Cloning and Expression Analysis of ATG8 (Autophagy-Related 8) Gene Family in Solanaceae
Abstract
1. Introduction
2. Results
2.1. Cloning of the ATG8 Gene Family
2.2. Bioinformatics Analysis of the ATG8 Gene Family
2.2.1. Prediction of the Physicochemical Properties and Subcellular Localisation of ATG8 Proteins
2.2.2. Secondary and Tertiary Structure Prediction of ATG8 Proteins
2.2.3. Analysis of ATG8 Protein Phosphorylation Sites, Transmembrane Structures, Signal Peptides, and Conformational Plausibility
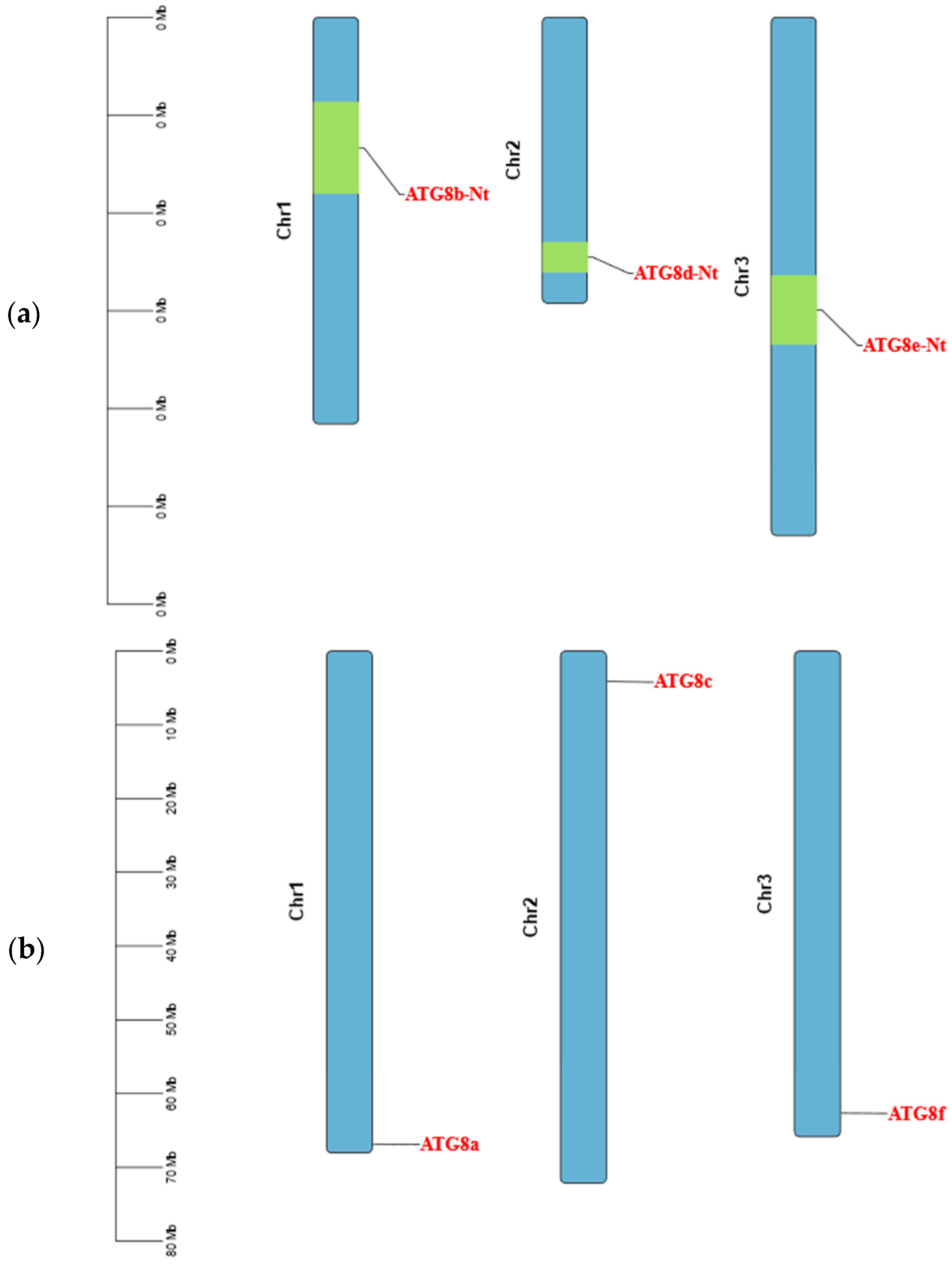
2.2.4. Analysis of ATG8 Gene Structures and Chromosomal Localisation
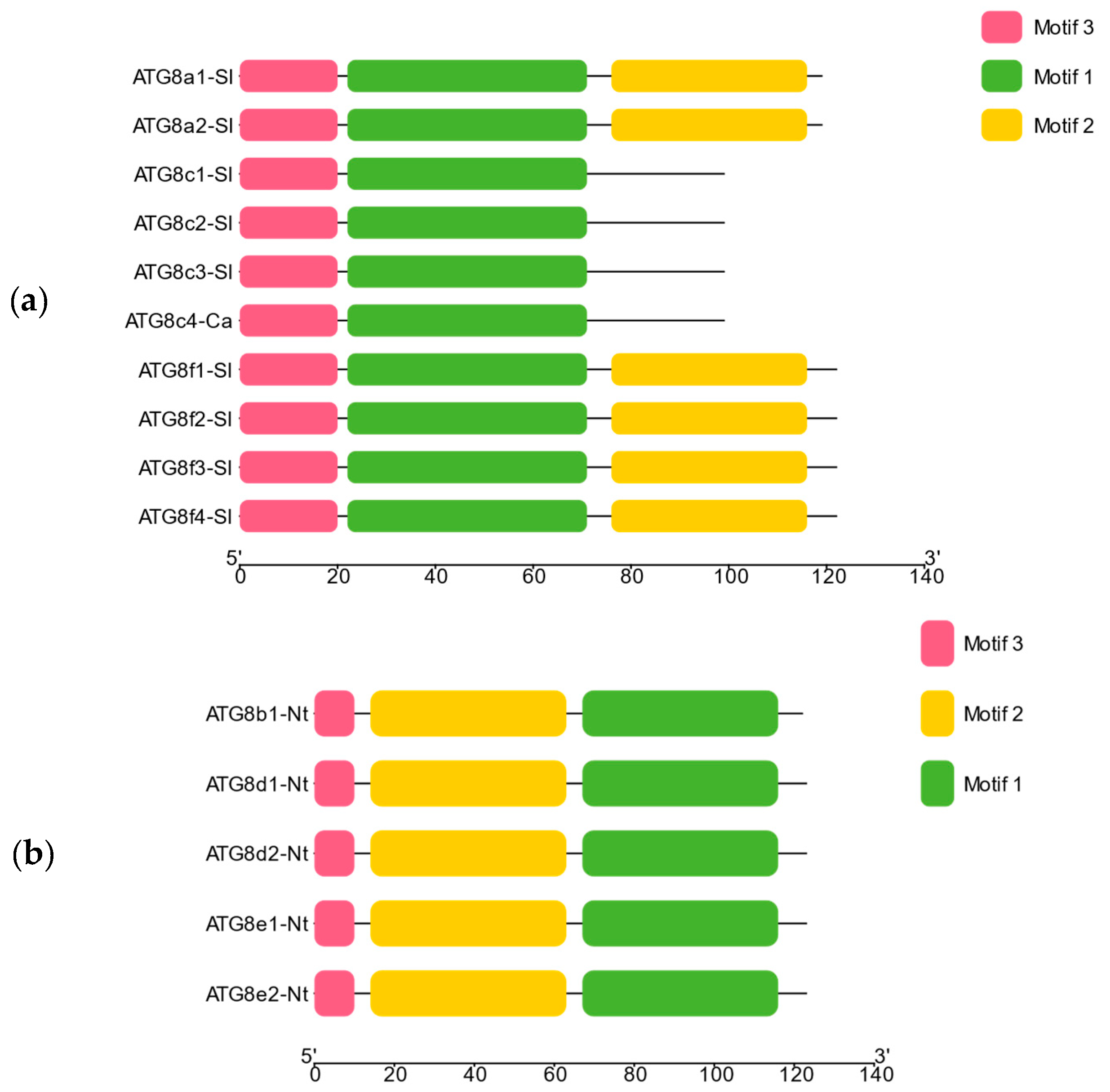
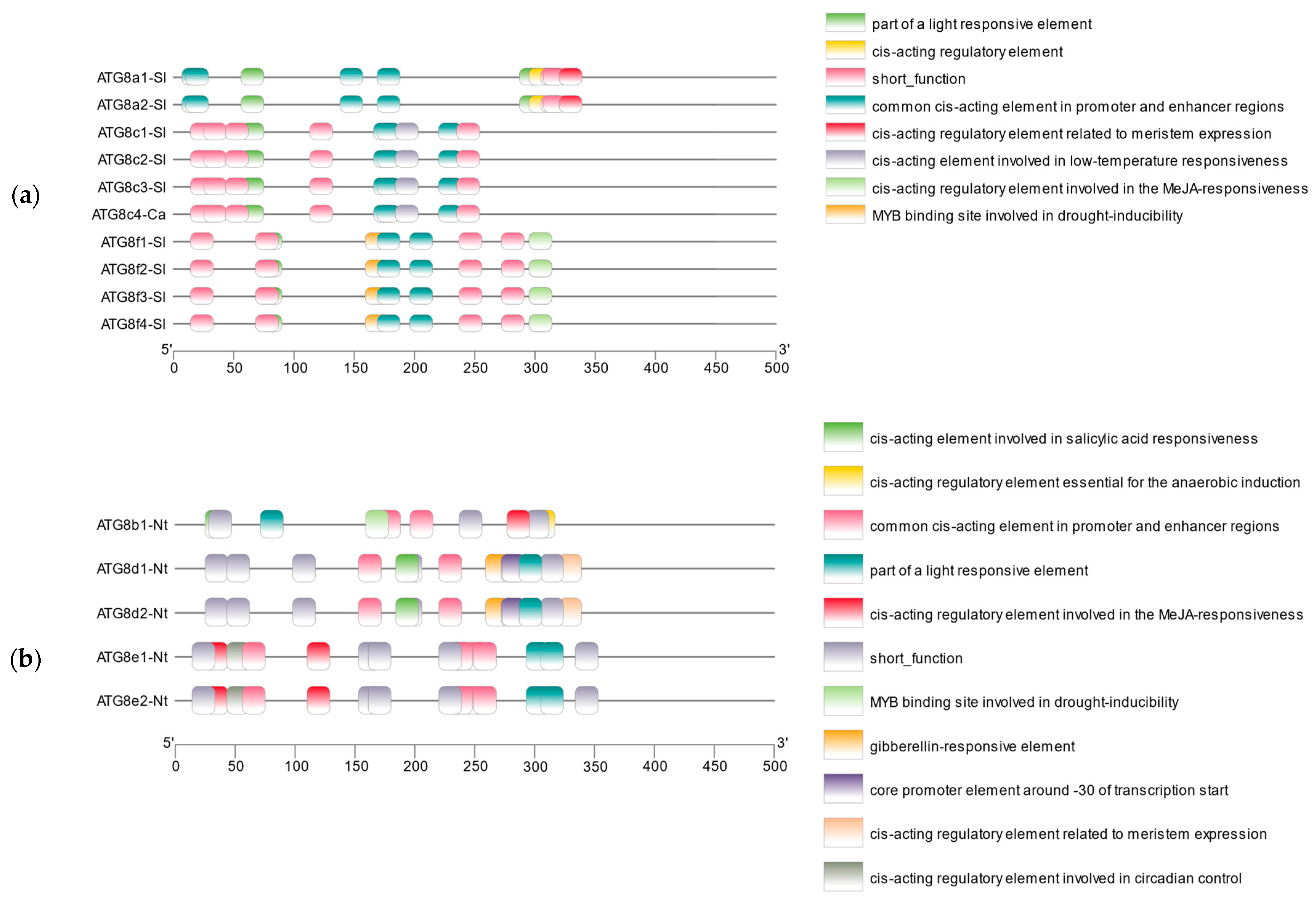
2.2.5. Analysis of Conserved Motifs and Cis-Acting Elements
2.2.6. Phylogenetic Analysis of ATG8 Gene Family
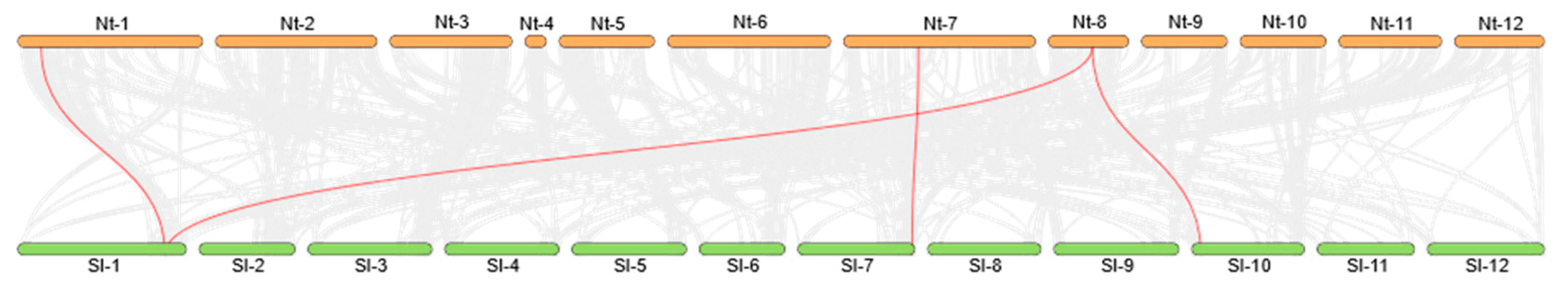
2.2.7. Synteny Analysis
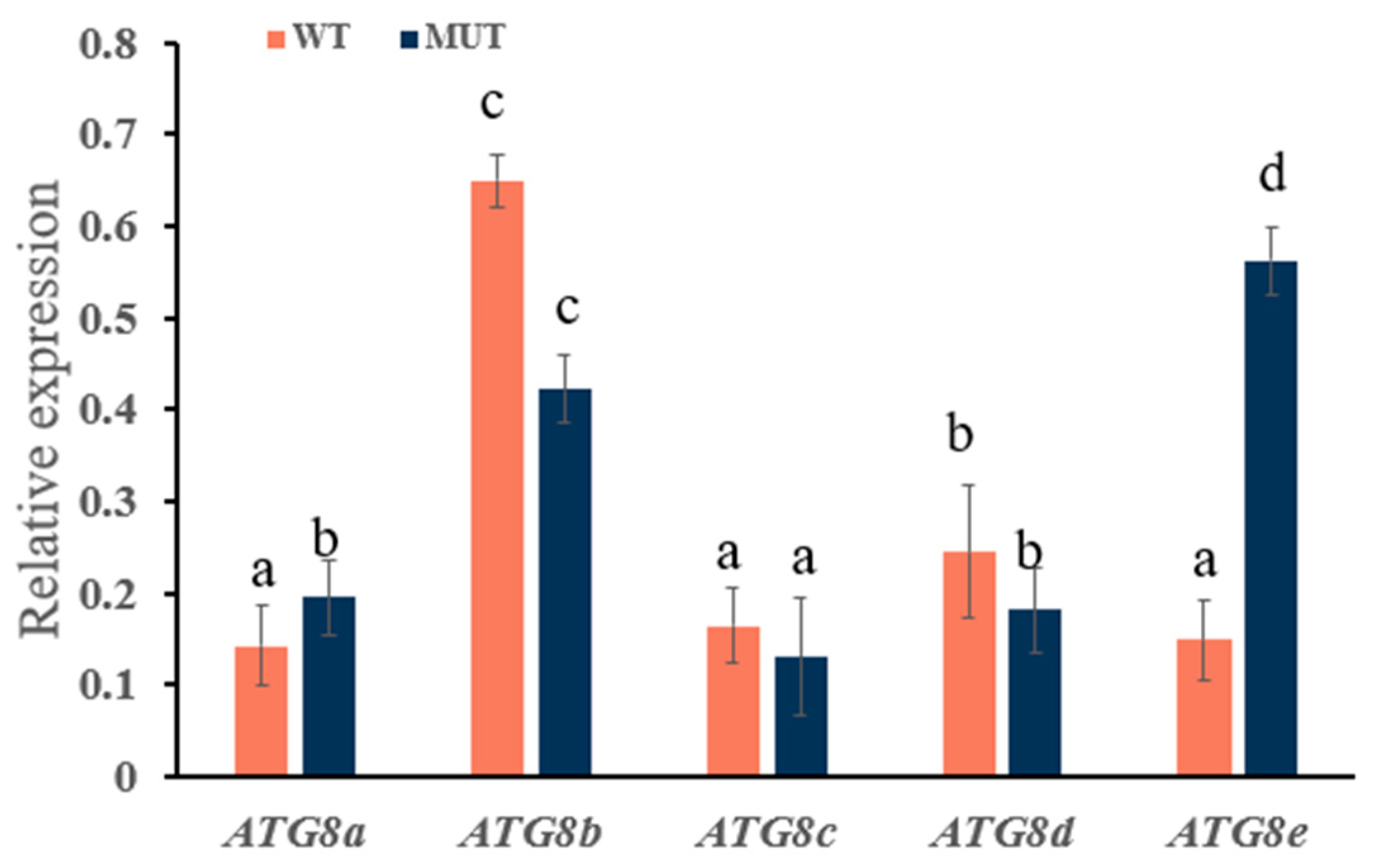
2.3. Analysis of ATG8 Expression in Osthole-Induced Disease Resistance in Pepper and Tomato
2.4. Effect of the Autophagy Inhibitor 3-MA on ATG8 Gene Family Expression
3. Discussion
4. Materials and Methods
4.1. Test Strains and Vectors
4.2. Test Plants
4.3. Reagents and Culture Media
4.4. Cloning of Target Genes
4.4.1. Total RNA Extraction
4.4.2. cDNA Synthesis
4.4.3. RT-PCR
4.5. Bioinformatics Analysis
4.6. ATG8 Expression During Osthole-Induced Disease Resistance in Pepper and Tomato
4.7. Effect of the Autophagy Inhibitor 3-MA on ATG8 Gene Expression
4.8. Statistical Analysis
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cao, J.; Zhou, J. Functions of plant autophagy and prospects for its agricultural applications. Science in China. Life Sci. 2023, 53, 304–321. [Google Scholar]
- Jing, Z.; Liu, N.; Zhang, Z.; Hou, X. Research progress on plant responses to stress combinations in the context of climate change. Plants 2024, 13, 469. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lai, M.; Liang, S.; Wang, X.; Gao, C.; Yang, C. Function and transcriptional regulation of autophagy-related genes in plants. Chin. Bull. Bot. 2021, 56, 201. [Google Scholar]
- Yang, M.; Liu, Y. Autophagy in plant viral infection. FEBS Lett. 2022, 596, 2152–2162. [Google Scholar]
- Liang, M. Functions and Mechanisms of Autophagy Related Gene CaATG6 in Pepper Response to Stresses. Ph.D. Dissertation, Northwest Agriculture and Forestry University, Yangling, China, 2023. [Google Scholar]
- Hu, R.; Yang, X.; Jia, L.; Xiang, Y.; Yue, J.; Wang, H. Virus-mediated expression of GFP-ATG8 for autophagy monitoring in wheat. Acta Agron. Sin. 2021, 47, 2371–2378. [Google Scholar]
- He, Y.; Fu, Q.; Sellick-Jaynes; Li, D.; Ge, S.H. Cloning, prokaryotic expression, purification and bioinformatics analysis of bovine ATG10 gene. Heilongjiang Anim. Husb. Vet. Med. 2023, 11, 15–20. [Google Scholar]
- Gao, W.; Liu, J.; Ma, X.; Shuai, P. Identification and bioinformatics analysis of Chinese fir NAC gene family. J. Cent. S For. Univ. 2022, 42, 108–118. [Google Scholar]
- Yao, Q.; Li, H. CfATG6 and CfATG14 regulate the autophagy and pathogenicity of Colletotrichum fructicola. J. Microbiol. 2024, 64, 1289–1305. [Google Scholar]
- Zhang, W.; Sun, H.; Xing, L.-P.; Wei, X.; Wang, H. Cloning of autophagy-related genes, ATG10s, in Wheat and Their Expression Characteristics Induced by Blumeria graminis. Acta Agron. Sin. 2014, 40, 1392–1402. [Google Scholar]
- Liao, H.; Byeon, I.; Tsai, M. Structure and function of a new phosphopeptide-binding domain containing the FHA2 of Rad53. J. Mol. Biol. 1999, 294, 1041–1049. [Google Scholar]
- Zhang, J.; Liu, J.; Zhou, X.; Liu, S.; Zhuang, Y.; Yang, Y. Identification and expression analysis of autophagy-related gene ATG8 family in Solanum melongena L. Northwest J. Bot. 2023, 43, 1621–1628. (In Chinese) [Google Scholar]
- Li, Y.; Cui, D.; Huang, C.; Sui, X.; Fan, Q.; Chu, X. Preparation of highly specific wheat ATG8 antibody and its application in the detection of autophagy. Crops 2022, 48, 2390–2399. [Google Scholar]
- He, S.; Xiao, Q.; Shi, M.; Li, W.; Peng, Z.; Cheng, J.; Wu, J. Preparation and application of polyclonal antibody against autophagy gene ATG8 in Aedes albopictus. Chin. J. Pathog. Biol. 2023, 18, 787–792. [Google Scholar]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, K.; Hanaoka, H.; Sato, S.; Kato, T.; Tabata, S.; Noda, T.; Ohsumi, Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 2004, 16, 2967–2983. [Google Scholar] [CrossRef]
- Liu, W.; Li, F.; Sun, H.; Wang, Y.; Yu, G.; Wang, F.; Qian, Y.; Yang, J. Tobacco mosaic virus infection on tobacco plants induces autophagy. J. Plant Pathol. 2016, 46, 759–766. [Google Scholar]
- Wang, H.; Ge, F.; Li, J.; Huang, X.; Li, Y.; Hui, G.; Wu, H.; Zhang, G. Applied study of supercritical CO2 fluid extraction in extracting volatile constituents of Cnidium monnieri Seeds. Chin. Med. Mater. 1996, 19, 84–85. [Google Scholar]
- Berkarda, B. Preliminary report on warfarin for the treatment of herpes simplex. Ir. Coll. Phys. Surg. 1978, 22, 56. [Google Scholar]
- Zhiqi, S. A Study on the Antifungal Activity of Osthole. Ph.D. Dissertation, Nanjing Agricultural University, Nanjing, China, 2008. [Google Scholar]
- Chen, Y.H.; Guo, D.S.; Lu, M.H.; Yue, J.Y.; Liu, Y.; Shang, C.M.; An, D.R.; Zhao, M.M. Inhibition effect of osthole from Cnidium monnieri on tobacco mosaic virus (TMV) infection in Nicotiana glutinosa. Molecules 2020, 25, 65. [Google Scholar] [CrossRef]
- Rana, R.M.; Dong, S.; Ali, Z.; Huang, J.; Zhang, H.S. Regulation of ATG6/Beclin-1 homologs by abiotic stresses and hormones in rice (Oryza sativa L.). Genet. Mol. Res. 2012, 11, 3676–3687. [Google Scholar] [CrossRef]
- Chen, Q.; Soulay, F.; Saudemont, B.; Elmayan, T.; Marmagne, A.; Masclaux-Daubresse, C.L. Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain Filling. Plant Cell Physiol. 2019, 60, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Yang, H. Analysis of Transcriptome and ATG Gene Family of Vicia faba at Germination Stage under Drought Stress. MD Dissertation, Changjiang University, Jinzhou, China, 2021. [Google Scholar]
- Wu, R. Identification and Functional Analysis of PmSUSs Gene in Masson Pine (Pinus massoniana Lamb). MD Dissertation, Guizhou University, Guiyang, China, 2022. [Google Scholar]
- Wu, Z.; Chen, W.; Zhao, Z.; Xu, H.; Li, H.; Peng, X.; Chen, D.; Zhang, M. Genome-wide identification and bioinformatics analysis of GRAS gene family in maize. China Agric. Sci. Technol. 2024, 26, 15–25. [Google Scholar]
- Li, F.F.; Zhang, M.; Zhang, C.; Zhou, X. Nuclear autophagy degrades a geminivirus nuclear protein to restrict viral infection in solanaceous plants. New Phytol. 2020, 225, 1746–1761. [Google Scholar] [CrossRef]
- Guo, D. Research on Honokiol and Bacillus velezensis for TMV Control. MD Dissertation, Northwest Agriculture and Forestry University, Yangling, China, 2020. [Google Scholar]
- Chen, X. Bioinformatics analysis of tomato OFP gene family. Jiangsu Agric. Sci. 2021, 49, 39–48. [Google Scholar]
- Cai, Y.; Dai, D.; Tan, H.; Wang, D.; Yang, F.; Wang, L.; Sheng, Y. Analysis of genetic structure and construction of gene editing vector of melon AMS gene. Xinjiang Agric. Sci. 2023, 60, 61–68. [Google Scholar]
- Wu, M.; Tan, X.; Zhou, R.; Zhan, W.; Hu, X. Cloning and sequence analysis of full-length cDNA of EMF2 gene in Camellia oleifera. Econ. For. Res. 2013, 31, 7–12. [Google Scholar]
- Zhang, C.; Deng, Y.; Huang, S.; Wu, Y.; Zhang, G.; Man, B.; Li, D. Cloning and expression of nitrite reductase gene HcNiR in kenaf. Fujian Agric. J. 2022, 37, 600–608. [Google Scholar]
- Shao, Y.; Zhong, J.; Lu, Y.; Song, M.; Chen, Y. Identification and molecular evolutionary analysis of viruses causing egg-plant mottle crinkle disease. Jiangsu J. Agric. Sci. 2023, 39, 674–682. [Google Scholar]
- Chen, T.; Yang, C.; Yu, G.; Luo, M.; Zhang, X.; Lyu, L.; Chen, Q. Effect of Autophagy-related Gene PlATG12 on the Growth, Development and Pathogenicity of Peronophythora litchii. J. Agric. Biotechnol. 2024, 32, 2137–2149. [Google Scholar]
- Zhang, Y.; Zhou, S.; Sun, X. Cloning and biological information analysis of tomato SlFHA1 gene. J. Shandong Agric. Univ. 2023, 54, 828–834. [Google Scholar]
- Zhou, S.; Hong, Q.; Li, Y.; Li, Q.; Wang, M. Autophagy contributes to regulate the ROS levels and PCD progress in TMV-infected tomatoes. Plant Sci. 2018, 269, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using Real Time Quantitive PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]




| Gene | Protein Length (KDa) | Molecular Weight (Mr/103) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathy | Subcellular Localisation |
|---|---|---|---|---|---|---|---|
| ATG8a1-Sl | 119 | 13,744.94 | 8.78 | 40.52 | 84.45 | −0.361 | Nucleus |
| ATG8a2-Sl | 119 | 13,744.94 | 8.78 | 40.52 | 84.45 | −0.361 | Nucleus |
| ATG8b1-Nt | 122 | 14,031.14 | 6.61 | 32.69 | 92.70 | −0.418 | Chloroplast |
| ATG8c1-Sl | 99 | 11,428.18 | 7.93 | 44.53 | 93.54 | −0.415 | Nucleus |
| ATG8c2-Sl | 99 | 11,428.18 | 7.93 | 44.53 | 93.54 | −0.415 | Nucleus |
| ATG8c3-Sl | 99 | 11,428.18 | 7.93 | 44.53 | 93.54 | −0.415 | Nucleus |
| ATG8c4-Ca | 99 | 11,428.18 | 7.93 | 44.53 | 93.54 | −0.415 | Nucleus |
| ATG8d1-Nt | 123 | 14,109.19 | 6.61 | 35.45 | 86.42 | −0.385 | Cytoplasm |
| ATG8d2-Nt | 123 | 14,109.19 | 6.61 | 35.45 | 86.42 | −0.385 | Plasma membrane |
| ATG8e1-Nt | 123 | 14,107.26 | 6.61 | 43.04 | 90.41 | −0.335 | Chloroplast |
| ATG8e2-Nt | 123 | 14,107.26 | 6.61 | 43.04 | 90.41 | −0.335 | Chloroplast |
| ATG8f1-Sl | 122 | 13,930.08 | 7.85 | 35.19 | 94.34 | −0.331 | Mitochondria Peroxisome |
| ATG8f2-Sl | 122 | 13,930.08 | 7.85 | 35.19 | 94.34 | −0.331 | Nucleus |
| ATG8f3-Sl | 122 | 13,930.08 | 7.85 | 35.19 | 94.34 | −0.331 | Nucleus |
| ATG8f4-Sl | 122 | 13,930.08 | 7.85 | 35.19 | 94.34 | −0.331 | Nucleus |
| Gene | Serine | Threonine | Tyrosine |
|---|---|---|---|
| ATG8a1-Sl | 4 | 1 | 1 |
| ATG8a2-Sl | 4 | 3 | 0 |
| ATG8b1-Nt | 4 | 1 | 0 |
| ATG8c1-Sl | 8 | 3 | 0 |
| ATG8c2-Sl | 8 | 3 | 0 |
| ATG8c3-Sl | 8 | 3 | 0 |
| ATG8c4-Ca | 15 | 3 | 0 |
| ATG8d1-Nt | 8 | 2 | 0 |
| ATG8d2-Nt | 7 | 3 | 0 |
| ATG8e1-Nt | 6 | 1 | 1 |
| ATG8e2-Nt | 5 | 2 | 1 |
| ATG8f1-Sl | 10 | 9 | 0 |
| ATG8f2-Sl | 11 | 0 | 0 |
| ATG8f3-Sl | 9 | 0 | 0 |
| ATG8f4-Sl | 9 | 0 | 0 |
| Gene | Most Favoured Regions (%) | Additional Allowed Regions (%) | Generously Allowed Regions (%) | Disallowed Regions (%) |
|---|---|---|---|---|
| ATG8a1-Sl | 88.0 | 12.0 | - | - |
| ATG8a2-Sl | 88.0 | 12.0 | - | - |
| ATG8b1-Nt | 95.5 | 4.5 | - | - |
| ATG8c1-Sl | 55.0 | 40.0 | 5.0 | - |
| ATG8c2-Sl | 55.0 | 40.0 | 5.0 | - |
| ATG8c3-Sl | 55.0 | 40.0 | 5.0 | - |
| ATG8c4-Ca | 92.7 | 2.8 | - | - |
| ATG8d1-Nt | 95.7 | 4.3 | - | - |
| ATG8d2-Nt | 94.6 | 5.4 | - | - |
| ATG8e1-Nt | 95.8 | 4.2 | - | - |
| ATG8e2-Nt | 96.9 | 3.1 | - | - |
| ATG8f1-Sl | 100.0 | - | - | - |
| ATG8f2-Sl | 88.6 | 8.6 | - | 2.9 |
| ATG8f3-Sl | 86.1 | 11.1 | 2.8 | - |
| ATG8f4-Sl | 86.1 | 11.1 | 2.8 | - |
| Name of Primer | Sequence | Gene ID |
|---|---|---|
| ATG8a-F | GGATGCTTTTCCACTC | JF304784 |
| ATG8a-R | GTGAAGAAACAGGATACCATC | |
| ATG8b-F | GTTAAGAGCTCATTCAAGCAGGA | KR336565 |
| ATG8b-R | TCCCCGAATGTGTTTTCTCCA | |
| ATG8c-F | GAGGAGGCAGGCAGAATCTT | MK189279 |
| ATG8c-R | ACCCAAATGTATTTTCGCCGC | |
| ATG8d-F | TGGCCAAGAGTTCTTTCAAGC | KR336567 |
| ATG8d-R | CCAAGCTCAAGGAACCCAAAAG | |
| ATG8e-F | TGAACACCCCATGGAGAGGA | KR336568 |
| ATG8e-R | GGAACCCAAATGTATTTTCT | |
| ATG8f-F | GGCTAAGAGCTCATTCAAGCA | NM_001247705 |
| ATG8f-R | CTACAGTTCGCTCAGGACCCCGAA |
| Name of Primer | Sequence | Reference |
|---|---|---|
| ATG8a-F | CCTGCTGATCTGACTGTGGG | [37] |
| ATG8a-R | CTGTCGGAGGAAGGATATTTTTC | |
| ATG8b-F | CAGTTGGGCAATTTGTCTATGTC | |
| ATG8b-R | TTCAGGTCCCCGAATGTGTT | |
| ATG8c-F | TATTCCCAACATTGACAAGAAAAAG | |
| ATG8c-R | TGACGTAGACAAACTGCCCCA | |
| ATG8d-F | CATCCGAGAGAAGTATCCCGA | |
| ATG8d-R | CAGACAACAGAGCAGCCGTG | |
| ATG8e-F | TTTGGAGAGGAGGCAGGCA | |
| ATG8e-R | CAGACATCAGAGCAGCAGTTGG | |
| ATG8a-F | CCTGCTGATCTGACTGTGGG | |
| ATG8a-R | CTGTCGGAGGAAGGATATTTTTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Lu, Y.; Dong, S.; Yang, C.; Yang, S. Cloning and Expression Analysis of ATG8 (Autophagy-Related 8) Gene Family in Solanaceae. Plants 2024, 13, 2924. https://doi.org/10.3390/plants13202924
Chen Y, Lu Y, Dong S, Yang C, Yang S. Cloning and Expression Analysis of ATG8 (Autophagy-Related 8) Gene Family in Solanaceae. Plants. 2024; 13(20):2924. https://doi.org/10.3390/plants13202924
Chicago/Turabian StyleChen, Yahan, Yunshuang Lu, Shibo Dong, Chengde Yang, and Shunyi Yang. 2024. "Cloning and Expression Analysis of ATG8 (Autophagy-Related 8) Gene Family in Solanaceae" Plants 13, no. 20: 2924. https://doi.org/10.3390/plants13202924
APA StyleChen, Y., Lu, Y., Dong, S., Yang, C., & Yang, S. (2024). Cloning and Expression Analysis of ATG8 (Autophagy-Related 8) Gene Family in Solanaceae. Plants, 13(20), 2924. https://doi.org/10.3390/plants13202924






