Genome-Wide Identification and Expression Analysis Reveal bZIP Transcription Factors Mediated Hormones That Functions during Early Somatic Embryogenesis in Dimocarpus longan
Abstract
:1. Introduction
2. Results
2.1. Identification of DlbZIP Family Members and Analysis of Basic Physicochemical Properties
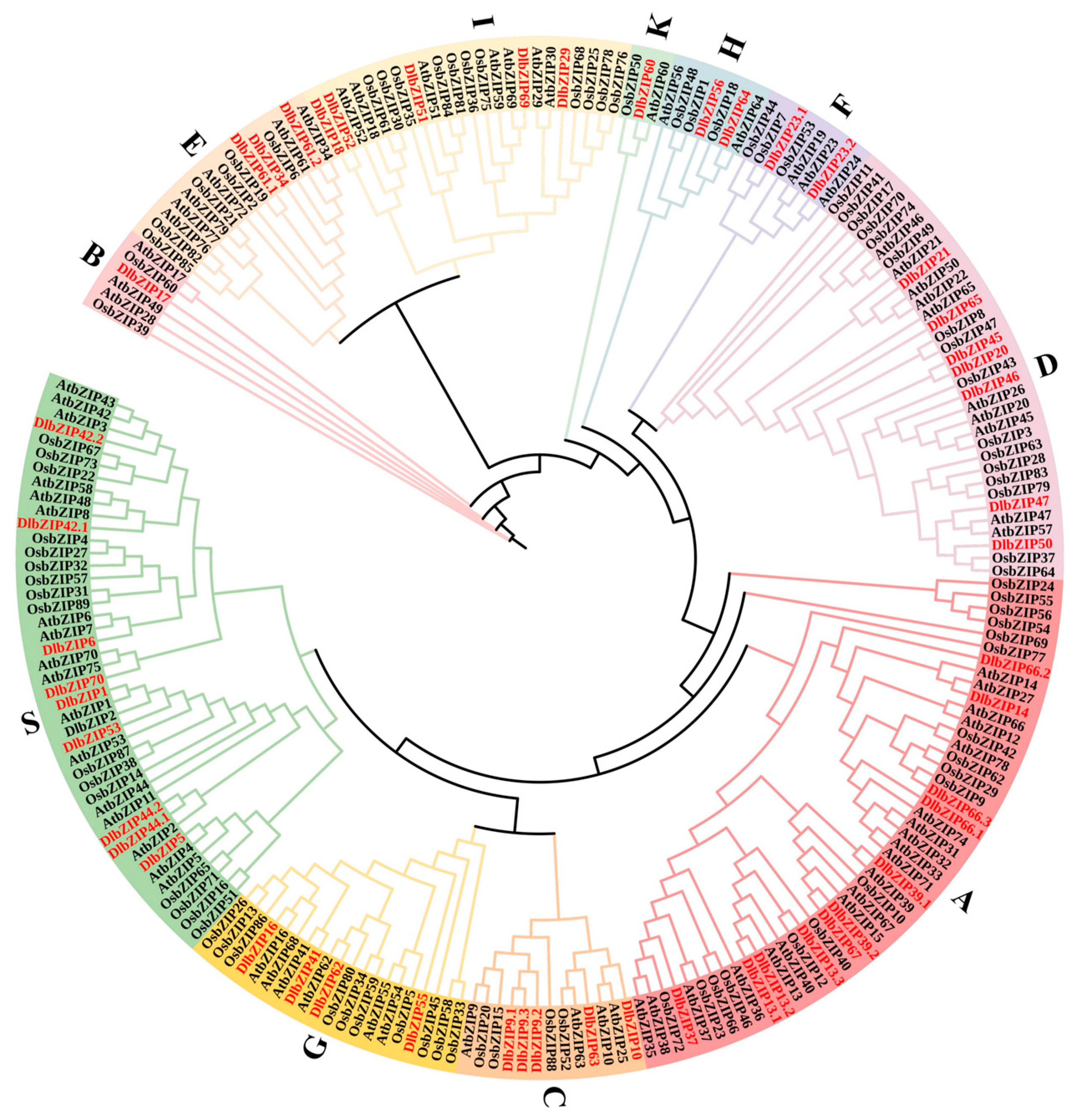
2.2. Phylogenetic Analysis of DlbZIP Family Members
2.3. Conserved Motif and Gene Structure of DlbZIP Family Members
2.4. Chromosomal Localization, Covariance Analysis and Cis-Acting Element of DlbZIP Family Members
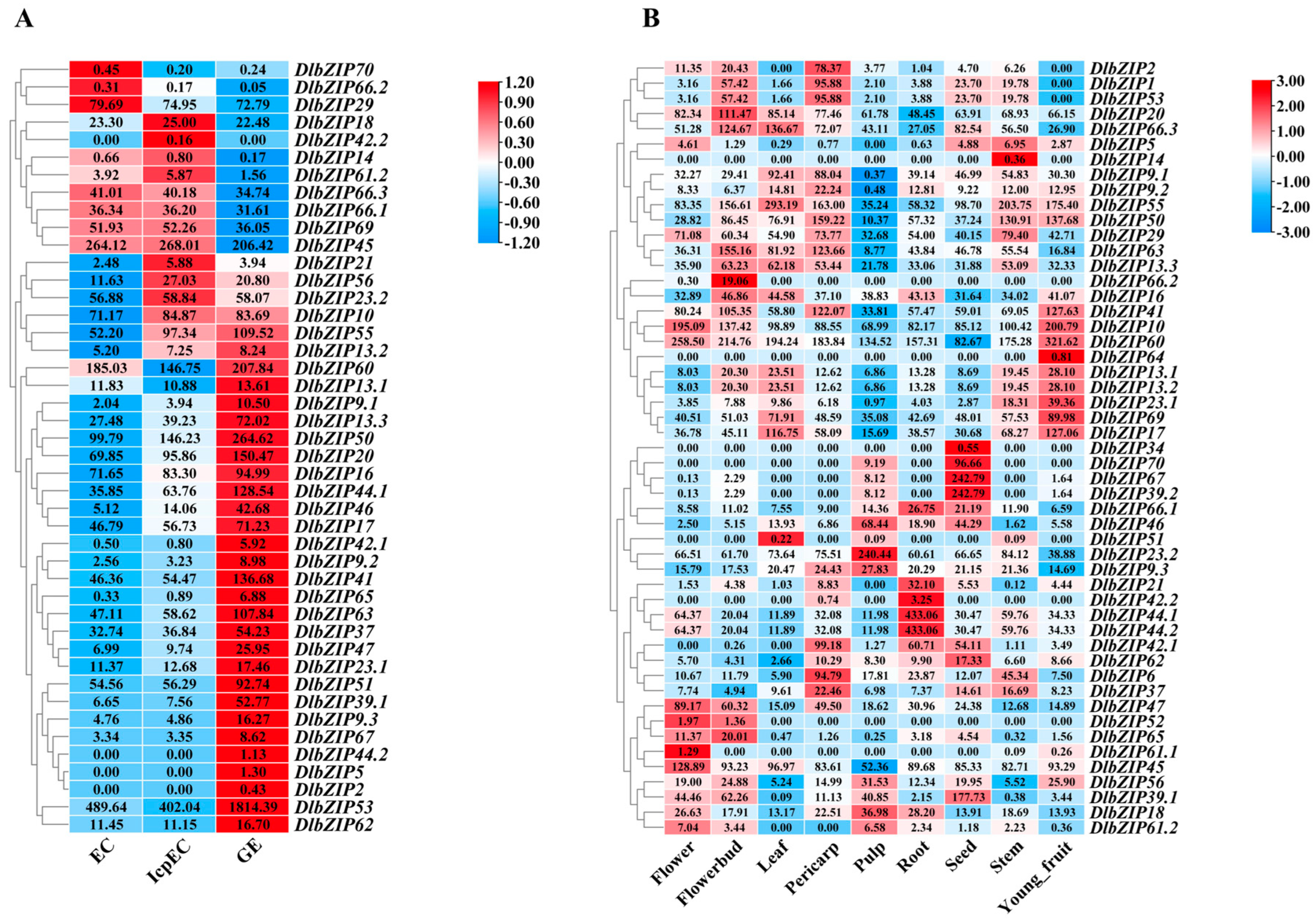
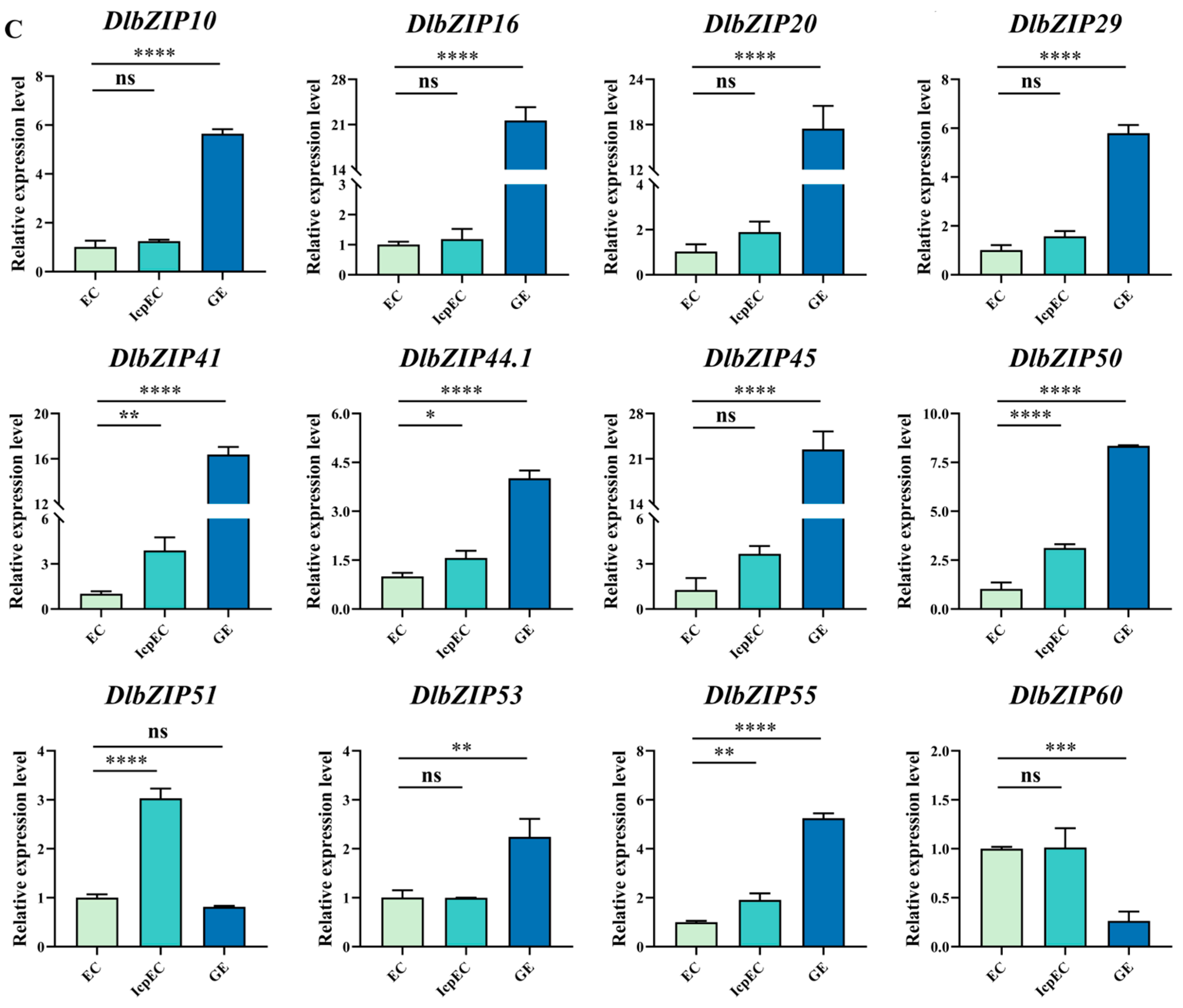
2.5. DlbZIP Family Members Respond to Early SE and Different Developmental Organs in D. longan
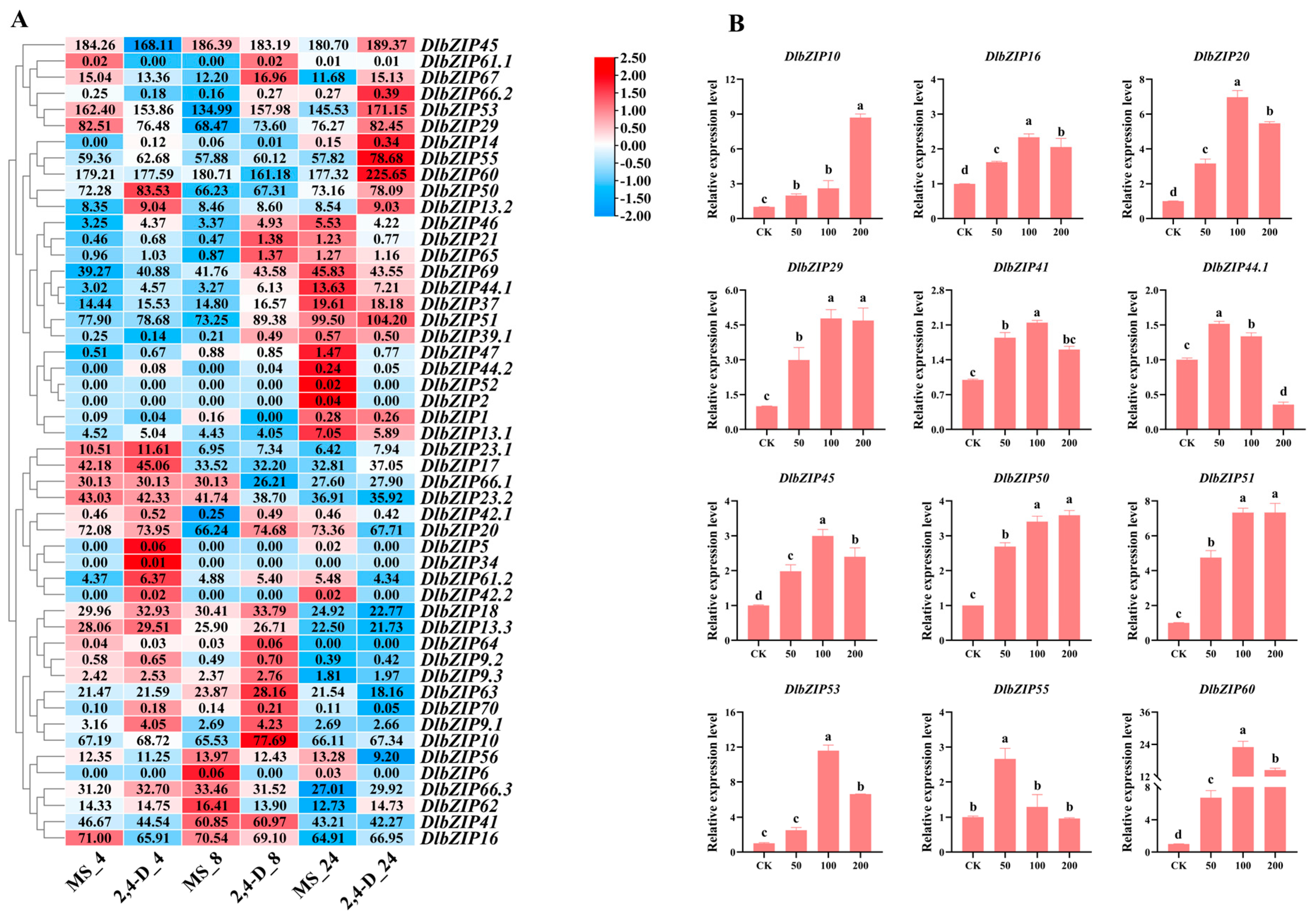
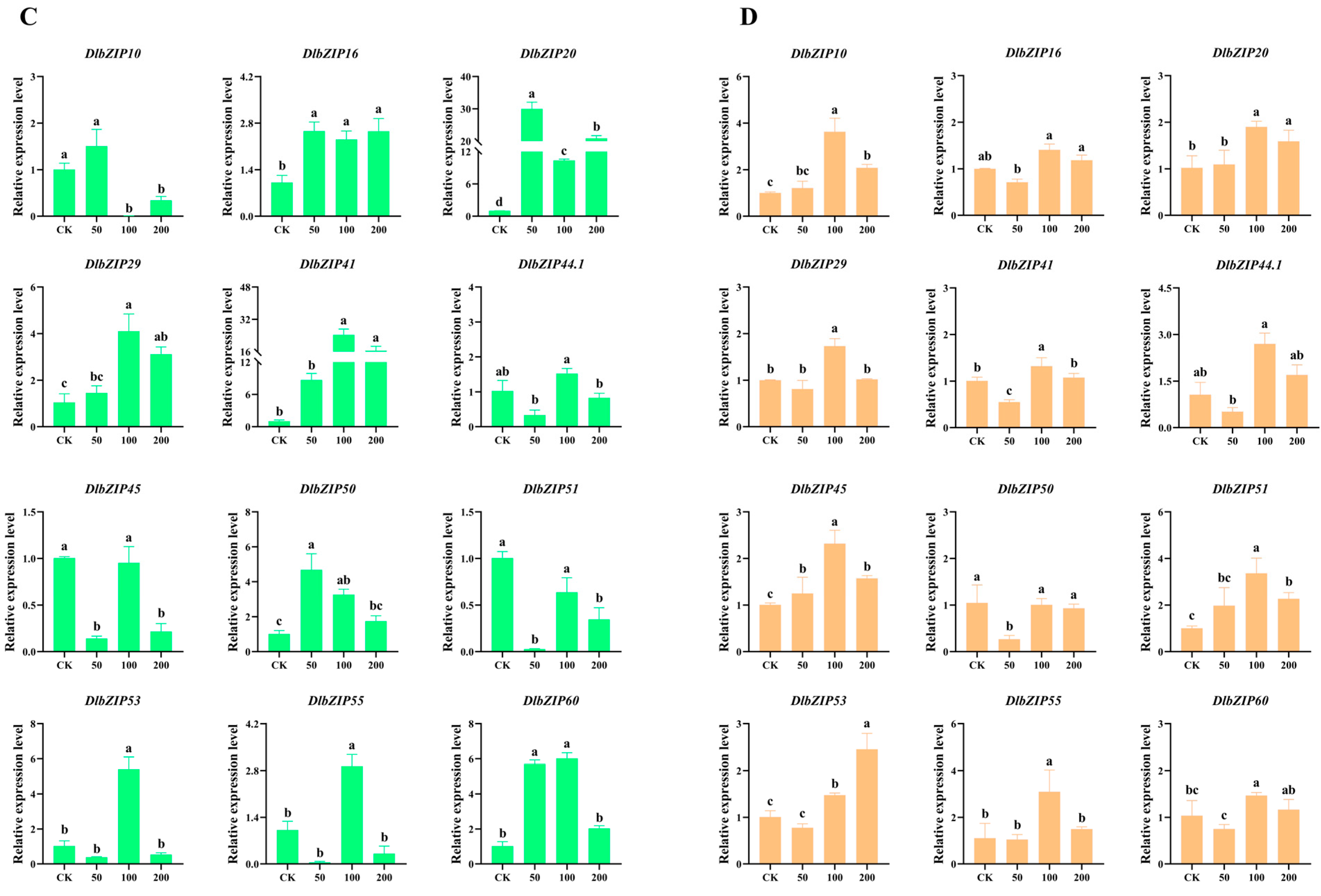
2.6. DlbZIP Family Members Are Involved in Multiple Hormone Transduction Pathways
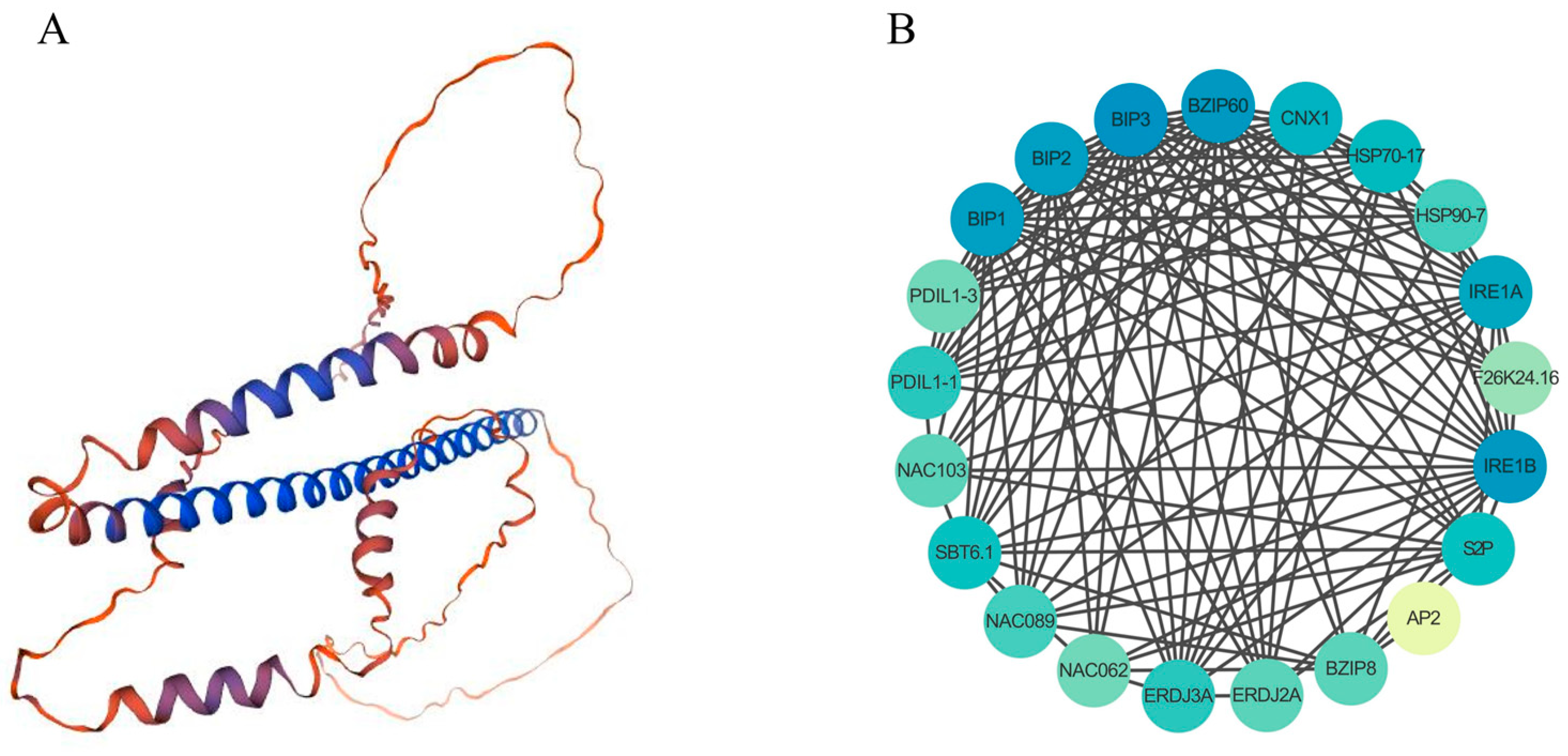
2.7. Tertiary Structure and Protein Interaction Networks of the DlbZIP60
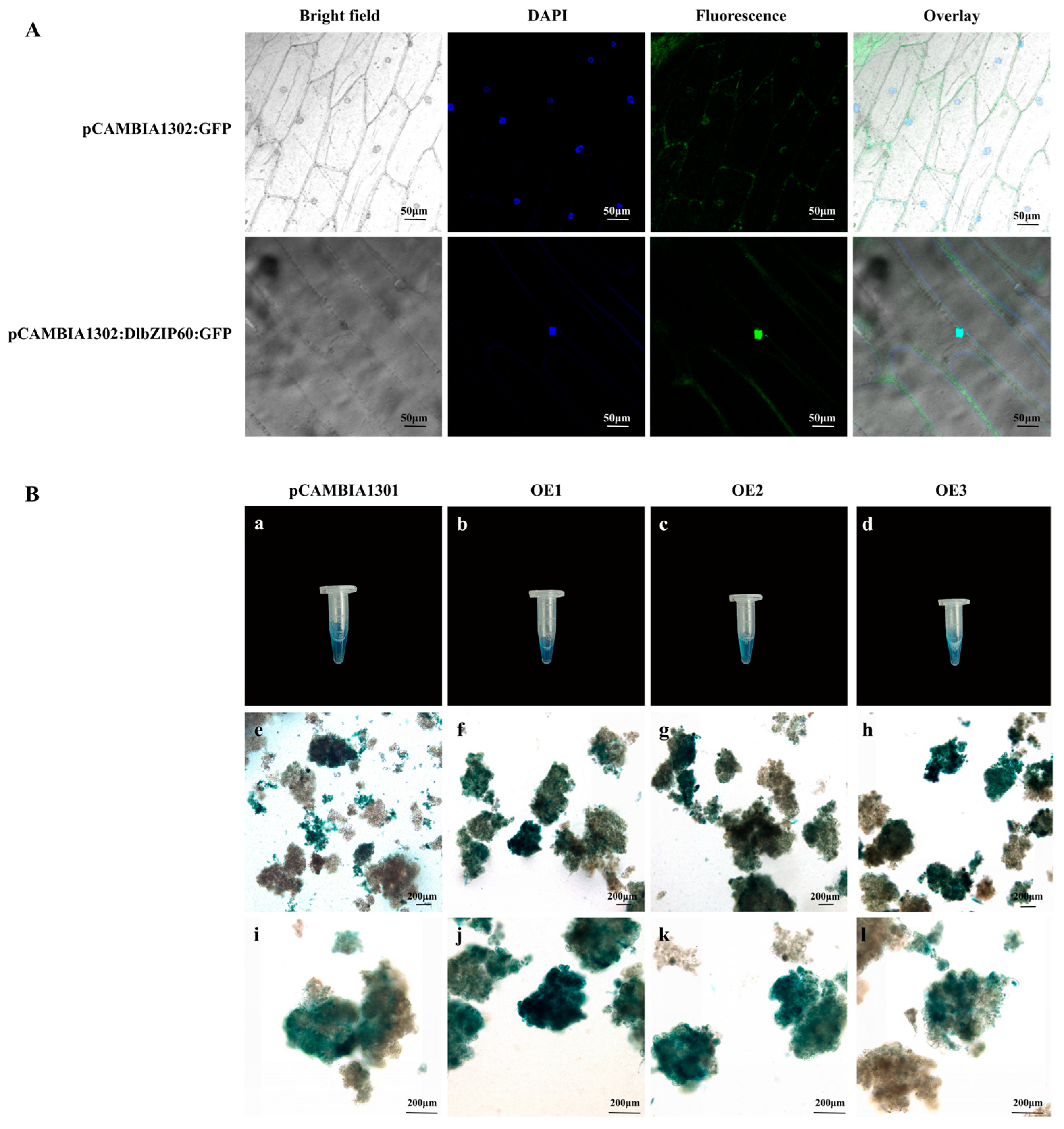
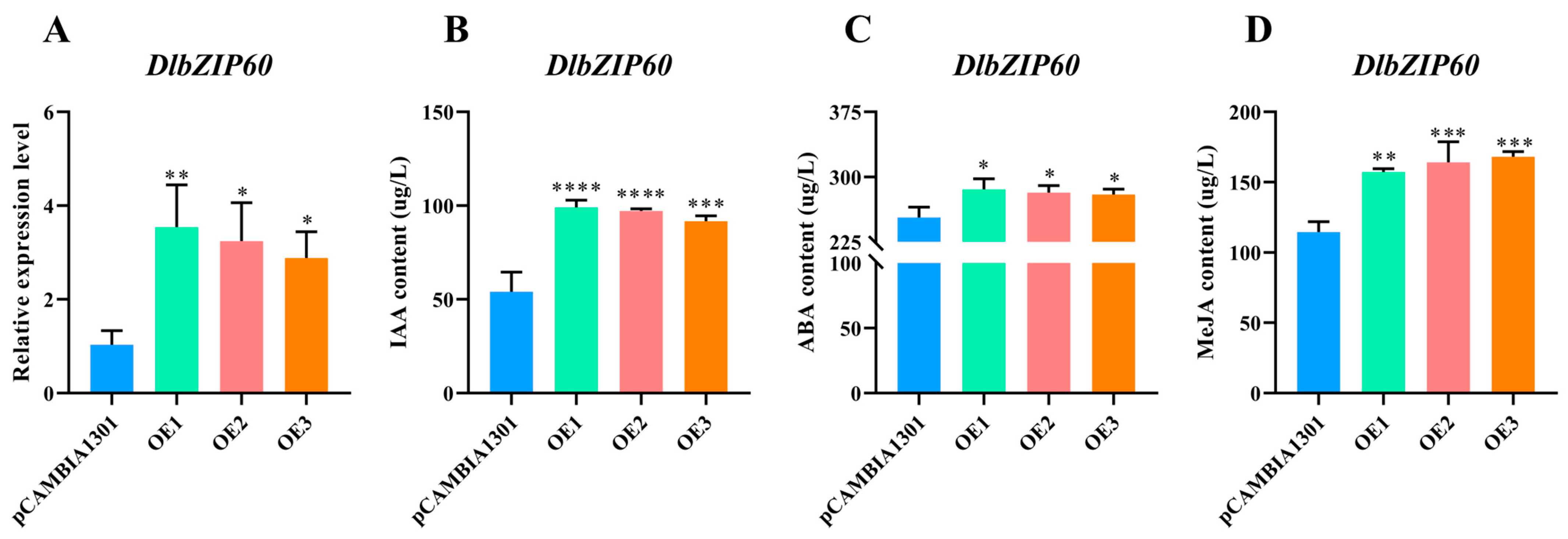
2.8. DlbZIP60 Is Located in the Nucleus and Affects the Content of Multiple Endogenous Hormones in D. longan EC
3. Discussion
3.1. DlbZIP Family Members Are Evolutionarily Conserved and Functionally Diverse
3.2. DlbZIP Family Members Involved in Early D. longan SE and Related Hormone Synthesis Pathways
3.3. Overexpression of DlbZIP60 May Be Involved in the Regulation of Embryogenesis in SE of D. longan
4. Materials and Methods
4.1. Plant Materials
4.2. Identification and Protein Physicochemical Properties of D. longan bZIP Family Members
4.3. Phylogenetic Tree, Conserved Motif and Gene Structure of DlbZIP Family Members
4.4. Chromosomal Localization, Covariance Analysis between Multiple Species and Cis-Acting Elements of DlbZIP Family Members
4.5. Analysis of Specific Expression of DlbZIP Family Members
4.6. The qRT-PCR Analysis of DlbZIP Family Members during Early SE, under Different Hormone Treatments and Transient Transformation
4.7. Tertiary Structure and Protein Interaction Networks of DlbZIP Family Members
4.8. Subcellular Localization Analysis
4.9. Transient Transformation of D. longan EC
4.10. Measurement of Endogenous Hormones in D. longan EC after Transient Transformation
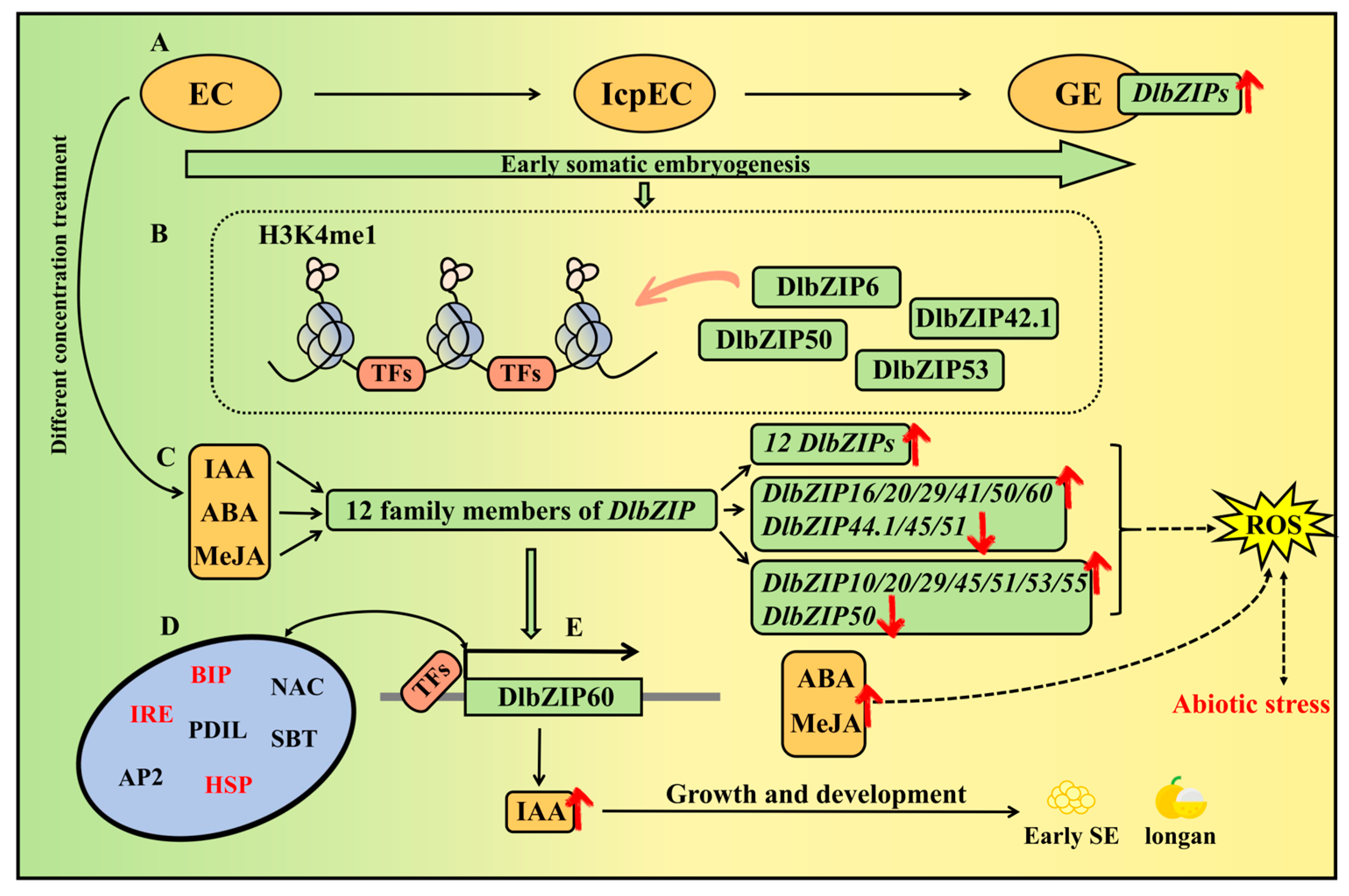
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Yu, Z.; Cheng, X.; Pan, W.; Yu, C.; Duan, Y. The ferroptosis activity is associated with neurological recovery following chronic compressive spinal cord injury. Neural. Regen. Res. 2023, 18, 2482–2488. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Han, H.; Zhang, B.; Wang, L.; Wu, J.; Chen, Z.; Lin, K.; Hao, J.; Jia, R.; Zhang, Y. Identification of Crucial Genes and Regulatory Pathways in Alfalfa against Fusarium Root Rot. Plants 2023, 12, 3634. [Google Scholar] [CrossRef] [PubMed]
- Droge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family—An update. Curr. Opin. Plant Biol. 2018, 45 Pt A, 36–49. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kumar, P.; Sharma, D.; Verma, S.K.; Halterman, D.; Kumar, A. Genome-wide identification and expression profiling of basic leucine zipper transcription factors following abiotic stresses in potato (Solanum tuberosum L.). PLoS ONE 2021, 16, e0247864. [Google Scholar] [CrossRef] [PubMed]
- Baloglu, M.C.; Eldem, V.; Hajyzadeh, M.; Unver, T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 2014, 9, e96014. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Fu, F.; Zhang, H.; Song, F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genom. 2015, 16, 771. [Google Scholar] [CrossRef]
- Wei, R.M.; Zheng, J.Y.; Liu, F.; Ma, Y.Q.; Li, X.F.; Yang, B.Z.; Zou, X.X.; Xie, L.L.; Dai, X.Z. Genome wide identification and expression snalysis of the bZIP gene family in Pepper. Acta Hortic. Sin. 2018, 45, 1535–1550. [Google Scholar]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef]
- Talanian, R.V.; Mcknight, C.J.; Kim, P.S. Sequence-specific DNA-binding by a short peptide dimer. Science 1990, 249, 769–771. [Google Scholar] [CrossRef]
- Kang, S.G.; Price, J.; Lin, P.C.; Hong, J.C.; Jang, J.C. The arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol. Plant 2010, 3, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Ellenberger, T.E.; Brandl, C.J.; Struhl, K.; Harrison, S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted a helices: Crystal structure of the protein-DNA complex. Cell 1992, 71, 1223–1237. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Wang, X.; Feng, T.; Sun, X.; Wang, Y.; Huang, L.; Gao, M.; Wang, Y.; Wang, X. Expression of a grape (Vitis vinifera) bZIP transcription factor, VlbZIP36, in Arabidopsis thaliana confers tolerance of drought stress during seed germination and seedling establishment. Plant Sci. 2016, 252, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.L.; Zang, X.S.; Wang, F.; Zhang, L.Y.; Tian, X.J.; Ni, Z.F.; Yao, Y.Y.; Hu, Y.R.; Xin, M.M.; Peng, H.R.; et al. Isolation and function analysis of heat stress related transcription factor gene TabZIP28 in wheat (Triticum aestivum). J. Agric. Biotechnol. 2016, 24, 157–167. [Google Scholar]
- Chuang, C.F.; Running, M.P.; Williams, R.W.; Meyerowitz, E.M. The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 1999, 13, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Silveira, A.B.; Gauer, L.; Tomaz, J.P.; Cardoso, P.R.; Carmello-Guerreiro, S.; Vincentz, M. The Arabidopsis AtbZIP9 protein fused to the VP16 transcriptional activation domain alters leaf and vascular development. Plant Sci. 2007, 172, 1148–1156. [Google Scholar] [CrossRef]
- Zhu, M.; Meng, X.; Cai, J.; Li, G.; Dong, T.; Li, Z. Basic leucine zipper transcription factor SlbZIP1 mediates salt and drought stress tolerance in tomato. BMC Plant Biol. 2018, 18, 83. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, Y.; Zhang, G.; Gan, Z.; Gao, M.; Lv, J.; Wu, T.; Zhang, X.; Xu, X.; Yang, S.; et al. Group-C/S1 bZIP heterodimers regulate MdIPT5b to negatively modulate drought tolerance in apple species. Plant J. 2021, 107, 399–417. [Google Scholar] [CrossRef]
- Geng, X.; Zang, X.; Li, H.; Liu, Z.; Zhao, A.; Liu, J.; Peng, H.; Yao, Y.; Hu, Z.; Ni, Z.; et al. Unconventional splicing of wheat TabZIP60 confers heat tolerance in transgenic Arabidopsis. Plant Sci. 2018, 274, 252–260. [Google Scholar] [CrossRef]
- Kaur, N.; Kaitheri, K.P. Tomato bZIP60 mRNA undergoes splicing in endoplasmic reticulum stress and in response to environmental stresses. Plant Physiol. Biochem. 2021, 160, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Yang, L.; Jia, G.; Yan, K.; Zhang, S.; Yang, G.; Wu, C.; Gai, Y.; Zheng, C.; Huang, J. Maize HEAT UP-REGULATED GENE 1 plays vital roles in heat stress tolerance. J. Exp. Bot. 2022, 73, 6417–6433. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Liao, X.; Li, X.; Wang, B.; Luo, Y.; Yang, X.; Tian, Y.; Zhang, L.; Zhang, F.; Pan, Y.; et al. DgbZIP3 interacts with DgbZIP2 to increase the expression of DgPOD for cold stress tolerance in chrysanthemum. Hortic. Res. 2022, 9, uhac105. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Jiao, Z.; Bian, L.; Wan, Y.; Yu, K.; Zhang, G.; Guo, D. Overexpression of Vitis vinifera VvbZIP60 enhances Arabidopsis resistance to powdery mildew via the salicylic acid signaling pathway. Sci. Hortic. 2019, 256, 108640–108646. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, X.; Zhang, X.; Bi, Y.; Gao, W. A bZIP transcription factor, PqbZIP1, is involved in the plant defense response of American ginseng. PeerJ 2022, 10, e12939. [Google Scholar] [CrossRef]
- Mei, Z.Q.; Fu, S.Y.; Yu, H.Q.; Yang, L.Q.; Duan, C.G.; Liu, X.Y. Genetic characterization and authentication of Dimocarpus longan lour. using an improved RAPD technique. Genet. Mol. Res. 2014, 13, 1447–1455. [Google Scholar] [CrossRef]
- Lai, Z.X. Study of Longan Biotechnology, 1st ed.; Fujian Science and Technology Press: Fuzhou, China, 2003. [Google Scholar]
- Lin, Y.; Min, J.; Lai, R.; Wu, Z.; Chen, Y.; Yu, L.; Cheng, C.; Jin, Y.; Tian, Q.; Liu, Q.; et al. Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. Gigascience 2017, 6, gix023. [Google Scholar] [CrossRef]
- Chen, Y.; Xie, D.; Ma, X.; Xue, X.; Liu, M.; Xiao, X.; Lai, C.; Xu, X.; Chen, X.; Chen, Y.; et al. Genome-wide high-throughput chromosome conformation capture analysis reveals hierarchical chromatin interactions during early somatic embryogenesis. Plant Physiol. 2023, 193, 555–577. [Google Scholar] [CrossRef]
- Li, Z.; Fu, Z.; Zhang, S.; Zhang, X.; Xue, X.; Chen, Y.; Zhang, Z.; Lai, Z.; Lin, Y. Genome-wide analysis of the GLP gene family and overexpression of GLP1-5-1 to promote lignin accumulation during early somatic embryo development in Dimocarpus longan. BMC Genom. 2023, 24, 138. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, X.; Xue, X.; Liu, M.; Zhang, X.; Xiao, X.; Lai, C.; Zhang, Z.; Lai, Z.; Lin, Y. Genome-wide analysis of the SAUR gene family and function exploration of DlSAUR32 during early longan somatic embryogenesis. Plant Physiol. Biochem. 2023, 195, 362–374. [Google Scholar] [CrossRef]
- Ma, X.; Chen, Y.; Liu, M.; Xue, X.; Zhang, X.; Xu, L.; Lai, Z.; Lin, Y. Genome-wide analysis of the XTH gene family and functional analysis of DlXTH23.5/25 during early longan somatic embryogenesis. Front. Plant Sci. 2022, 13, 1043464. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, X.; Shen, X.; Chen, R.; Zhu, C.; Zhang, Z.; Chen, Y.; Lin, W.; Xu, X.; Lin, Y.; et al. Genome-wide identification and characterization of DEAD-box helicase family associated with early somatic embryogenesis in Dimocarpus longan Lour. J. Plant Physiol. 2021, 258–259, 153364. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, Y.; Zhang, S.; Su, L.; Xu, X.; Chen, X.; Lai, Z.; Lin, Y. Genome-wide identification and expression analyses of Sm genes reveal their involvement in early somatic embryogenesis in Dimocarpus longan Lour. PLoS ONE 2020, 15, e0230795. [Google Scholar] [CrossRef] [PubMed]
- Chern, M.S.; Bobb, A.J.; Bustos, M.M. The regulator of MAT2 (ROM2) protein binds to early maturation promoters and represses PvALF-activated transcription. Plant Cell 1996, 8, 305–321. [Google Scholar] [PubMed]
- Kim, S.Y.; Chung, H.J.; Thomas, T.L. Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 1997, 11, 1237–1251. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Ren, H.; Xie, H.; Ma, Z.; Chen, F. Identification and characterization of bZIP-type transcription factors involved in carrot (Daucus carota L.) somatic embryogenesis. Plant J. 2009, 60, 207–217. [Google Scholar] [CrossRef]
- Liu, J.X.; Howell, S.H. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar] [CrossRef]
- Kim, B.; Shim, S.; Zhang, H.; Lee, C.; Jang, S.; Jin, Z.; Seo, J.; Kwon, S.W.; Koh, H.J. Dynamic transcriptome changes driven by the mutation of OsCOP1 underlie flavonoid biosynthesis and embryogenesis in the developing rice seed. J. Plant Growth Regul. 2023, 42, 4436–4452. [Google Scholar] [CrossRef]
- Collin, A.; Daszkowska-Golec, A.; Kurowska, M.; Szarejko, I. Barley ABI5 (Abscisic acid INSENSITIVE 5) Is involved in abscisic acid-dependent drought response. Front. Plant Sci. 2020, 11, 1138. [Google Scholar] [CrossRef]
- Liu, W.; Zhao, C.; Liu, L.; Huang, D.; Ma, C.; Li, R.; Huang, L. Genome-wide identification of the TGA gene family in kiwifruit (Actinidia chinensis spp.) and revealing its roles in response to Pseudomonas syringae pv. actinidiae (Psa) infection. Int. J. Biol. Macromol. 2022, 222 Pt A, 101–113. [Google Scholar] [CrossRef]
- Deng, Y.; Humbert, S.; Liu, J.X.; Srivastava, R.; Rothstein, S.J.; Howell, S.H. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7247–7252. [Google Scholar] [CrossRef]
- Zhang, H.; Ding, X.; Wang, H.; Chen, H.; Dong, W.; Zhu, J.; Wang, J.; Peng, S.; Dai, H.; Mei, W. Systematic evolution of bZIP transcription factors in Malvales and functional exploration of AsbZIP14 and AsbZIP41 in Aquilaria sinensis. Front. Plant Sci. 2023, 14, 1243323. [Google Scholar] [CrossRef]
- Zhou, R.; Zhao, G.; Zheng, S.; Xie, S.; Lu, C.; Liu, S.; Wang, Z.; Niu, J. Comprehensive functional analysis of the bZIP family in Bletilla striata Reveals that BsbZIP13 could respond to multiple abiotic stresses. Int. J. Mol. Sci. 2023, 24, 15202. [Google Scholar] [CrossRef]
- Lai, Z.; Pan, L.; Chen, Z. Establishment and maintenance of longan embryogenic cell lines. J. Fujian Agric. Univ. 1997, 2, 33–40. [Google Scholar]
- Philippe, L.; Berardini, T.Z.; Li, D.; David, S.; Christopher, W.; Rajkumar, S.; Robert, M.; Kate, D.; Alexander, D.L.; Margarita, G.H. The arabidopsis information resource (TAIR): Improved gene annotation and new tools, D1. Nucleic Acids Res. 2011, 40, D1202–D1240. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big Biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.; Tsirigos, K.D.; Brunak, S.; von Heijne, G. A Brief History of Protein Sorting Prediction. Protein. J. 2019, 38, 200–216. [Google Scholar] [CrossRef]
- Bolser, D.; Staines, D.M.; Pritchard, E.; Kersey, P. Ensembl Plants: Integrating tools for visualizing, mining, and analyzing plant genomics data. Methods Mol. Biol. 2016, 1374, 115–140. [Google Scholar] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Liu, C.; Xie, T.; Chen, C.; Luan, A.; Long, J.; Li, C.; Ding, Y.; He, Y. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genom. 2017, 18, 503. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30 (Suppl. S1), S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M.; et al. STRING 8—A global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37, D412–D416. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, T.; Lan, S.; Xv, L.; Zhang, X.; Ma, X.; Li, Z.; Gao, J.; Chen, Y.; Lai, Z.; Lin, Y. Genome-Wide Identification and Expression Analysis Reveal bZIP Transcription Factors Mediated Hormones That Functions during Early Somatic Embryogenesis in Dimocarpus longan. Plants 2024, 13, 662. https://doi.org/10.3390/plants13050662
Zhai T, Lan S, Xv L, Zhang X, Ma X, Li Z, Gao J, Chen Y, Lai Z, Lin Y. Genome-Wide Identification and Expression Analysis Reveal bZIP Transcription Factors Mediated Hormones That Functions during Early Somatic Embryogenesis in Dimocarpus longan. Plants. 2024; 13(5):662. https://doi.org/10.3390/plants13050662
Chicago/Turabian StyleZhai, Tingkai, Shuoxian Lan, Luzhen Xv, Xueying Zhang, Xiangwei Ma, Zhuoyun Li, Jie Gao, Yukun Chen, Zhongxiong Lai, and Yuling Lin. 2024. "Genome-Wide Identification and Expression Analysis Reveal bZIP Transcription Factors Mediated Hormones That Functions during Early Somatic Embryogenesis in Dimocarpus longan" Plants 13, no. 5: 662. https://doi.org/10.3390/plants13050662
APA StyleZhai, T., Lan, S., Xv, L., Zhang, X., Ma, X., Li, Z., Gao, J., Chen, Y., Lai, Z., & Lin, Y. (2024). Genome-Wide Identification and Expression Analysis Reveal bZIP Transcription Factors Mediated Hormones That Functions during Early Somatic Embryogenesis in Dimocarpus longan. Plants, 13(5), 662. https://doi.org/10.3390/plants13050662







