Transcriptomic Profiling Unravels the Disruption of Photosynthesis Apparatuses and Induction of Immune Responses by a Bipartite Begomovirus in Tomato Plants
Abstract
:1. Introduction
2. Results
2.1. Phenotypes of Control and TYLCTHV-Infected Plants
2.2. RNA-Seq and Genome Mapping
2.3. Analysis and Functional Annotation of Differentially Expressed Genes (DEGs)
2.4. Functional Enrichment of DEGs
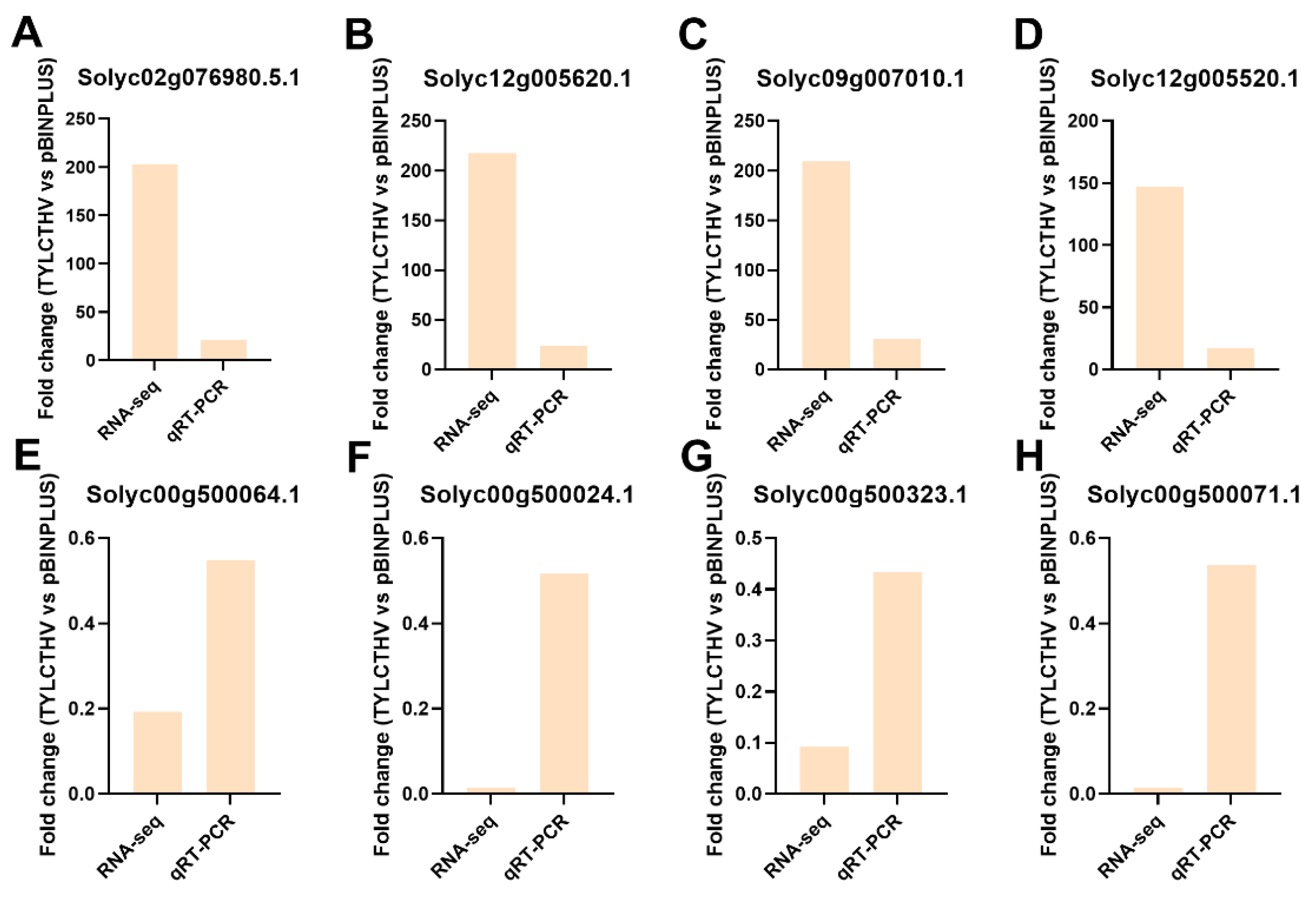
2.5. Qrt-PCR Verification of Transcriptomic Data
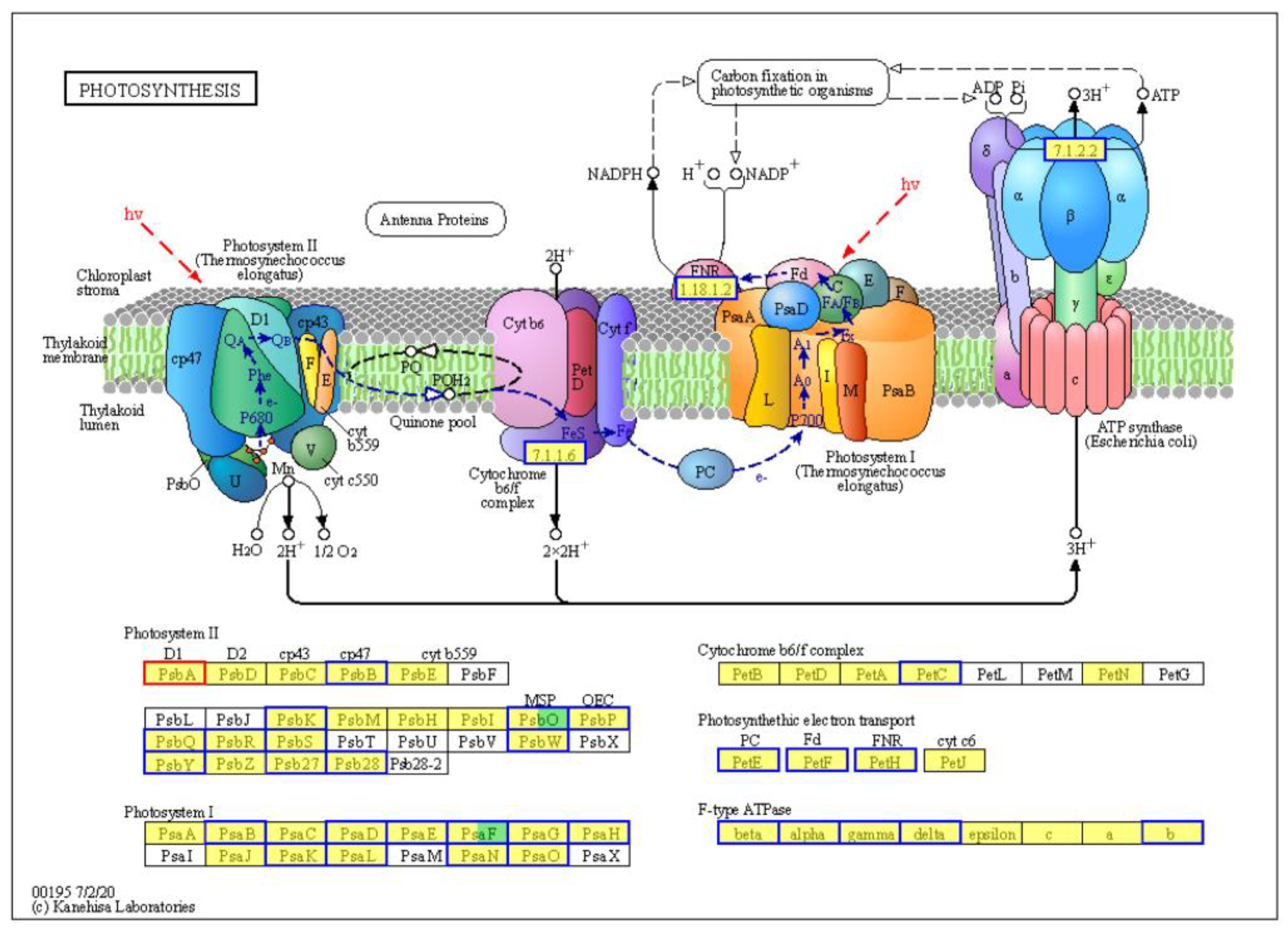
2.6. Interference of Plant Photosynthesis Processes by TYLCTHV
2.7. Induction of Plant Defense Responses by TYLCTHV
3. Discussion
4. Materials and Methods
4.1. Plants and Virus
4.2. CDNA Library Preparation and Illumina Sequencing
4.3. Quality Control and Read Mapping
4.4. Transcript Assembly and Analysis of Gene Expression Level
4.5. Functional Enrichment Analysis of DEGs
4.6. Quantitative Reverse-Transcription PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lefeuvre, P.; Martin, D.P.; Elena, S.F.; Shepherd, D.N.; Roumagnac, P.; Varsani, A. Evolution and ecology of plant viruses. Nat. Rev. Microbiol. 2019, 17, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Tatineni, S.; Hein, G.L. Plant viruses of agricultural importance: Current and future perspectives of virus disease management strategies. Phytopathology 2023, 113, 117–141. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Navas-Castillo, J. Begomoviruses: What is the secret(s) of their success? Trends Plant Sci. 2023, 28, 715–727. [Google Scholar] [CrossRef]
- Ascencio-Ibáñez, J.T.; Sozzani, R.; Lee, T.J.; Chu, T.M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef] [PubMed]
- Wang, A. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu. Rev. Phytopathol. 2015, 53, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, N.; Sahu, P.P.; Prasad, M.; Chakraborty, S. Chilli leaf curl virus infection highlights the differential expression of genes involved in protein homeostasis and defense in resistant chilli plants. Appl. Microbiol. Biotechnol. 2015, 99, 4757–4770. [Google Scholar] [CrossRef]
- Yan, Z.; Wolters, A.M.A.; Navas-Castillo, J.; Bai, Y.L. The global dimension of tomato yellow leaf curl disease: Current status and breeding perspectives. Microorganisms 2021, 9, 740. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Lett, J.M.; Martin, D.P.; Roumagnac, P.; Varsani, A.; Zerbini, F.M.; Navas-Castillo, J. ICTV Report Consortium. 2021. ICTV virus taxonomy profile: Geminiviridae 2021. J. Gen. Virol 2021, 102, 001696. [Google Scholar]
- Prasad, A.; Sharma, N.; Hari-Gowthem, G.; Muthamilarasan, M.; Prasad, M. Tomato yellow leaf curl virus: Impact, challenges, and management. Trends Plant Sci. 2020, 25, 897–911. [Google Scholar] [CrossRef]
- Attathom, S.; Chiemsombat, P.; Kositratana, W.; Sae-Ung, N. Complete nucleotide sequence and genome analysis of bipartite Tomato yellow leaf curl virus in Thailand. Kasetsart J. Nat. Sci. 1994, 28, 632–639. [Google Scholar]
- Attathom, S.; Chiemsombat, P.; Sutabutra, T.; Pongpanitanond, R. Characterization of nucleic acid of tomato yellow leaf curl virus. Kasetsart J. Nat. Sci. 1990, 24, 1–5. [Google Scholar]
- Sawangjit, S.; Chatchawankanphanich, O.; Chiemsombat, P.; Attathom, T.; Dale, J.; Attathom, S. Molecular characterization of tomato-infecting begomoviruses in Thailand. Virus Res. 2005, 109, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, W.H.; Mou, D.F.; Hsieh, C.K.; Weng, S.H.; Tsai, W.S.; Tsai, C.W. Vector Transmission of tomato yellow leaf curl Thailand virus by the whitefly Bemisia tabaci: Circulative or propagative? Insects 2021, 12, 181. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, L.; Tsai, W.S.; Shih, S.L.; Lee, L.M. Emergence and diversity of begomoviruses infecting solanaceous crops in East and Southeast Asia. Virus Res. 2014, 186, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.S.; Shih, S.L.; Kenyon, L.; Green, S.K.; Jan, F.J. Temporal distribution and pathogenicity of the predominant tomato-infecting begomoviruses in Taiwan. Plant Pathol. 2011, 60, 787–799. [Google Scholar] [CrossRef]
- Charoenvilaisiri, S.; Seepiban, C.; Phironrit, N.; Phuangrat, B.; Yoohat, K.; Deeto, R.; Chatchawankanphanich, O.; Gajanandana, O. Occurrence and distribution of begomoviruses infecting tomatoes, peppers and cucurbits in Thailand. Crop Prot. 2020, 127, 104948. [Google Scholar] [CrossRef]
- Kwak, H.R.; Hong, S.B.; Byun, H.S.; Park, B.; Choi, H.S.; Myint, S.S.; Kyaw, M.M. Incidence and molecular identification of begomoviruses infecting tomato and pepper in Myanmar. Plants 2022, 11, 1031. [Google Scholar] [CrossRef]
- Lestari, S.M.M.; Hidayat, S.H.H.; Hidayat, P.; Kil, E.J.; Lee, S.; Kim, S.M.; Lee, K.Y. Identification of begomoviruses associated with the insect vector Bemisia tabaci and various host plants on Java Island, Indonesia. Arch. Insect Biochem. Physiol. 2023, 112, 21984. [Google Scholar] [CrossRef]
- Kushwaha, N.K. Chilli leaf curl virus infection downregulates the expression of the genes encoding chloroplast proteins and stress-related proteins. Physiol. Mol. Biol. Plants 2019, 25, 1185–1196. [Google Scholar] [CrossRef]
- Song, L.; Wang, Y.; Zhao, L.; Zhao, T. Transcriptome profiling unravels the involvement of phytohormones in tomato resistance to the tomato yellow leaf curl virus (TYLCV). Horticulturae 2022, 8, 143. [Google Scholar] [CrossRef]
- Romero-Rodríguez, B.; Petek, M.; Jiao, C.; Križnik, M.; Zagorščak, M.; Fei, Z.J.; Bejarano, E.R.; Gruden, K.; Castillo, A.G. Transcriptional and epigenetic changes during tomato yellow leaf curl virus infection in tomato. BMC Plant Biol. 2023, 23, 651. [Google Scholar] [CrossRef] [PubMed]
- Stirbet, A.; Lazár, D.; Guo, Y.; Govindjee, G. Photosynthesis: Basics, history and modelling. Ann. Bot. 2020, 126, 511–537. [Google Scholar] [CrossRef] [PubMed]
- Ngou, B.P.M.; Jones, J.D.G.; Ding, P. Plant immune networks. Trends Plant Sci. 2022, 27, 255–273. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, L.L.; Zhang, X.; Wu, X.J.; Fang, R.X. Plant defense networks against insect-borne pathogens. Trends Plant Sci. 2021, 26, 272–287. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.M.; Zhou, T.; Carr, J.P. An update on salicylic acid biosynthesis, its induction and potential exploitation by plant viruses. Curr. Opin. Virol. 2020, 42, 8–17. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]






| Sample | Raw Reads | Clean Reads | Error% | Q20% | Q30% | GC% |
|---|---|---|---|---|---|---|
| pBINPLUS_1 | 49,891,168 | 49,476,800 | 0.0123 | 98.57 | 95.56 | 43 |
| pBINPLUS_2 | 64,534,292 | 63,907,418 | 0.0123 | 98.57 | 95.53 | 43.42 |
| pBINPLUS_3 | 54,661,946 | 54,296,048 | 0.0122 | 98.59 | 95.6 | 43.11 |
| pBINPLUS_4 | 48,012,992 | 47,627,718 | 0.0124 | 98.52 | 95.38 | 42.43 |
| TYLCTHV_1 | 56,556,462 | 56,007,018 | 0.0123 | 98.54 | 95.45 | 41.75 |
| TYLCTHV_2 | 58,147,208 | 57,713,032 | 0.0122 | 98.59 | 95.61 | 41.64 |
| TYLCTHV_3 | 43,688,846 | 43,341,960 | 0.0123 | 98.55 | 95.47 | 42.17 |
| TYLCTHV_4 | 49,327,424 | 48,968,828 | 0.0123 | 98.57 | 95.53 | 42.35 |
| Sample | Total Reads | Total Mapped | Multiple Mapped | Uniquely Mapped |
|---|---|---|---|---|
| pBINPLUS_1 | 49,476,800 | 48,600,635(98.23%) | 1,889,678(3.82%) | 46,710,957(94.41%) |
| pBINPLUS_2 | 63,907,418 | 62,747,944(98.19%) | 2,603,437(4.07%) | 60,144,507(94.11%) |
| pBINPLUS_3 | 54,296,048 | 53,260,364(98.09%) | 1,922,290(3.54%) | 51,338,074(94.55%) |
| pBINPLUS_4 | 47,627,718 | 46,783,258(98.23%) | 1,539,095(3.23%) | 45,244,163(95.00%) |
| TYLCTHV_1 | 56,007,018 | 53,991,001(96.40%) | 1,560,371(2.79%) | 52,430,630(93.61%) |
| TYLCTHV_2 | 57,713,032 | 54,568,886(94.55%) | 2,173,070(3.77%) | 52,395,816(90.79%) |
| TYLCTHV_3 | 43,341,960 | 41,758,846(96.35%) | 1,147,672(2.65%) | 40,611,174(93.70%) |
| TYLCTHV_4 | 48,968,828 | 47,174,688(96.34%) | 1,473,859(3.01%) | 45,700,829(93.33%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, W.-Z.; Rong, T.; Liu, X.-Y.; Rao, Q. Transcriptomic Profiling Unravels the Disruption of Photosynthesis Apparatuses and Induction of Immune Responses by a Bipartite Begomovirus in Tomato Plants. Plants 2024, 13, 3198. https://doi.org/10.3390/plants13223198
He W-Z, Rong T, Liu X-Y, Rao Q. Transcriptomic Profiling Unravels the Disruption of Photosynthesis Apparatuses and Induction of Immune Responses by a Bipartite Begomovirus in Tomato Plants. Plants. 2024; 13(22):3198. https://doi.org/10.3390/plants13223198
Chicago/Turabian StyleHe, Wen-Ze, Ting Rong, Xun-Yue Liu, and Qiong Rao. 2024. "Transcriptomic Profiling Unravels the Disruption of Photosynthesis Apparatuses and Induction of Immune Responses by a Bipartite Begomovirus in Tomato Plants" Plants 13, no. 22: 3198. https://doi.org/10.3390/plants13223198
APA StyleHe, W.-Z., Rong, T., Liu, X.-Y., & Rao, Q. (2024). Transcriptomic Profiling Unravels the Disruption of Photosynthesis Apparatuses and Induction of Immune Responses by a Bipartite Begomovirus in Tomato Plants. Plants, 13(22), 3198. https://doi.org/10.3390/plants13223198






