ND-FISH with New Oligo Probes for Chromosome Identification of Cichorium intybus Revealing Karyotypic Variation and Divergence of Asteraceae Species
Abstract
:1. Introduction
2. Results
2.1. Genomic Distribution of TRs in Chicory and Lettuce Genomes
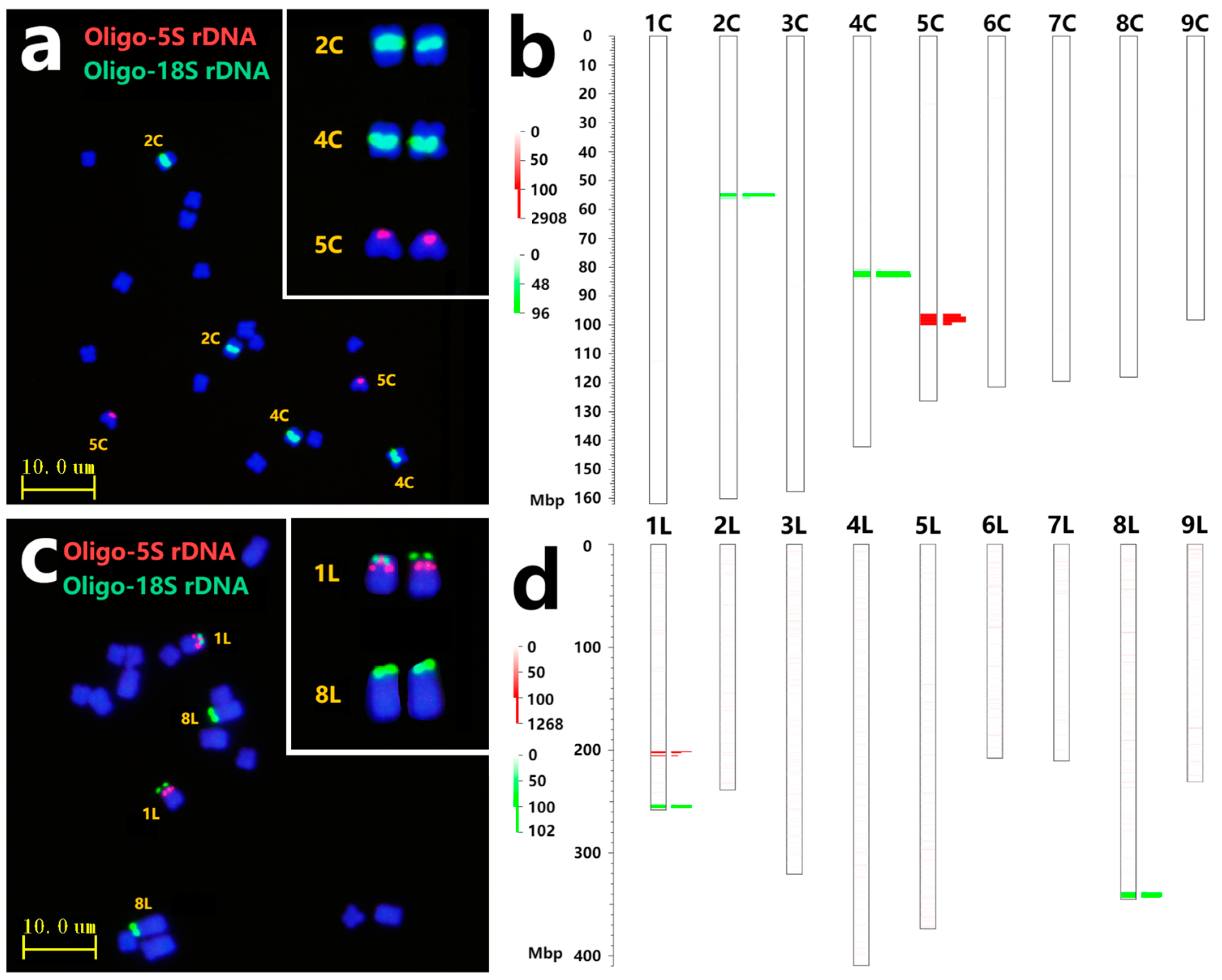
2.2. Chromosomal Location of rDNA Site and FISH Validation
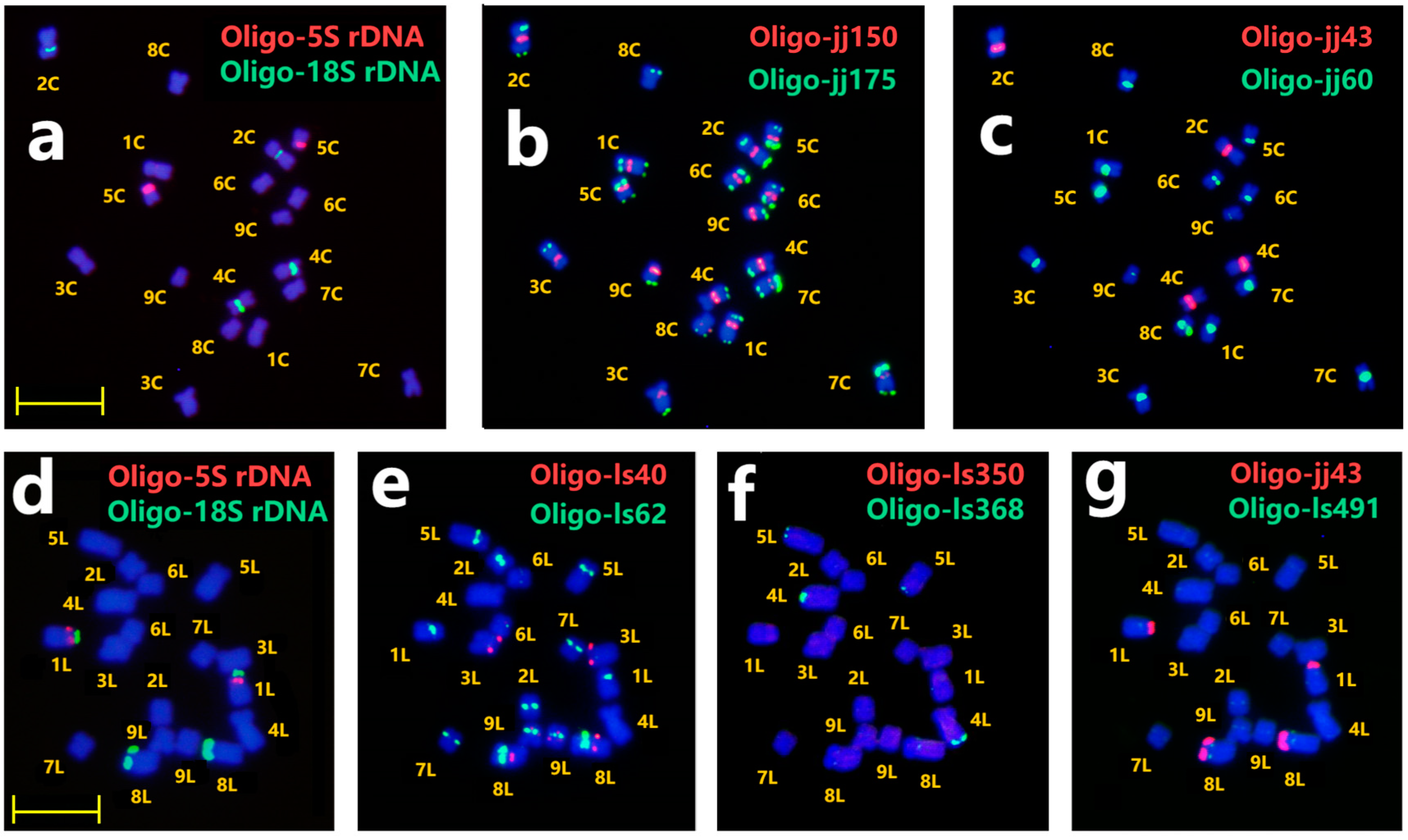
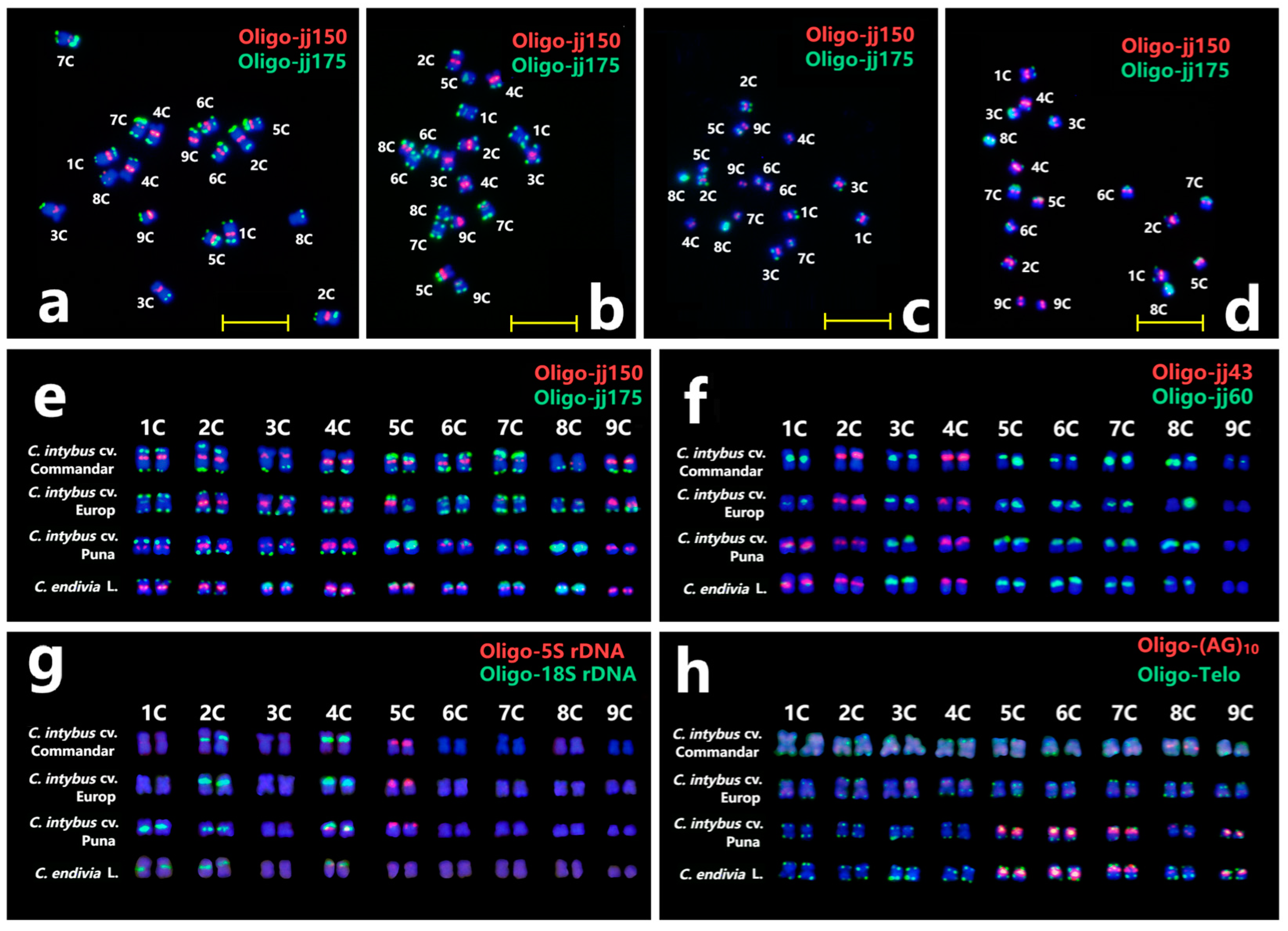
2.3. ND-FISH of New TR Probes for Chromosome Identification in Cichorium intybus cv. Commander and Lettuce
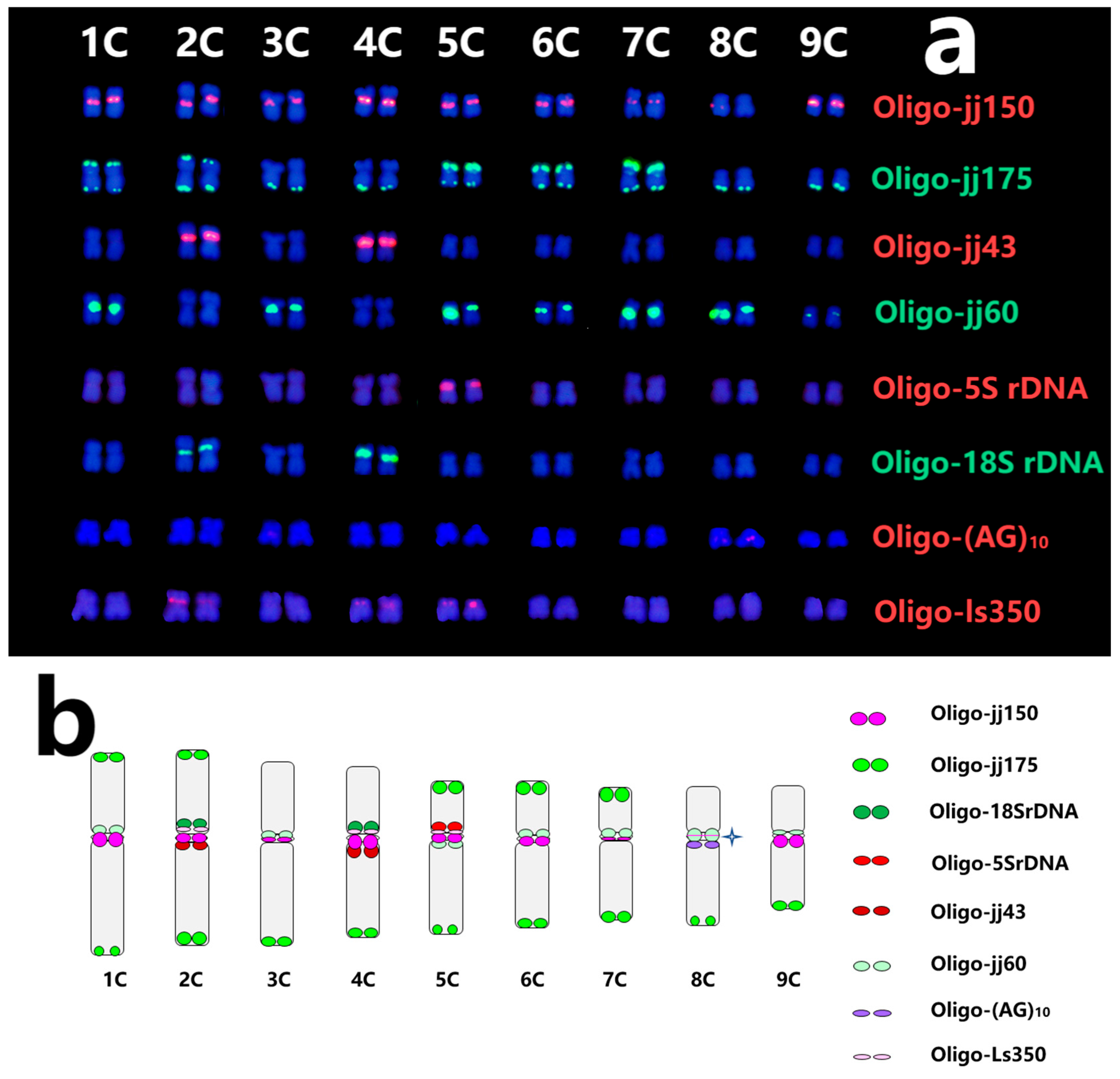
2.4. Standard Karyotype of C. intybus cv. Commander and Lettuce
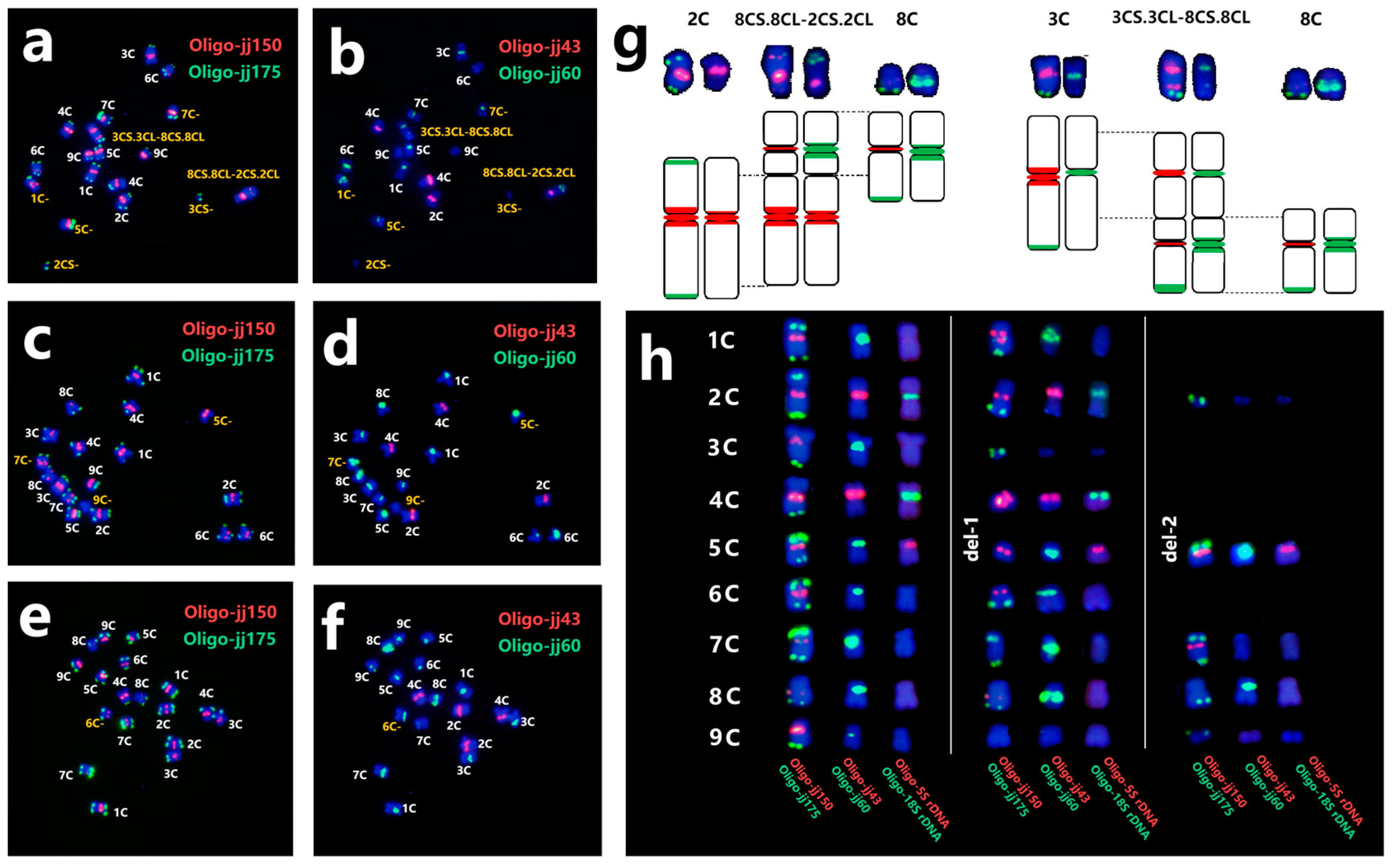
2.5. Chromosome Variation in C. intybus cv. Commander
2.6. Comparative Karyotypic Analysis of Several Cichorium-Related Species
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Bioinformatic Analysis of Tandem Repeats in Chicory and Lettuce Genomes
4.3. Chromosome Preparation and FISH Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Song, C.; Liu, Y.F.; Song, A.P.; Chen, S.L. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of chrysanthemum flowers and medicinal traits. Mol. Plant 2018, 11, 1482–1491. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Qin, Y.J.; Wang, R.; Yang, X.Z. Comparative genomics reveals a unique nitrogen-carbon balance system in Asteraceae. Nat. Commun. 2023, 14, 4334. [Google Scholar] [CrossRef]
- Garcia-Jacas, N.; Susanna, A. Cardueae (Carduoideae). In Systematics, Evolution, and Biogeography of Compositae; Funk, V., Ed.; International Association for Plant Taxonomy: Vienna, Austria, 2009; pp. 293–313. [Google Scholar]
- Bernardes, E.C.S.; Vasconcelos, S.; Carvalho, R.; Brasileiro-Vidal, A.C. Intra- and interspecific chromosome polymorphisms in cultivated Cichorium L. species (Asteraceae). Genet. Mol. Biol. 2013, 36, 357–364. [Google Scholar] [CrossRef]
- Can, M.; Ayan, İ.; Acar, Z.; Başaran, U. Forage Chicory (Cichorium intybus L.): A review. In Innovative Approaches in Meadow-Rangeland and Forage Crops; Seydosoglu, S., Ed.; Iksad Publications: Golbasi, Turkey, 2020; pp. 47–84. [Google Scholar]
- Duda, Ł.; Kłosiński, K.K.; Budryn, G.; Jaśkiewicz, A.; Kołat, D. Medicinal use of chicory (Cichorium intybus L.). Sci. Pharm. 2024, 92, 31. [Google Scholar] [CrossRef]
- Qadia, I.; Ovais, S.; Zargar, M.I.; Rehman, M.U. Cichorium intybus: A comprehensive review on its pharmacological activity and phytochemistry. In Edible Plants in Health and Diseases; Masoodi, M.H., Rehman, M.U., Eds.; Springer: Singapore, 2022; pp. 373–398. [Google Scholar]
- Rick, C.M. Hybridization between chicory and endive. Proc. Am. Soc. Hortic. Sci. 1953, 61, 459–466. [Google Scholar]
- Matoba, H.; Mizutani, T.; Yoshikazu, H.; Uchiyama, H. Chromosomal study of lettuce and its allied species (Lactuca spp., Asteraceae) by means of karyotype analysis and fluorescence in situ hybridization. Hereditas 2007, 144, 235–243. [Google Scholar] [CrossRef]
- Ge, R.C.; Zhao, M.L.; Gao, H.W.; Li, G.L. Study on C-banding and karyotype analysis of Puna chicory. Acta Agrestia Sin. 2002, 10, 190–193. [Google Scholar] [CrossRef]
- Imani, S.; Rahbarian, R.; Masoumi, A.; Khavari Khorasani, S. Cytogenetical study of different ecotypes of Cichorium intybus L. Crop Biotechnol. 2014, 3, 25–34. [Google Scholar]
- Jiang, J.; Gill, B.S. Nonisotopic in situ hybridization and plant genome mapping: The first 10 years. Genome 1994, 37, 717–725. [Google Scholar] [CrossRef]
- Kato, A.; Lamb, J.C.; Birchler, J.A. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 2004, 101, 13554–13559. [Google Scholar] [CrossRef]
- Younis, A.; Ramzan, F.; Hwang, Y.-J.; Lim, K.-B. FISH and GISH: Molecular cytogenetic tools and their applications in ornamental plants. Plant Cell Rep. 2015, 34, 1477–1488. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J. Fluorescence in situ hybridization in plants recent developments and future applications. Chromosome Res. 2019, 27, 153–165. [Google Scholar] [CrossRef]
- Lang, T.; Li, G.R.; Wang, H.J.; Yu, Z.J.; Chen, Q.H. Physical location of tandem repeats in the wheat genome and application for chromosome identification. Planta 2018, 249, 663–675. [Google Scholar] [CrossRef]
- Tang, S.Y.; Tang, Z.X.; Qiu, L.; Yang, Z.J.; Li, G.R. Developing new oligo probes to distinguish specific chromosomal segments and the A, B, D genomes of wheat (Triticum aestivum L.) using ND-FISH. Front. Plant Sci. 2018, 9, 1104. [Google Scholar] [CrossRef]
- Fan, W.; Wang, S.; Wang, H.C.; Wang, A.Q.; Jiang, F. The genomes of chicory, endive, great burdock and yacon provide insights into Asteraceae palaeo-polyploidization history and plant inulin production. Mol. Ecol. Resour. 2022, 22, 3124–3140. [Google Scholar] [CrossRef]
- Wang, K.; Jin, J.Y.; Wang, J.X.; Wang, X.R.; Guo, L. The complete telomere-to-telomere genome assembly of lettuce. Plant Commun. 2024, 5, 101011. [Google Scholar] [CrossRef]
- Cao, S.; Sawettalake, N.; Shen, L.S. Gapless genome assembly and epigenetic profiles reveal gene regulation of whole-genome triplication in lettuce. GigaScience 2024, 13, giae043. [Google Scholar] [CrossRef]
- Waegneer, E.; Rombauts, S.; Baert, J.; Ruttink, T. Industrial chicory genome gives insights into the molecular timetable of anther development and male sterility. Front. Plant Sci. 2023, 14, 1181529. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, Z.W.; Han, X.Y.; Li, D.Y. The chromosome-scale assembly of endive (Cichorium endivia) genome provides insights into the sesquiterpenoid biosynthesis. Genomics 2022, 114, 110400. [Google Scholar] [CrossRef]
- Jiang, W.X.; Jiang, C.Z.; Yuan, W.G.; Yang, Z.J. A universal karyotypic system for hexaploid and diploid Avena species brings oat cytogenetics into the genomics era. BMC Plant Biol. 2021, 21, 213. [Google Scholar] [CrossRef]
- Gupta, R.C.; Bala, S.; Suruchi. Cytogenetical studies in seven ornamental species of Chrysanthemum (Asteraceae). Cytologia 2013, 78, 439–448. [Google Scholar] [CrossRef]
- Miranda Zanetti, J.; Greizerstein, E.; Camadro, E.; Poverene, M. Genomic relationships between hexaploid Helianthus resinosus and diploid Helianthus annuus (Asteraceae). Plant Syst. Evol. 2014, 300, 1071–1078. [Google Scholar] [CrossRef]
- Jiang, J.M.; Gill, B.S. Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 2006, 49, 1057–1068. [Google Scholar] [CrossRef]
- Pathak, D.; Ali, S. Repetitive DNA: A tool to explore animal genomes/transcriptomes. In Functional Genomics; Germana, M., Francesca, P., Eds.; Intech: Rijeka, Croatia, 2012; pp. 157–180. [Google Scholar] [CrossRef]
- Martinez-Zapater, J.M.; Estelle, M.A.; Somerville, C.R. A highly repeated DNA sequence in Arabidopsis thaliana. Mol. Gen. Genet. 1986, 204, 417–423. [Google Scholar] [CrossRef]
- Cheng, Z.K.; Dong, F.G.; Langdon, T.; Jiang, J.M. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 2002, 14, 1691–1704. [Google Scholar] [CrossRef]
- Lee, H.R.; Zhang, W.L.; Langdon, T.; Jiang, J.M. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. USA 2005, 102, 11793–11798. [Google Scholar] [CrossRef]
- Zhao, J.; Hao, W.W.; Tang, C.G.; Zhang, X.Y. Plasticity in Triticeae centromere DNA sequences: A wheat × tall wheatgrass (decaploid) model. Plant J. 2019, 100, 314–327. [Google Scholar] [CrossRef]
- Wang, Y.X.; Jha, A.K.; Chen, R.J.; Yang, M. Polyploidy-associated genomic instability in Arabidopsis thaliana. Genesis 2010, 48, 254–263. [Google Scholar] [CrossRef]
- Selmecki, A.M.; Maruvka, Y.E.; Richmond, P.A.; Pellman, D. Polyploidy can drive rapid adaptation in yeast. Nature 2015, 519, 349–352. [Google Scholar] [CrossRef]
- Mota, L.; Torices, R.; Loureiro, J. The evolution of haploid chromosome numbers in the sunflower family. Genome Biol. Evol. 2016, 8, 3516–3528. [Google Scholar] [CrossRef]
- He, Y.; He, J.; Zhao, Y.; Chen, F.D. Divergence of 10 satellite repeats in Artemisia (Asteraceae Anthemideae) based on sequential fluorescence in situ hybridization analysis evidence for species identification and evolution. Chromosome Res. 2024, 32, 5. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Sung, S.; Kim, J.S.; Lee, J. Telomeres reforged with non-telomeric sequences in mouse embryonic stem cells. Nat. Commun. 2021, 12, 1097. [Google Scholar] [CrossRef]
- Tan, K.T.; Slevin, M.K.; Leibowitz, M.L.; Meyerson, M. Neotelomeres and telomere-spanning chromosomal arm fusions in cancer genomes revealed by long-read sequencing. Cell Genom. 2024, 4, 100588. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Cuadrado, Á.; Jouve, N. Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 2010, 119, 495–503. [Google Scholar] [CrossRef]
- Sýkorová, E.; Lim, K.Y.; Fajkus, J. The signature of the Cestrum genome suggests an evolutionary response to the loss of (TTTAGGG)n telomeres. Chromosoma 2003, 112, 164–172. [Google Scholar] [CrossRef]
- Han, F.P.; Lamb, J.C.; Birchler, J.A. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. USA 2006, 103, 3238–3243. [Google Scholar] [CrossRef]
- Fu, S.L.; Chen, L.; Wang, Y.Y.; Tang, Z.X. Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci. Rep. 2015, 5, 10552. [Google Scholar] [CrossRef]
| Chromosome | TR Total Length (bp) | Chromosome Length (bp) | TR Content (%) | Chromosome | TR Total Length (bp) | Chromosome Length (bp) | TR Content (%) |
|---|---|---|---|---|---|---|---|
| 1C | 10,840,173 | 161,702,566 | 6.7 | 1L | 4,077,116 | 258,130,948 | 1.58 |
| 2C | 9,312,424 | 159,989,680 | 5.82 | 2L | 629,559 | 238,535,665 | 0.26 |
| 3C | 9,318,956 | 157,584,665 | 5.91 | 3L | 598,133 | 320,657,391 | 0.19 |
| 4C | 9,394,122 | 142,005,817 | 6.61 | 4L | 2,718,271 | 409,415,085 | 0.66 |
| 5C | 10,118,225 | 126,222,701 | 8.02 | 5L | 2,281,982 | 373,650,124 | 0.61 |
| 6C | 6,982,906 | 121,338,966 | 5.75 | 6L | 351,418 | 207,794,748 | 0.17 |
| 7C | 9,970,721 | 119,386,547 | 8.35 | 7L | 641,683 | 210,595,814 | 0.3 |
| 8C | 10,811,047 | 117,937,712 | 9.17 | 8L | 1,531,263 | 345,107,305 | 0.44 |
| 9C | 7,606,651 | 98,217,271 | 7.74 | 9L | 1,174,833 | 229,575,853 | 0.51 |
| Total | 84,355,225 | 1,204,385,925 | 7.00 | Total | 14,004,258 | 2,593,462,933 | 0.54 |
| Oligo Probes | Sequences | Reference |
|---|---|---|
| Oligo-(AG)10 | AGAGAGAGAGAGAGAGAGAG | [39] |
| Oligo-Telo | TTTAGGGTTTAGGGTTTAGGG | [40] |
| Oligo-5S rDNA | TCAGAACTCCGAAGTTAAGCGTGCTTGGGCGAGAGTAGTAC | [16] |
| Oligo-18S rDNA | GGGCAAGTCTGGTGCCAGCAGCCGCGGT | [16] |
| Oligo-jj43 | GACCCATGGACGACCAGGTGAACAAGGCACCGTGCCATACACA | This study |
| Oligo-jj60 | ACCCTAATACATCATATGAAAACTAAAAAACCCTAATCCATCATATG | This study |
| Oligo-jj150 | GATGGGTTATGGTCAACCAGGTAGGGTTGTGATGGGTTATGGTCAACC | This study |
| Oligo-jj175 | TACACCAGAAATGTTTAAAATGATCACAAAGTGTAATTATAGGTAATTTAT | This study |
| Oligo-ls40 | CACAACCAACACCCATACATGACCTAATGAGGTCCAAACC | This study |
| Oligo-ls62 | AATTTAGCAAAGCTTTGATGAGCATTGTTTAATTCTTGAGTTCCAC | This study |
| Oligo-ls350 | GTACGCGGGGCGTGGGACCCACGTGACCGATCGGCTCCGTTCC | This study |
| Oligo-ls368 | AGAATGAGAACATAATAATGTTGATATAAAGTTGCTAA | This study |
| Oligo-ls491 | ATCATGAGTTCCTTACGCAAATATCGCCATAATTGTTGT | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.; Jiang, C.; Huang, D.; Zheng, Z.; Yang, W.; Li, G.; Fu, C.; Liao, H.; Long, W.; Yang, Z.; et al. ND-FISH with New Oligo Probes for Chromosome Identification of Cichorium intybus Revealing Karyotypic Variation and Divergence of Asteraceae Species. Plants 2024, 13, 3135. https://doi.org/10.3390/plants13223135
Chen M, Jiang C, Huang D, Zheng Z, Yang W, Li G, Fu C, Liao H, Long W, Yang Z, et al. ND-FISH with New Oligo Probes for Chromosome Identification of Cichorium intybus Revealing Karyotypic Variation and Divergence of Asteraceae Species. Plants. 2024; 13(22):3135. https://doi.org/10.3390/plants13223135
Chicago/Turabian StyleChen, Meiling, Chengzhi Jiang, Doudou Huang, Zhiqiang Zheng, Wenzhuo Yang, Guangrong Li, Chun Fu, Hong Liao, Wencong Long, Zujun Yang, and et al. 2024. "ND-FISH with New Oligo Probes for Chromosome Identification of Cichorium intybus Revealing Karyotypic Variation and Divergence of Asteraceae Species" Plants 13, no. 22: 3135. https://doi.org/10.3390/plants13223135
APA StyleChen, M., Jiang, C., Huang, D., Zheng, Z., Yang, W., Li, G., Fu, C., Liao, H., Long, W., Yang, Z., & Yang, Y. (2024). ND-FISH with New Oligo Probes for Chromosome Identification of Cichorium intybus Revealing Karyotypic Variation and Divergence of Asteraceae Species. Plants, 13(22), 3135. https://doi.org/10.3390/plants13223135







