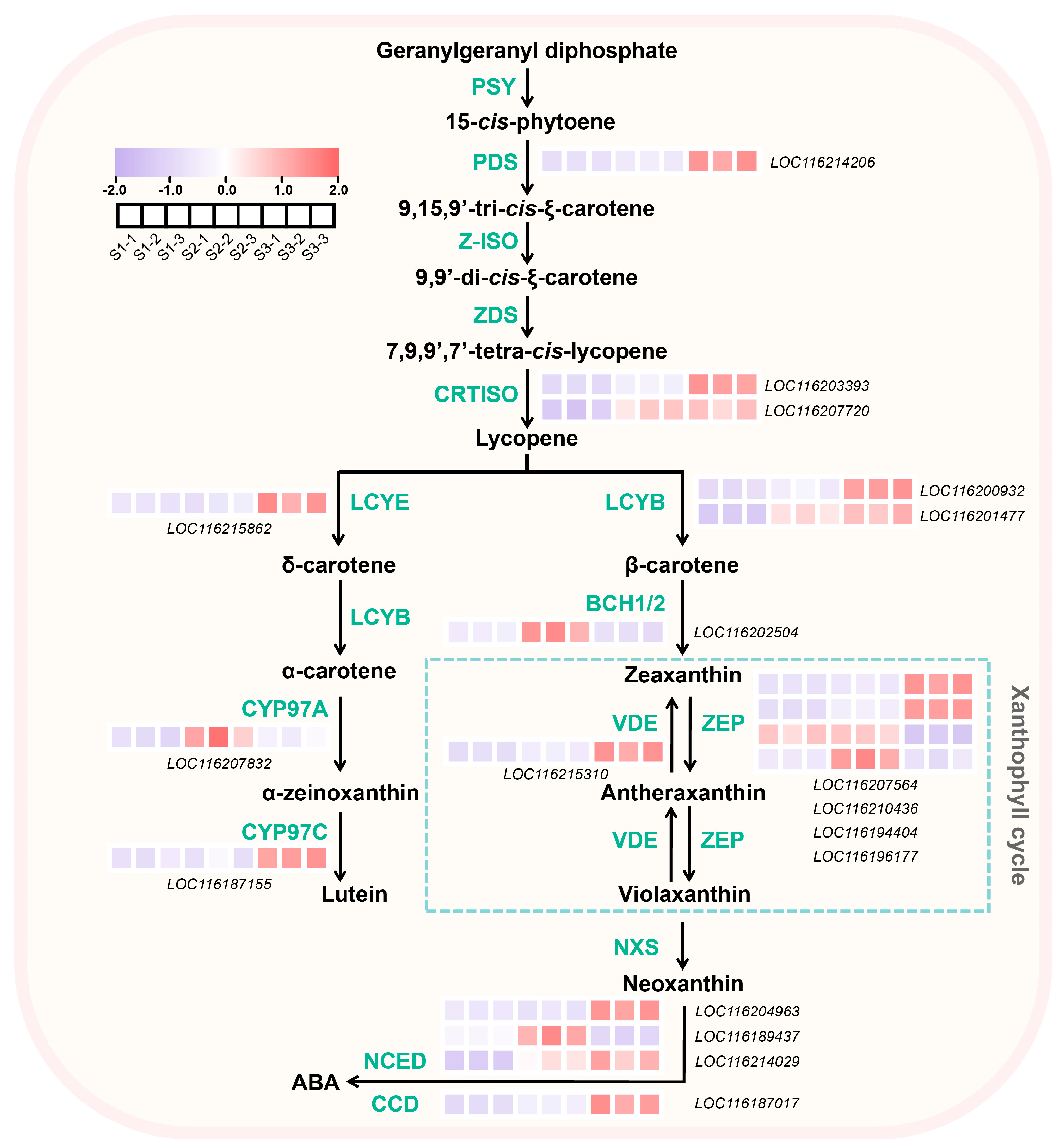
2.5. DEGs Associated with Carotenoids Biosynthesis in Pomegranate Peel During Fruit Development
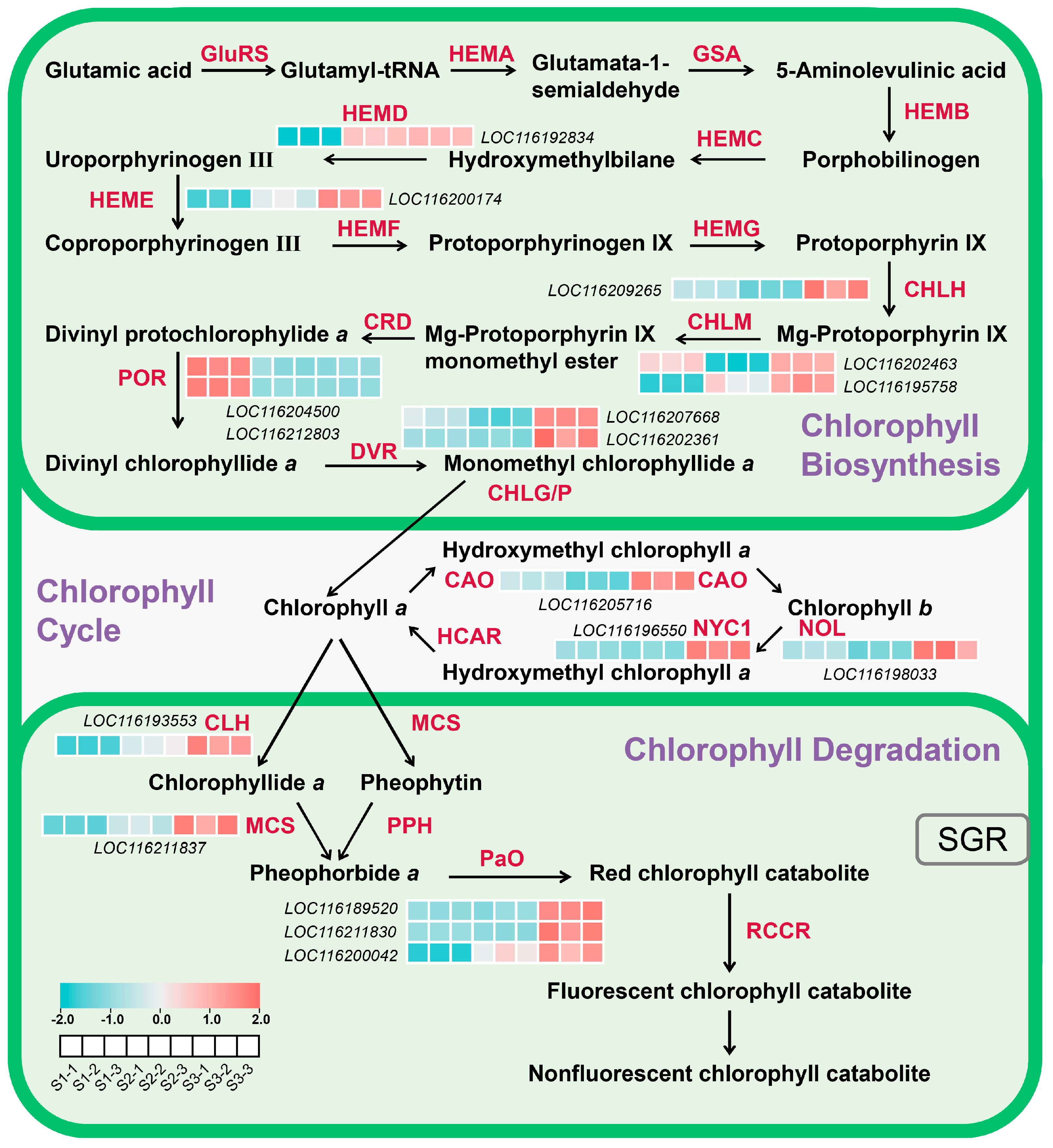
2.6. DEGs Associated with Chlorophyll Biosynthesis and Degradation in Pomegranate Peel During Fruit Development
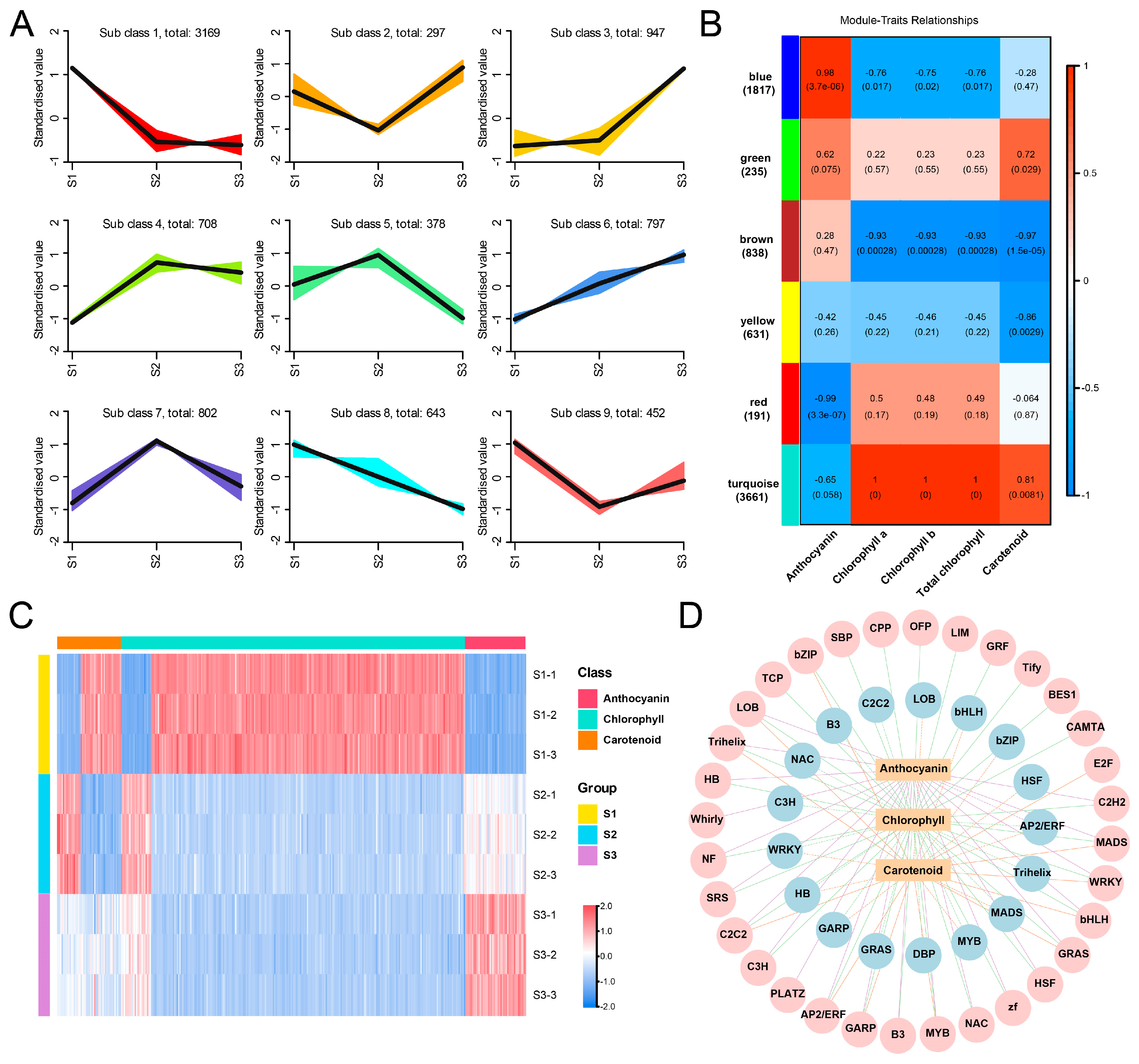
2.7. Screening Pigments Metabolism-Related TFs Based on K-Means and Weighted Gene Co-Expression Network Analysis (WGCNA)
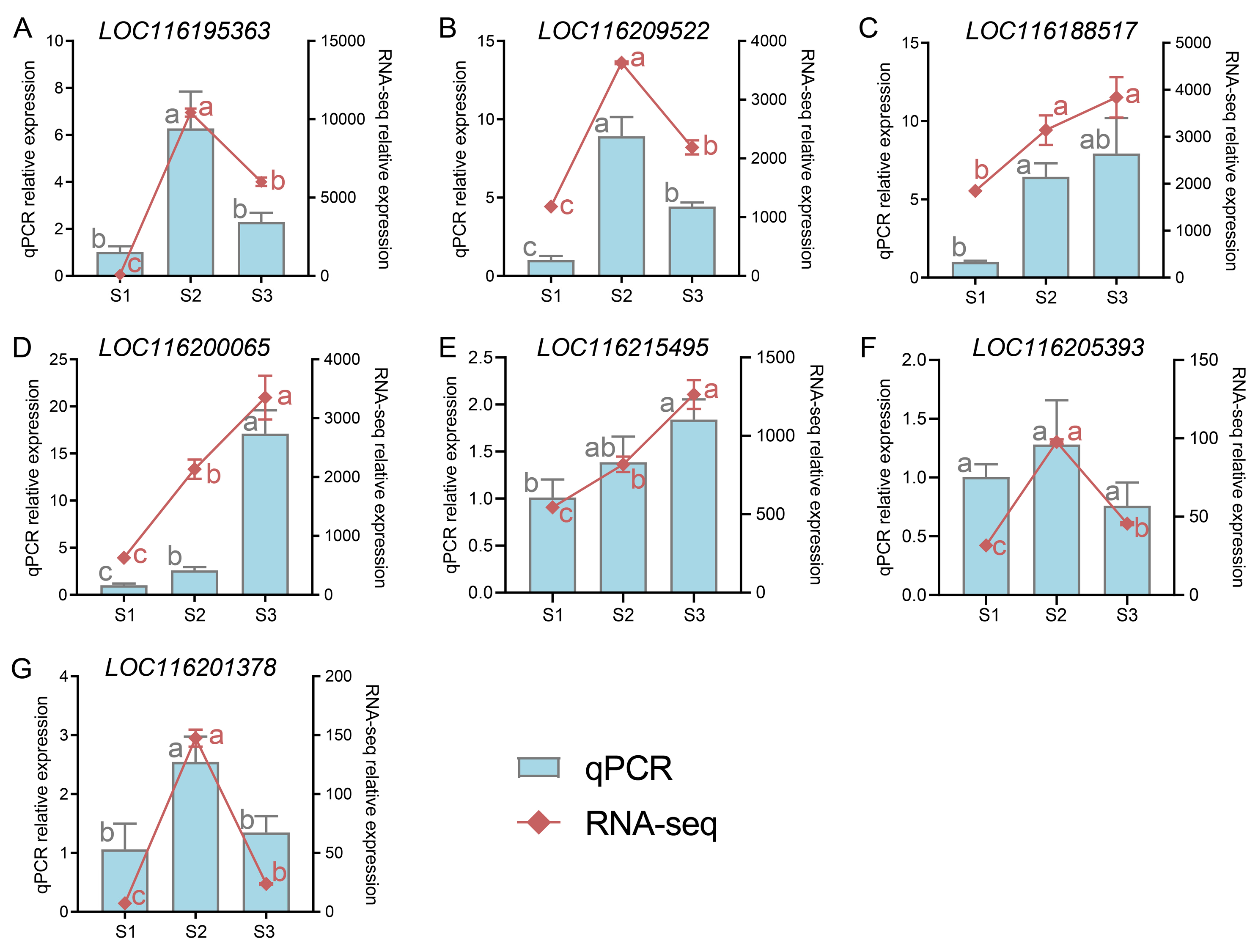
2.8. Validation of DEGs by Quantitative Polymerase Chain Reaction (q-PCR)
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement of Anthocyanin, Chlorophyll and Carotenoid Contents
4.3. RNA Extraction, Library Construction and RNA-Seq
4.4. cDNA Synthesis and q-PCR
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Da Silva, J.A.T.; Rana, T.S.; Narzary, D.; Verma, N.; Meshram, D.T.; Ranade, S.A. Pomegranate biology and biotechnology: A review. Sci. Hortic. 2013, 160, 85–107. [Google Scholar] [CrossRef]
- Giménez-Bastida, J.A.; Ávila-Gálvez, M.Á.; Espín, J.C.; González-Sarrías, A. Evidence for health properties of pomegranate juices and extracts beyond nutrition: A critical systematic review of human studies. Trends Food Sci. Technol. 2021, 114, 410–423. [Google Scholar] [CrossRef]
- Holland, D.; Hatib, K.; Bar-Ya’akov, I. Pomegranate: Botany, horticulture, breeding. Hortic. Rev. 2009, 35, 127–191. [Google Scholar] [CrossRef]
- Cömert, E.D.; Mogol, B.A.; Gökmen, V. Relationship between color and antioxidant capacity of fruits and vegetables. Curr. Res. Food Sci. 2020, 2, 1–10. [Google Scholar] [CrossRef]
- Ding, R.; Che, X.; Shen, Z.; Zhang, Y. Metabolome and transcriptome profiling provide insights into green apple peel reveals light- and UV-B-responsive pathway in anthocyanins accumulation. BMC Plant Biol. 2021, 21, 351. [Google Scholar] [CrossRef]
- Campbell, B.L.; Nelson, R.G.; Ebel, R.C.; Dozier, W.A.; Adrian, J.L.; Hockema, B.R. Fruit quality characteristics that affect consumer preferences for satsuma mandarins. HortScience 2004, 39, 1664–1669. [Google Scholar] [CrossRef]
- Wang, Y.; Tu, H.; Zhang, J.; Wang, H.; Liu, Z.; Zhou, J.; He, W.; Lin, Y.; Zhang, Y.; Li, M.; et al. Identifying potential anthocyanin biosynthesis regulator in Chinese cherry by comprehensive genome-wide characterization of the R2R3-MYB transcription factor gene family. BMC Genom. 2024, 25, 784. [Google Scholar] [CrossRef]
- Abbott, J.A. Quality measurement of fruits and vegetables. Postharvest Biol. Technol. 1999, 15, 207–225. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, X.; Wu, R.; Tang, X.; Yang, Y.; Fan, X.; Gong, H.; Grierson, D.; Yin, X.; Li, J.; et al. Integrated metabolomic and transcriptomic analyses provide comprehensive new insights into the mechanism of chitosan delay of kiwifruit postharvest ripening. Postharvest Biol. Technol. 2024, 210, 112746. [Google Scholar] [CrossRef]
- Sun, T.; Rao, S.; Zhou, X.; Li, L. Plant carotenoids: Recent advances and future perspectives. Mol. Hortic. 2022, 2, 3. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Sun, T.; Yuan, H.; Cao, H.; Yazdani, M.; Tadmor, Y.; Li, L. Carotenoid Metabolism in Plants: The Role of Plastids. Mol. Plant 2018, 11, 58–74. [Google Scholar] [CrossRef]
- Tanaka, A.; Tanaka, R. Chlorophyll metabolism. Curr. Opin. Plant Biol. 2006, 9, 248–255. [Google Scholar] [CrossRef]
- Yu, Q.; Shen, Y.; Wang, Q.; Wang, X.; Fan, L.; Wang, Y.; Zhang, S.; Liu, Z.; Zhang, M. Light deficiency and waterlogging affect chlorophyll metabolism and photosynthesis in Magnolia sinostellata. Trees 2019, 33, 11–22. [Google Scholar] [CrossRef]
- Yang, Y.-n.; Yao, G.-f.; Zheng, D.; Zhang, S.-l.; Wang, C.; Zhang, M.-y.; Wu, J. Expression differences of anthocyanin biosynthesis genes reveal regulation patterns for red pear coloration. Plant Cell Rep. 2015, 34, 189–198. [Google Scholar] [CrossRef]
- Qin, G.; Xu, C.; Ming, R.; Tang, H.; Guyot, R.; Kramer, E.M.; Hu, Y.; Yi, X.; Qi, Y.; Xu, X.; et al. The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 2017, 91, 1108–1128. [Google Scholar] [CrossRef]
- Yuan, Z.; Fang, Y.; Zhang, T.; Fei, Z.; Han, F.; Liu, C.; Liu, M.; Xiao, W.; Zhang, W.; Wu, S.; et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2018, 16, 1363–1374. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Yuan, Z.H.; Yin, Y.L.; Feng, L.J. Patterns of Pigment Changes in Pomegranate (Punica granatum L.) Peel during Fruit Ripening. Acta Hortic. 2015, 1089, 83–89. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Yuan, Z.H.; Fang, Y.M.; Yin, Y.L.; Feng, L.J. Characterization and evaluation of major anthocyanins in pomegranate (Punica granatum L.) peel of different cultivars and their development phases. Eur. Food Res. Technol. 2013, 236, 109–117. [Google Scholar] [CrossRef]
- Mo, R.; Zhang, N.; Li, J.; Jin, Q.; Zhu, Z.; Dong, Z.; Li, Y.; Zhang, C.; Yu, C. Transcriptomic Analysis Provides Insights into Anthocyanin Accumulation in Mulberry Fruits. Horticulturae 2022, 8, 920. [Google Scholar] [CrossRef]
- Shi, B.; Wu, H.; Zheng, B.; Qian, M.; Gao, A.; Zhou, K. Analysis of Light-Independent Anthocyanin Accumulation in Mango (Mangifera indica L.). Horticulturae 2021, 7, 423. [Google Scholar] [CrossRef]
- Qin, L.; Xie, H.; Xiang, N.; Wang, M.; Han, S.; Pan, M.; Guo, X.; Zhang, W. Dynamic Changes in Anthocyanin Accumulation and Cellular Antioxidant Activities in Two Varieties of Grape Berries during Fruit Maturation under Different Climates. Molecules 2022, 27, 384. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Pan, D.-L.; Jia, Z.-H.; Wang, T.; Wang, G.; Guo, Z.-R. Chlorophyll, carotenoid and vitamin C metabolism regulation in Actinidia chinensis ‘Hongyang’ outer pericarp during fruit development. PLoS ONE 2018, 13, e0194835. [Google Scholar] [CrossRef]
- Singh, K.; Kumar, A.; Kajal, M.; Singh, B. Characterization and expression analysis of chalcone synthase and chalcone isomerase genes in Phyllanthus emblica (L.). J. Plant Biochem. Biotechnol. 2019, 28, 105–113. [Google Scholar] [CrossRef]
- Cui, Z.; Gu, J.; Li, J.; Zhao, A.; Fu, Y.; Wang, T.; Li, T.; Li, X.; Sheng, Y.; Zhao, Y.; et al. Tyrosine promotes anthocyanin biosynthesis in pansy (Viola × wittrockiana) by inducing ABA synthesis. Trop. Plants 2022, 1, 9. [Google Scholar] [CrossRef]
- Kaur, R.; Aslam, L.; Kapoor, N.; Mahajan, R. Identification and comparative expression analysis of chalcone synthase, flavanone 3-hydroxylase and dihydroflavonol 4-reductase genes in wild pomegranate (Punica granatum L.) organs. Braz. J. Bot. 2020, 43, 883–896. [Google Scholar] [CrossRef]
- Fei, X.; Wei, Y.; Qi, Y.; Luo, Y.; Hu, H.; Wei, A. Integrated LC-MS/MS and Transcriptome Sequencing Analysis Reveals the Mechanism of Color Formation during Prickly Ash Fruit Ripening. Front. Nutr. 2022, 9, 847823. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, L.; Liu, Z.; Zhao, Z.; Zhao, J.; Wang, Z.; Zhou, G.; Liu, P.; Liu, M. Transcriptome and metabolome profiling unveil the mechanisms of Ziziphus jujuba Mill. peel coloration. Food Chem. 2020, 312, 125903. [Google Scholar] [CrossRef]
- Zheng, X.-T.; Chen, Y.-L.; Zhang, X.-H.; Cai, M.-L.; Yu, Z.-C.; Peng, C.-L. ANS-deficient Arabidopsis is sensitive to high light due to impaired anthocyanin photoprotection. Funct. Plant Biol. 2019, 46, 756–765. [Google Scholar] [CrossRef]
- Kobayashi, S.; Ishimaru, M.; Ding, C.; Yakushiji, H.; Goto, N. Comparison of UDP-glucose: Flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Sci. 2001, 160, 543–550. [Google Scholar] [CrossRef]
- Kaur, R.; Kapoor, N.; Aslam, L.; Mahajan, R. Molecular characterization of PgUFGT gene and R2R3-PgMYB transcription factor involved in flavonoid biosynthesis in four tissues of wild pomegranate (Punica granatum L.). J. Genet. 2019, 98, 94. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Qi, Y.; Zhang, A.; Liu, Z.; Ren, X. Identification and characterization of AcUFGT6b, a xylosyltransferase involved in anthocyanin modification in red-fleshed kiwifruit (Actinidia chinensis). Plant Cell Tissue Organ Cult. 2019, 138, 257–271. [Google Scholar] [CrossRef]
- Wang, H.; Yin, L.; Liu, J.; Jia, L.; Ding, X.; Shen, D.; Feng, K.; Xu, Z.; Xiong, A. The carotenoid cleavage dioxygenases gene AgCCD4 regulates the pigmentation of celery tissues with different colors. Sci. Agric. Sin. 2021, 54, 3279–3294. [Google Scholar] [CrossRef]
- Fan, J.; Wu, J.; Li, Y.; Bie, H.; Wang, J.; Guo, J.; Cao, K.; Wang, L. Analysis of the Expression and Promoter Activity of the Key Gene PpCCD4 Involved in Carotenoid Synthesis in Peach. J. Fruit Sci. 2020, 37, 1271–1280. [Google Scholar] [CrossRef]
- Dang, Z.; Zhu, M.; Chen, H.; Zhang, Y.; Gao, A.; Ma, W.; Chen, Y.; Wei, Y.; Zhang, H. MiMYB10 transcription factor regulates biosynthesis and accumulation of carotenoid involved genes in mango fruit. Int. J. Biol. Macromol. 2023, 253, 127665. [Google Scholar] [CrossRef]
- Ma, X.; Luo, X.; Wei, Y.; Bai, T.; Shi, J.; Zheng, B.; Xu, W.; Li, L.; Wang, S.; Zhang, J.; et al. Chromosome-Scale Genome and Comparative Transcriptomic Analysis Reveal Transcriptional Regulators of β-Carotene Biosynthesis in Mango. Front. Plant Sci. 2021, 12, 749108. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, Y.; Zhu, K.; Yang, W.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 2018, 176, 2657–2676. [Google Scholar] [CrossRef]
- Wang, M.; Wang, G.; Ji, J.; Wang, J. The effect of pds gene silencing on chloroplast pigment composition, thylakoid membrane structure and photosynthesis efficiency in tobacco plants. Plant Sci. 2009, 177, 222–226. [Google Scholar] [CrossRef]
- Kang, C.; Zhai, H.; Xue, L.; Zhao, N.; He, S.; Liu, Q. A lycopene β-cyclase gene, IbLCYB2, enhances carotenoid contents and abiotic stress tolerance in transgenic sweetpotato. Plant Sci. 2018, 272, 243–254. [Google Scholar] [CrossRef]
- Zhang, J.; He, L.; Dong, J.; Zhao, C.; Wang, Y.; Tang, R.; Wang, W.; Ji, Z.; Cao, Q.; Xie, H.E. Integrated metabolic and transcriptional analysis reveals the role of carotenoid cleavage dioxygenase 4 (IbCCD4) in carotenoid accumulation in sweetpotato tuberous roots. Biotechnol. Biofuels Bioprod. 2023, 16, 45. [Google Scholar] [CrossRef]
- Römer, S.; Lübeck, J.; Kauder, F.; Steiger, S.; Adomat, C.; Sandmann, G. Genetic engineering of a zeaxanthin-rich potato by antisense inactivation and co-suppression of carotenoid epoxidation. Metab. Eng. 2002, 4, 263–272. [Google Scholar] [CrossRef]
- Masuda, T.; Fusada, N.; Oosawa, N.; Takamatsu, K.i.; Yamamoto, Y.Y.; Ohto, M.; Nakamura, K.; Goto, K.; Shibata, D.; Shirano, Y. Functional analysis of isoforms of NADPH: Protochlorophyllide oxidoreductase (POR), PORB and PORC, in Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 963–974. [Google Scholar] [CrossRef]
- Yang, Y.-L.; Xu, J.; Rao, Y.-C.; Zeng, Y.-J.; Liu, H.-J.; Zheng, T.-T.; Zhang, G.-H.; Hu, J.; Guo, L.-B.; Qian, Q. Cloning and functional analysis of pale-green leaf (PGL10) in rice (Oryza sativa L.). Plant Growth Regul. 2016, 78, 69–77. [Google Scholar] [CrossRef]
- Huang, H.; He, W. Application of exogenous cytokinin regulates cytokinin oxidase and antioxidant activity to maintain chlorophyll pigment during ripening of banana fruit. Food Biosci. 2023, 55, 102998. [Google Scholar] [CrossRef]
- Pružinská, A.; Tanner, G.; Anders, I.; Roca, M.; Hörtensteiner, S. Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron–sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 2003, 100, 15259–15264. [Google Scholar] [CrossRef]
- Gomez-Lobato, M.E.; Civello, P.M.; Martínez, G.A. Effects of ethylene, cytokinin and physical treatments on BoPaO gene expression of harvested broccoli. J. Sci. Food Agric. 2012, 92, 151–158. [Google Scholar] [CrossRef]
- Zou, S.-C.; Zhuo, M.-G.; Abbas, F.; Hu, G.-B.; Wang, H.-C.; Huang, X.-M. Transcription factor LcNAC002 coregulates chlorophyll degradation and anthocyanin biosynthesis in litchi. Plant Physiol. 2023, 192, 1913–1927. [Google Scholar] [CrossRef]
- Jin, Y.; Li, C.; Zhang, S.; Liu, J.; Wang, M.; Guo, Y.; Xu, H.; Ge, Y. Sucrose, cell wall, and polyamine metabolisms involve in preserving postharvest quality of ‘Zaosu’ pear fruit by L-glutamate treatment. Plant Physiol. Biochem. 2024, 208, 108455. [Google Scholar] [CrossRef]
- Onkokesung, N.; Reichelt, M.; van Doorn, A.; Schuurink, R.C.; van Loon, J.J.; Dicke, M. Modulation of flavonoid metabolites in Arabidopsis thaliana through overexpression of the MYB75 transcription factor: Role of kaempferol-3, 7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae. J. Exp. Bot. 2014, 65, 2203–2217. [Google Scholar] [CrossRef]
- Jian, W.; Cao, H.; Yuan, S.; Liu, Y.; Lu, J.; Lu, W.; Li, N.; Wang, J.; Zou, J.; Tang, N. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 2019, 6, 22. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Z.; Li, L.X.; Wang, H.B.; Zhou, H.; Chen, X.S.; Feng, S.Q. Apple MdMYB306-like inhibits anthocyanin synthesis by directly interacting with MdMYB17 and MdbHLH33. Plant J. 2022, 110, 1021–1034. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.; Huang, D.; Xing, W.; Wu, B.; Wei, Q.; Xu, Y.; Zhan, R.; Ma, F.; Song, S. Research progress on the MYB transcription factors in tropical fruit. Trop. Plants 2022, 1, 5. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef]
- Zhou, H.; Peng, Q.; Zhao, J.; Owiti, A.; Ren, F.; Liao, L.; Wang, L.; Deng, X.; Jiang, Q.; Han, Y. Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front. Plant Sci. 2016, 7, 1557. [Google Scholar] [CrossRef]
- Ben-Simhon, Z.; Judeinstein, S.; Nadler-Hassar, T.; Trainin, T.; Bar-Ya’akov, I.; Borochov-Neori, H.; Holland, D. A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 2011, 234, 865–881. [Google Scholar] [CrossRef]
- An, J.P.; Zhang, X.W.; You, C.X.; Bi, S.Q.; Wang, X.F.; Hao, Y.J. MdWRKY 40 promotes wounding-induced anthocyanin biosynthesis in association with Md MYB 1 and undergoes Md BT 2-mediated degradation. New Phytol. 2019, 224, 380–395. [Google Scholar] [CrossRef]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.; Zhang, H.; Liu, J.; Zhang, K. The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef]
- Fournier-Level, A.; Lacombe, T.; Le Cunff, L.; Boursiquot, J.-M.; This, P. Evolution of the VvMybA gene family, the major determinant of berry colour in cultivated grapevine (Vitis vinifera L.). Heredity 2010, 104, 351–362. [Google Scholar] [CrossRef]
- Wang, W.Q.; Moss, S.M.; Zeng, L.; Espley, R.V.; Wang, T.; Lin-Wang, K.; Fu, B.L.; Schwinn, K.E.; Allan, A.C.; Yin, X.R. The red flesh of kiwifruit is differentially controlled by specific activation–repression systems. New Phytol. 2022, 235, 630–645. [Google Scholar] [CrossRef]
- Sun, Q.; Jiang, S.; Zhang, T.; Xu, H.; Fang, H.; Zhang, J.; Su, M.; Wang, Y.; Zhang, Z.; Wang, N. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 2019, 289, 110286. [Google Scholar] [CrossRef]
- Gong, J.; Zeng, Y.; Meng, Q.; Guan, Y.; Li, C.; Yang, H.; Zhang, Y.; Ampomah-Dwamena, C.; Liu, P.; Chen, C. Red light-induced kumquat fruit coloration is attributable to increased carotenoid metabolism regulated by FcrNAC22. J. Exp. Bot. 2021, 72, 6274–6290. [Google Scholar] [CrossRef]
- Dang, Q.; Sha, H.; Nie, J.; Wang, Y.; Yuan, Y.; Jia, D. An apple (Malus domestica) AP2/ERF transcription factor modulates carotenoid accumulation. Hortic. Res. 2021, 8, 223. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, Y.-S.; Han, S.-H.; Lee, B.-D.; Paek, N.-C. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 2015, 27, 1771–1787. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Thrimawithana, A.H.; Dejnoprat, S.; Lewis, D.; Espley, R.V.; Allan, A.C. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019, 221, 309–325. [Google Scholar] [CrossRef]
- Wu, M.; Xu, X.; Hu, X.; Liu, Y.; Cao, H.; Chan, H.; Gong, Z.; Yuan, Y.; Luo, Y.; Feng, B.; et al. SlMYB72 Regulates the Metabolism of Chlorophylls, Carotenoids, and Flavonoids in Tomato Fruit. Plant Physiol. 2020, 183, 854–868. [Google Scholar] [CrossRef]
- Wei, W.; Yang, Y.-Y.; Lakshmanan, P.; Kuang, J.-F.; Lu, W.-J.; Pang, X.-Q.; Chen, J.-Y.; Shan, W. Proteasomal degradation of MaMYB60 mediated by the E3 ligase MaBAH1 causes high temperature-induced repression of chlorophyll catabolism and green ripening in banana. Plant Cell 2023, 35, 1408–1428. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Jeong, J.; Kang, M.-Y.; Kim, J.; Paek, N.-C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef]
- Tak, H.; Negi, S.; Gupta, A.; Ganapathi, T. A stress associated NAC transcription factor MpSNAC67 from banana (Musa x paradisiaca) is involved in regulation of chlorophyll catabolic pathway. Plant Physiol. Biochem. 2018, 132, 61–71. [Google Scholar] [CrossRef]
- Zhang, D.; Quantick, P.C. Effects of chitosan coating on enzymatic browning and decay during postharvest storage of litchi (Litchi chinensis Sonn.) fruit. Postharvest Biol. Technol. 1997, 12, 195–202. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Analysis 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Luo, X.; Li, H.; Wu, Z.; Yao, W.; Zhao, P.; Cao, D.; Yu, H.; Li, K.; Poudel, K.; Zhao, D. The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft-and hard-seeded cultivars. Plant Biotechnol. J. 2020, 18, 955–968. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Shi, B.; Wu, H.; Zhu, W.; Zheng, B.; Wang, S.; Zhou, K.; Qian, M. Genome-Wide Identification and Expression Analysis of WRKY Genes during Anthocyanin Biosynthesis in the Mango (Mangifera indica L.). Agriculture 2022, 12, 821. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| Sample | Raw Reads | Clean Reads | Clean Base (G) | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| S1-1 | 52,424,972 | 51,682,184 | 7.75 | 0.03 | 97.95 | 94.01 | 49.81 |
| S1-2 | 51,211,690 | 50,438,940 | 7.57 | 0.03 | 97.89 | 93.9 | 49.87 |
| S1-3 | 53,622,346 | 52,811,144 | 7.92 | 0.03 | 97.86 | 93.82 | 49.9 |
| S2-1 | 57,381,414 | 55,067,352 | 8.26 | 0.03 | 97.99 | 94.41 | 50.65 |
| S2-2 | 49,502,618 | 48,759,030 | 7.31 | 0.03 | 97.86 | 93.81 | 49.95 |
| S2-3 | 53,041,172 | 50,933,314 | 7.64 | 0.03 | 97.94 | 94.33 | 50.94 |
| S3-1 | 45,436,078 | 44,890,314 | 6.73 | 0.03 | 97.96 | 94.02 | 50 |
| S3-2 | 54,455,452 | 52,331,886 | 7.85 | 0.02 | 98.06 | 94.52 | 50.57 |
| S3-3 | 42,335,952 | 41,696,868 | 6.25 | 0.03 | 98.01 | 94.12 | 49.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Yang, C.; Zhu, W.; Weng, Z.; Li, F.; Teng, Y.; Zhou, K.; Qian, M.; Deng, Q. Transcriptomic Analysis Reveals the Mechanism of Color Formation in the Peel of an Evergreen Pomegranate Cultivar ‘Danruo No.1’ During Fruit Development. Plants 2024, 13, 2903. https://doi.org/10.3390/plants13202903
Wang X, Yang C, Zhu W, Weng Z, Li F, Teng Y, Zhou K, Qian M, Deng Q. Transcriptomic Analysis Reveals the Mechanism of Color Formation in the Peel of an Evergreen Pomegranate Cultivar ‘Danruo No.1’ During Fruit Development. Plants. 2024; 13(20):2903. https://doi.org/10.3390/plants13202903
Chicago/Turabian StyleWang, Xiaowen, Chengkun Yang, Wencan Zhu, Zhongrui Weng, Feili Li, Yuanwen Teng, Kaibing Zhou, Minjie Qian, and Qin Deng. 2024. "Transcriptomic Analysis Reveals the Mechanism of Color Formation in the Peel of an Evergreen Pomegranate Cultivar ‘Danruo No.1’ During Fruit Development" Plants 13, no. 20: 2903. https://doi.org/10.3390/plants13202903
APA StyleWang, X., Yang, C., Zhu, W., Weng, Z., Li, F., Teng, Y., Zhou, K., Qian, M., & Deng, Q. (2024). Transcriptomic Analysis Reveals the Mechanism of Color Formation in the Peel of an Evergreen Pomegranate Cultivar ‘Danruo No.1’ During Fruit Development. Plants, 13(20), 2903. https://doi.org/10.3390/plants13202903









