Glycolipids Derived from the Korean Endemic Plant Aruncus aethusifolius Inducing Glucose Uptake in Mouse Skeletal Muscle C2C12 Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Bioassay-Guided Isolation of Compounds
2.2. Structural Elucidation of Compounds
2.3. Effect of Compounds on Glucose Uptake in Skeletal Muscle Cells
2.4. Effect of Compounds on Protein Expression of p-IRS-1, IRS-1, p-AMPK, AMPK, PPARγ, and GLUT4
2.5. Molecular Docking of Compounds on IRS-1, AMPK, and PPARγ
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Plant Material and Extraction
4.3. Isolation and Structural Characterization of Compounds
4.3.1. Gingerglycolipid A (1)
4.3.2. (2S)-3-Linolenoylglycerol-O-β-d-Galactopyranoside (2)
4.4. Acid Hydrolysis
4.5. Cell Culture
4.6. Cell Viability Assay
4.7. Glucose Uptake Assay
4.8. Western Blotting Analysis
4.9. Molecular Docking Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- da Silva, J.A.; de Souza, E.C.F.; Echazú Böschemeier, A.G.; da Costa, C.C.M.; Bezerra, H.S.; Feitosa, E.E.L.C. Diagnosis of diabetes mellitus and living with a chronic condition: Participatory study. BMC Public Health 2018, 18, 699. [Google Scholar] [CrossRef] [PubMed]
- Nobre, L.N.; da Silva, E. Health education and type 2 diabetes. Front. Nutr. 2023, 10, 1325517. [Google Scholar] [CrossRef] [PubMed]
- Fjodorova, N.; Novič, M.; Venko, K.; Drgan, V.; Rasulev, B.; Saçan, M.T.; Erdem, S.S.; Tugcu, G.; Toropova, A.P.; Toropov, A.A. How fullerene derivatives (FDs) act on therapeutically important targets associated with diabetic diseases. Comput. Struct. Biotechnol. J. 2022, 20, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Peiffer, C. Signalling aspects of insulin resistance in skeletal muscle: Mechanisms induced by lipid oversupply. Cell. Signal. 2000, 12, 583–594. [Google Scholar] [CrossRef]
- Merz, K.E.; Thurmond, D.C. Role of skeletal muscle in insulin resistance and glucose uptake. Compr. Physiol. 2011, 10, 785–809. [Google Scholar]
- Lee, J.O.; Lee, S.K.; Kim, J.H.; Kim, N.; You, G.Y.; Moon, J.W.; Kim, S.J.; Park, S.H.; Kim, H.S. Metformin regulates glucose transporter 4 (GLUT4) translocation through AMP-activated protein kinase (AMPK)-mediated Cbl/CAP signaling in 3T3-L1 preadipocyte cells. J. Biol. Chem. 2012, 287, 44121–44129. [Google Scholar] [CrossRef]
- He, J.-H.; Chen, L.-X.; Li, H. Progress in the discovery of naturally occurring anti-diabetic drugs and in the identification of their molecular targets. Fitoterapia 2019, 134, 270–289. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Kim, Y.H.; Min, B.-S.; Min, B.K.; Woo, M.H. Monoterpenoids from the aerial parts of Aruncus dioicus var. kamtschaticus and their antioxidant and cytotoxic activities. Bioorg. Med. Chem. Lett. 2011, 21, 3252–3256. [Google Scholar]
- Kim, D.-H.; Moon, Y.-S.; An, B.-J.; Son, J.-H. Potent anti-aging activity of Aruncus dioicus, a native plant of Ulleung-do, South Korea, in CCD-986sk fibroblasts via suppression of matrix metalloproteinases. J. Nat. Med. 2012, 66, 631–636. [Google Scholar] [CrossRef]
- Shin, J.-W.; Lee, S.-I.; Woo, M.-H.; Kim, S.-D. Effect of ethanol extracts of goat’s beard on streptozotocin induced diabetic symptoms and oxidative stress in rats. J. East Asian Soc. Diet. Life 2008, 18, 939–948. [Google Scholar]
- Kim, J.-I.; Yun, J.-A.; Jeong, Y.-K.; Baek, H.-J. Hypoglycemic and hypolipidemic effects of samnamul (shoot of Aruncus dioicus var. kamtschaticus Hara) in mice fed a high-fat/high-sucrose diet. Food Sci. Biotechnol. 2018, 27, 1467–1473. [Google Scholar] [PubMed]
- Jongsun, P.; Hwa-Jung, S.; Sang-Hun, O. The complete chloroplast genome of Aruncus aethusifolius (Rosaceae), a species endemic to Korea. Korean J. PI. Taxon. 2022, 52, 118–122. [Google Scholar]
- Ko, J.-H.; Cho, S.M.; Joo, S.-W.; Kim, H.-G.; Lee, Y.-G.; Kang, S.C.; Baek, N.-I. Glycosyl glycerides from the aerial parts of Malva verticillata and their chemopreventive effects. Bioorg. Chem. 2018, 78, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.; Bae, M.; Szamosvári, D.; Cassilly, C.D.; Bolze, A.S.; Jackson, D.R.; Xavier, R.J.; Clardy, J. Collinsella aerofaciens produces a pH-responsive lipid immunogen. J. Am. Chem. Soc. 2023, 145, 7071–7074. [Google Scholar] [CrossRef] [PubMed]
- Eck, M.J.; Dhe-Paganon, S.; Trüb, T.; Nolte, R.T.; Shoelson, S.E. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell 1996, 85, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Gampe, R.T.; Montana, V.G.; Lambert, M.H.; Miller, A.B.; Bledsoe, R.K.; Milburn, M.V.; Kliewer, S.A.; Willson, T.M.; Xu, H.E. Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol. Cell 2000, 5, 545–555. [Google Scholar] [CrossRef]
- Najmi, A.; Javed, S.A.; Al Bratty, M.; Alhazmi, H.A. Modern approaches in the discovery and development of plant-based natural products and their analogues as potential therapeutic agents. Molecules 2022, 27, 349. [Google Scholar] [CrossRef]
- Drucker, D.J.; Sherman, S.I.; Gorelick, F.S.; Bergenstal, R.M.; Sherwin, R.S.; Buse, J.B. Incretin-based therapies for the treatment of type 2 diabetes: Evaluation of the risks and benefits. Diabetes Care 2010, 33, 428. [Google Scholar] [CrossRef]
- Thulé, P.M. Mechanisms of current therapies for diabetes mellitus type 2. Adv. Physiol. Educ. 2012, 36, 275–283. [Google Scholar] [CrossRef]
- Giacoman-Martínez, A.; Alarcón-Aguilar, F.J.; Zamilpa, A.; Huang, F.; Romero-Nava, R.; Román-Ramos, R.; Almanza-Pérez, J.C. α-Amyrin induces GLUT4 translocation mediated by AMPK and PPARδ/γ in C2C12 myoblasts. Can. J. Physiol. Pharmacol. 2021, 99, 935–942. [Google Scholar] [CrossRef]
- Feng, S.-Y.; Wu, S.-J.; Chang, Y.-C.; Ng, L.-T.; Chang, S.-J. Stimulation of GLUT4 glucose uptake by anthocyanin-rich extract from black rice (Oryza sativa L.) via PI3K/Akt and AMPK/p38 MAPK signaling in C2C12 cells. Metabolites 2022, 12, 856. [Google Scholar] [CrossRef]
- Lee, D.; Kim, J.-Y.; Kwon, H.C.; Kwon, J.; Jang, D.S.; Kang, K.S. Dual beneficial effects of α-spinasterol isolated from Aster pseudoglehnii on glucose uptake in skeletal muscle cells and glucose-stimulated insulin secretion in pancreatic β-cells. Plants 2022, 11, 658. [Google Scholar] [CrossRef]
- Kawai, M.; Rosen, C.J. PPARγ: A circadian transcription factor in adipogenesis and osteogenesis. Nat. Rev. Endocrinol. 2010, 6, 629–636. [Google Scholar] [CrossRef]
- DeFronzo, R.A.; Tripathy, D. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care 2009, 32 (Suppl. S2), S157. [Google Scholar] [CrossRef] [PubMed]
- Mihaylova, M.M.; Shaw, R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011, 13, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Shaw, R.J. AMPK: Mechanisms of cellular energy sensing and restoration of metabolic balance. Mol. Cell 2017, 66, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Stocks, B.; Zierath, J.R. Post-translational modifications: The signals at the intersection of exercise, glucose uptake, and insulin sensitivity. Endocr. Rev. 2022, 43, 654–677. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.; Lee, D.; Dai Son, J.; Baek, J.G.; Lee, H.-S.; Park, I.; Kim, D.H.; Lee, S.K.; Kim, W.K.; Kwon, H.C.; et al. Chemical constituents isolated from Actinidia polygama and their α-glucosidase inhibitory activity and insulin secretion effect. Bioorg. Chem. 2023, 134, 106466. [Google Scholar] [CrossRef] [PubMed]
- Dhe-Paganon, S.; Ottinger, E.A.; Nolte, R.T.; Eck, M.J.; Shoelson, S.E. Crystal structure of the pleckstrin homology-phosphotyrosine binding (PH-PTB) targeting region of insulin receptor substrate 1. Proc. Natl. Acad. Sci. USA 1999, 96, 8378–8383. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhou, X.E.; Novick, S.J.; Shaw, S.J.; Li, Y.; Brunzelle, J.S.; Hitoshi, Y.; Griffin, P.R.; Xu, H.E.; Melcher, K. Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states. J. Biol. Chem. 2019, 294, 953–967. [Google Scholar] [CrossRef]
- Handa, N.; Takagi, T.; Saijo, S.; Kishishita, S.; Takaya, D.; Toyama, M.; Terada, T.; Shirouzu, M.; Suzuki, A.; Lee, S. Structural basis for compound C inhibition of the human AMP-activated protein kinase α2 subunit kinase domain. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 480–487. [Google Scholar] [CrossRef] [PubMed]

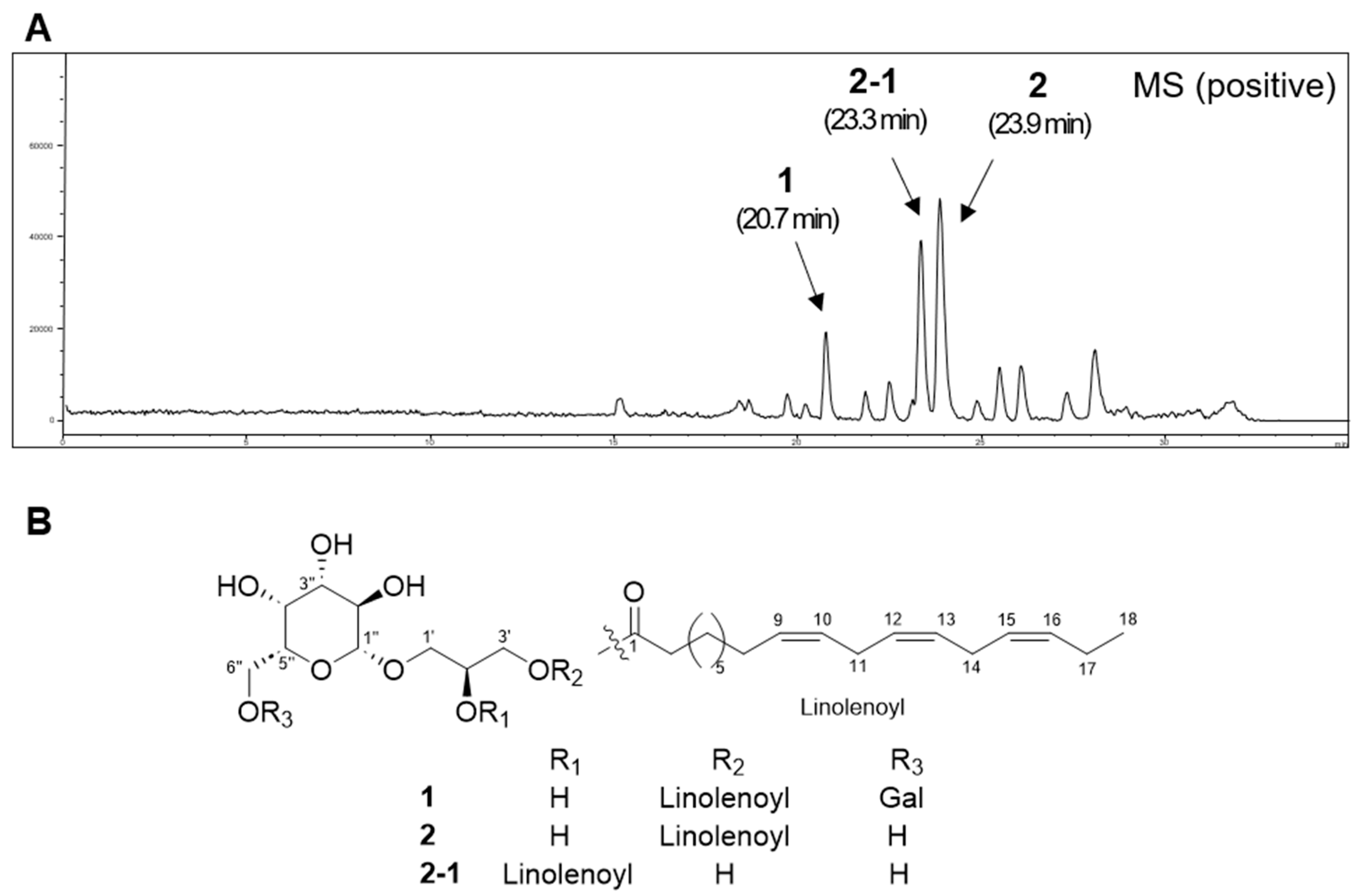


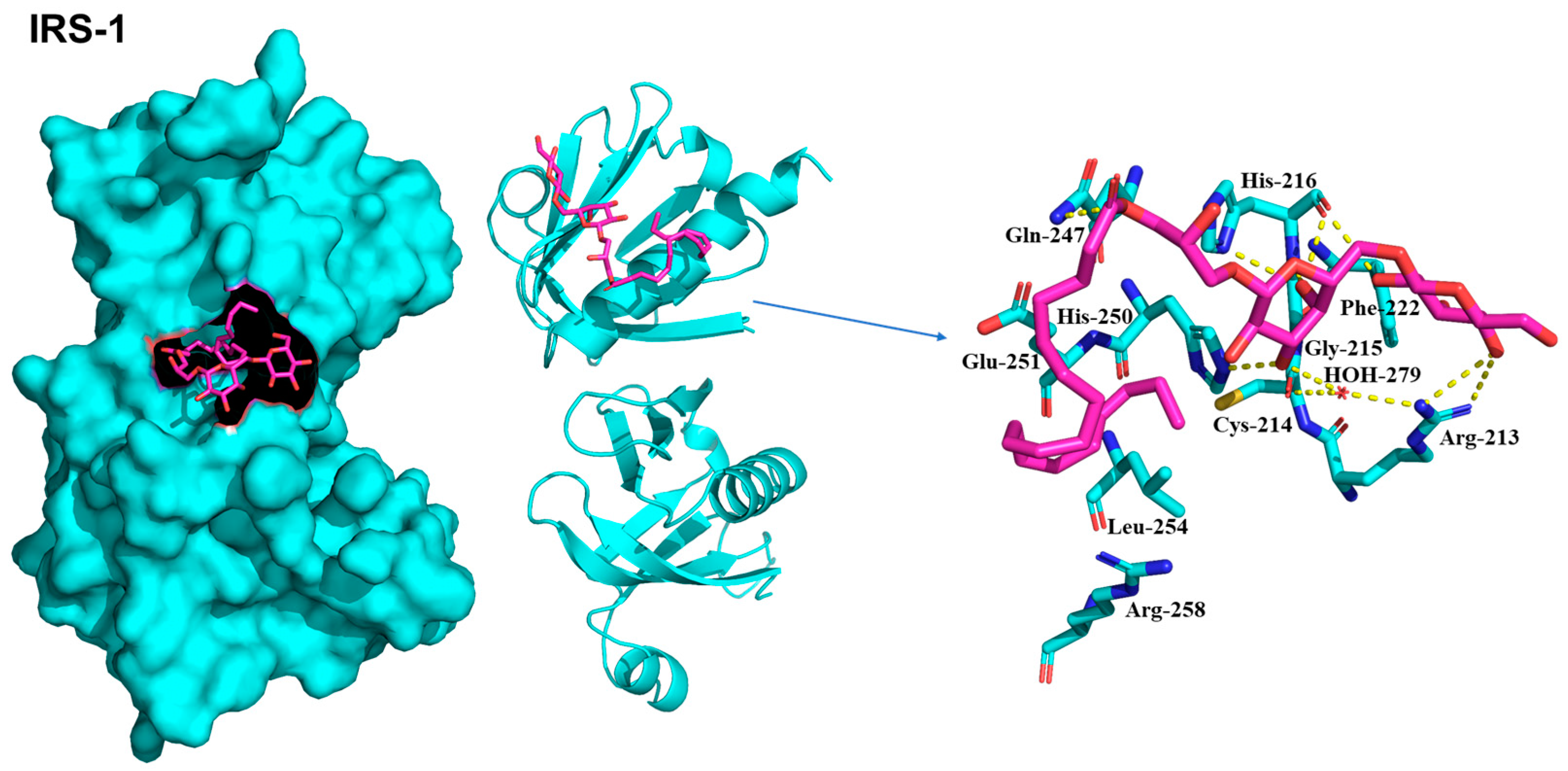
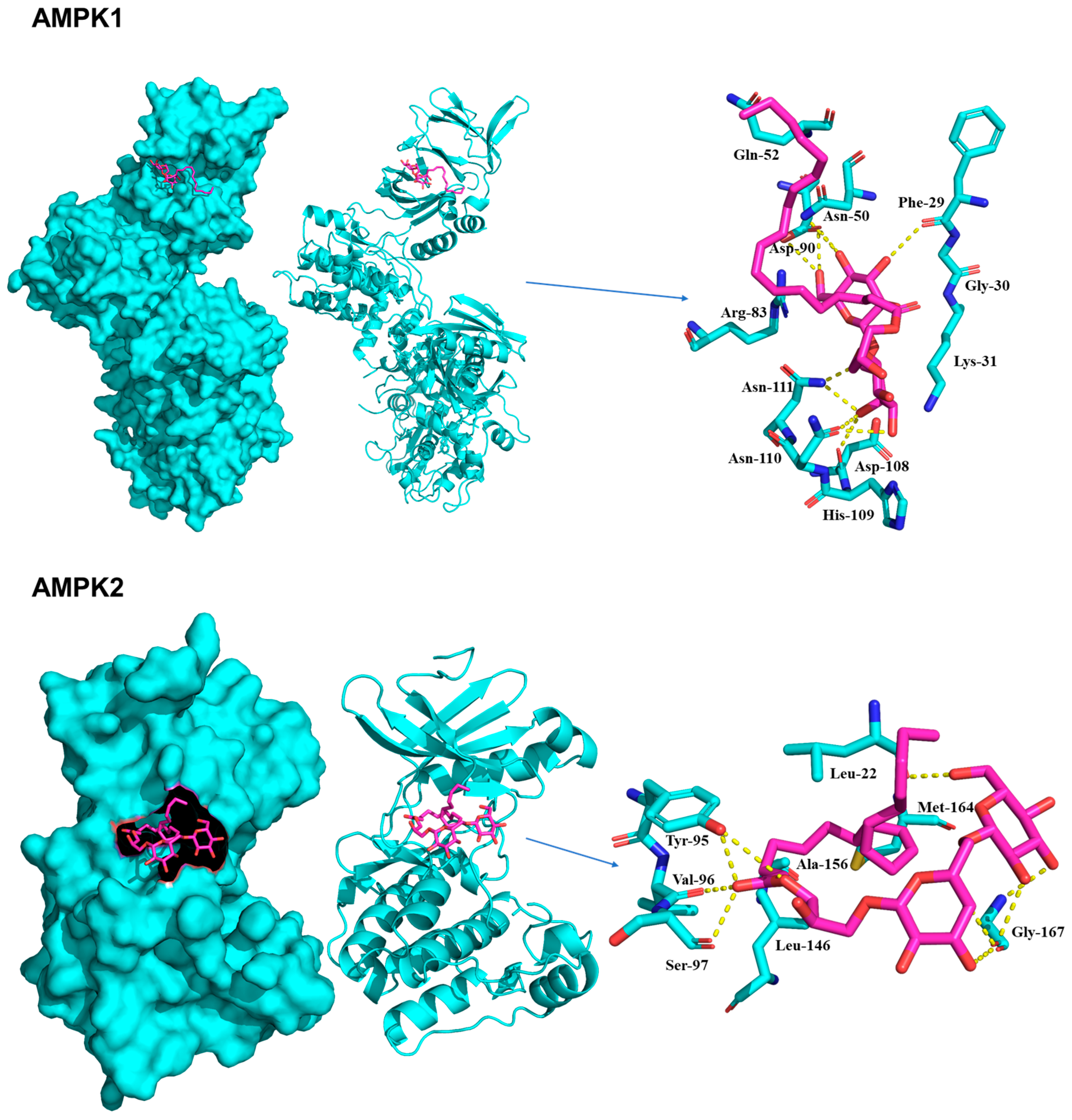
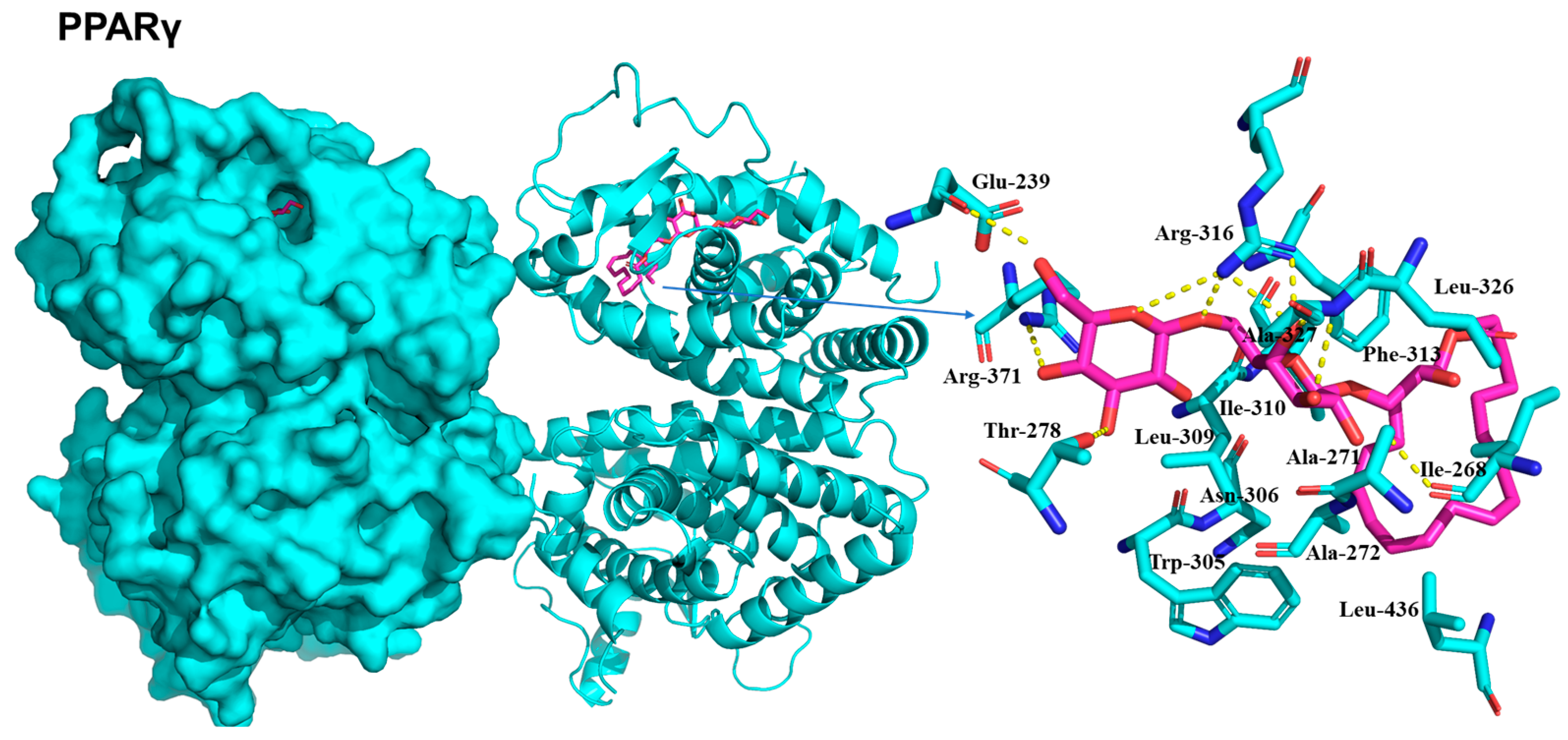
| No. | 1 | 2 | 2-1 | |||
|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | |
| 1 | 175.5 | 175.4 | 175.2 | |||
| 2 | 34.9 | 2.36, t (7.5) | 34.9 | 2.35, t (7.4) | 35.1 | 2.36, t (7.5) |
| 3 | 26.0 | 1.62, m | 26.0 | 1.62, m | 26 | 1.61, m |
| 4 | 30.7 a | 1.34 * | 30.7 | 1.34 * | 30.7 a | 1.34 * |
| 5 | 30.2 a | 1.34 * | 30.2–30.3 a | 1.34 * | 30.2 a | 1.34 * |
| 6 | 30.2 a | 1.34 * | 30.2–30.3 a | 1.34 * | 30.2 a | 1.34 * |
| 7 | 30.3 a | 1.34 * | 30.2–30.3 a | 1.34 * | 30.3 a | 1.34 * |
| 8 | 28.2 | 2.07, m | 28.2 | 2.09, m | 28.2 | 2.08, m |
| 9 | 131.1 | 5.31–5.40 * | 128.3 b | 5.30–5.39 * | 131.3 | 5.31–5.37 * |
| 10 | 128.9 d | 5.31–5.40 * | 128.9 b | 5.30–5.39 * | 128.9 b | 5.31–5.37 * |
| 11 | 26.4 | 2.81, m | 26.4 | 2.81, m | 26.4 | 2.81, m |
| 12 | 129.2 d | 5.31–5.40 * | 129.2 b | 5.30–5.39 * | 129.2 b | 5.31–5.37 * |
| 13 | 129.2 d | 5.31–5.40 * | 129.2 b | 5.30–5.39 * | 129.2 b | 5.31–5.37 * |
| 14 | 26.5 | 2.81, m | 26.5 | 2.81, m | 26.5 | 2.81, m |
| 15 | 128.2 d | 5.31–5.40 * | 131.1 b | 5.30–5.39 * | 128.3 b | 5.31–5.37 * |
| 16 | 132.7 | 5.31–5.40 * | 132.7 | 5.30–5.39 * | 132.7 | 5.31–5.37 * |
| 17 | 21.5 | 2.09, m | 21.5 | 2.10, m | 21.5 | 2.09, m |
| 18 | 14.7 | 0.98, t (7.6) | 14.7 | 0.98, t (7.6) | 14.7 | 0.98, t (7.5) |
| 1′ | 72.1 | 3.87, m | 71.9 | 3.92, dd (10.5, 5.1) | 68.8 | 3.97, dd (10.8, 5.5) |
| 3.67, m | 3.65, dd (10.5, 4.6) | 3.75, m | ||||
| 2′ | 69.7 | 4.00, m | 69.6 | 3.99, m | 74.7 | 5.05, p (5.5) |
| 3′ | 66.6 | 4.15, m | 66.6 | 4.15, m | 61.7 | 3.74, m |
| 1″ | 105.3 | 4.25, d (7.4) | 105.3 | 4.23, d (7.6) | 105.3 | 4.23, d (7.4) |
| 2″ | 72.5 b | 3.53, m | 72.6 | 3.54, m | 72.4 | 3.52, m |
| 3″ | 74.6 | 3.49, m | 74.8 | 3.47, dd (3.4, 9.8) | 74.9 | 3.46, dd (9.6, 3.3) |
| 4″ | 70.1 c | 3.87, m | 70.3 | 3.82, d (3.5) | 70.3 | 3.82, d (3.3) |
| 5″ | 74.7 | 3.75, m | 76.8 | 3.52, m | 76.8 | 3.50, m |
| 6″ | 67.8 | 3.90, m | 62.5 | 3.75, m | 62.5 | 3.72, m |
| 3.67, m | ||||||
| 1‴ | 100.5 | 4.86 * | ||||
| 2‴ | 70.2 c | 3.77, m | ||||
| 3‴ | 71.5 | 3.74, m | ||||
| 4‴ | 71.1 | 3.89, m | ||||
| 5‴ | 72.6 b | 3.86, m | ||||
| 6‴ | 62.7 | 3.71, m | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, J.G.; Park, D.H.; Vu, N.K.; Muvva, C.; Hwang, H.; Song, S.; Lee, H.-S.; Kim, T.-J.; Kwon, H.C.; Park, K.; et al. Glycolipids Derived from the Korean Endemic Plant Aruncus aethusifolius Inducing Glucose Uptake in Mouse Skeletal Muscle C2C12 Cells. Plants 2024, 13, 608. https://doi.org/10.3390/plants13050608
Baek JG, Park DH, Vu NK, Muvva C, Hwang H, Song S, Lee H-S, Kim T-J, Kwon HC, Park K, et al. Glycolipids Derived from the Korean Endemic Plant Aruncus aethusifolius Inducing Glucose Uptake in Mouse Skeletal Muscle C2C12 Cells. Plants. 2024; 13(5):608. https://doi.org/10.3390/plants13050608
Chicago/Turabian StyleBaek, Jong Gwon, Do Hwi Park, Ngoc Khanh Vu, Charuvaka Muvva, Hoseong Hwang, Sungmin Song, Hyeon-Seong Lee, Tack-Joong Kim, Hak Cheol Kwon, Keunwan Park, and et al. 2024. "Glycolipids Derived from the Korean Endemic Plant Aruncus aethusifolius Inducing Glucose Uptake in Mouse Skeletal Muscle C2C12 Cells" Plants 13, no. 5: 608. https://doi.org/10.3390/plants13050608
APA StyleBaek, J. G., Park, D. H., Vu, N. K., Muvva, C., Hwang, H., Song, S., Lee, H.-S., Kim, T.-J., Kwon, H. C., Park, K., Kang, K. S., & Kwon, J. (2024). Glycolipids Derived from the Korean Endemic Plant Aruncus aethusifolius Inducing Glucose Uptake in Mouse Skeletal Muscle C2C12 Cells. Plants, 13(5), 608. https://doi.org/10.3390/plants13050608







