OsUGT88C3 Encodes a UDP-Glycosyltransferase Responsible for Biosynthesis of Malvidin 3-O-Galactoside in Rice
Abstract
:1. Introduction
2. Results
2.1. Identification of Rice Varieties with Different Leaf Color Patterns
2.2. Expression Levels of Anthocyanin Biosynthesis Genes Were Positively Correlated with Most Anthocyanin Contents
2.3. Differentially Expressed Genes (DEGs) Were Enriched in Phenylpropanoid and Flavonoid Pathways
2.4. Exploration of UGT Genes Involved in Anthocyanin Biosynthesis
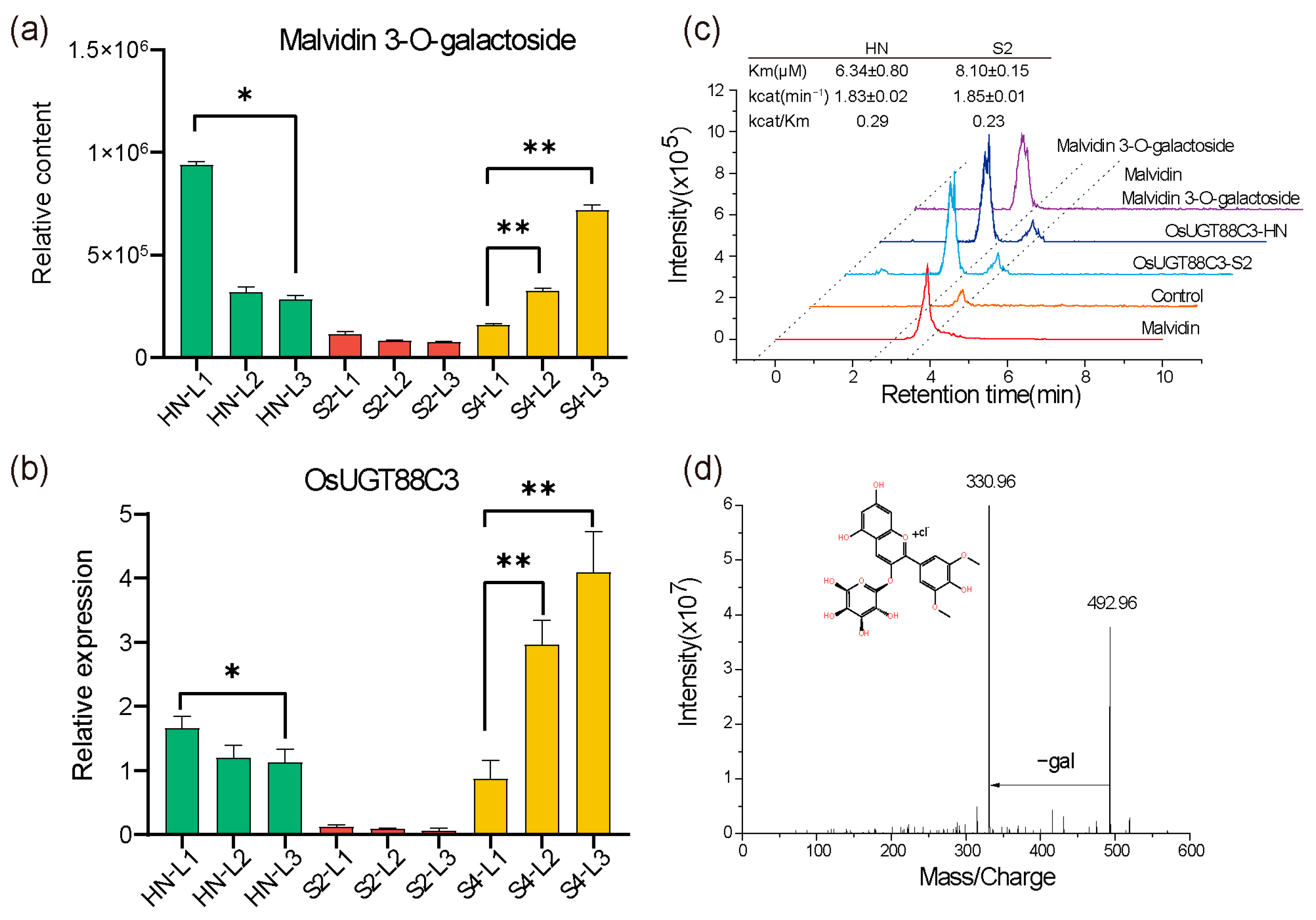
2.5. OsUGT88C3 Is Responsible for Malvidin Galactosylation
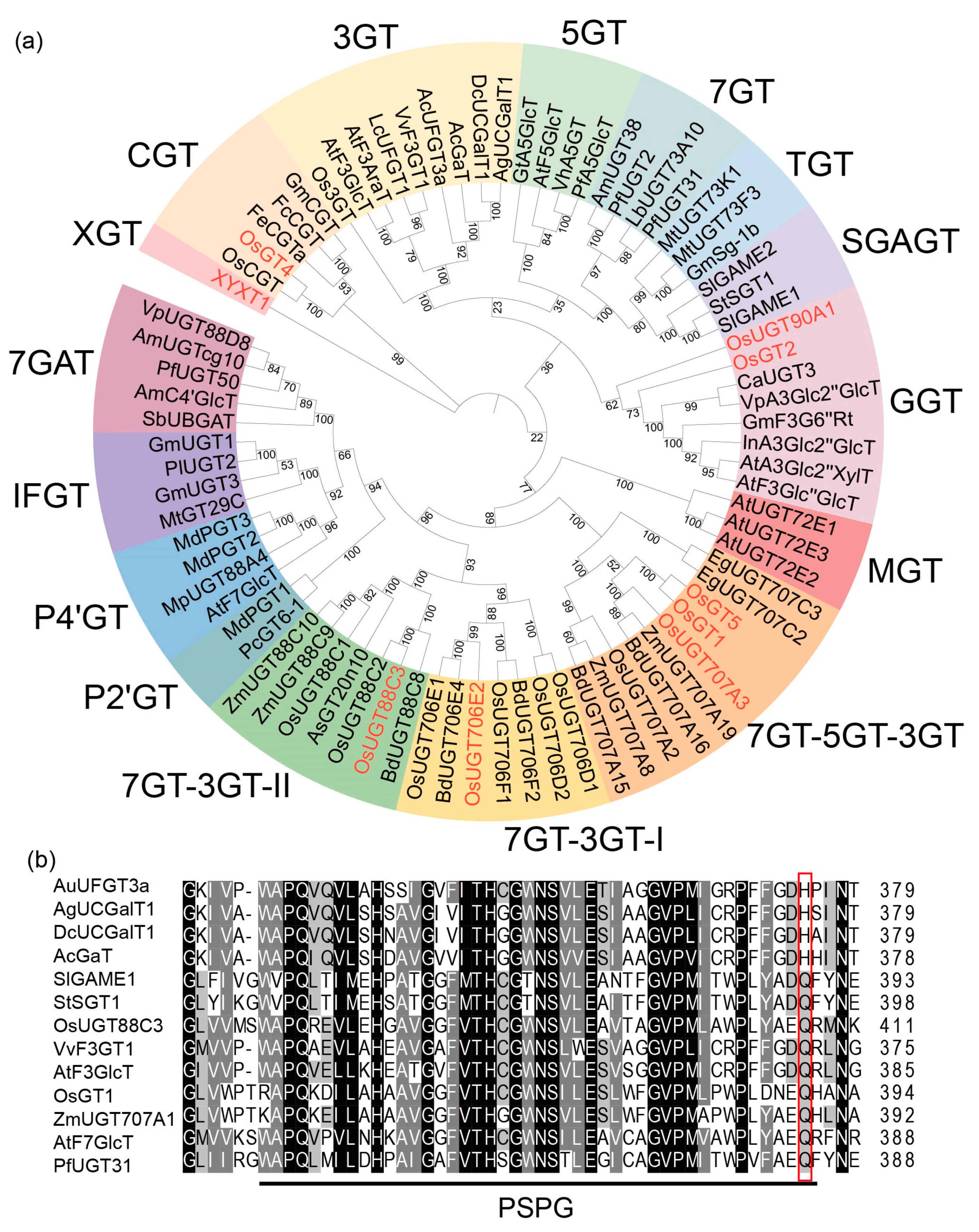
2.6. OsUGT88C3 Is a Unique Enzyme Distinct from Other UDP-Glycosyltransferase
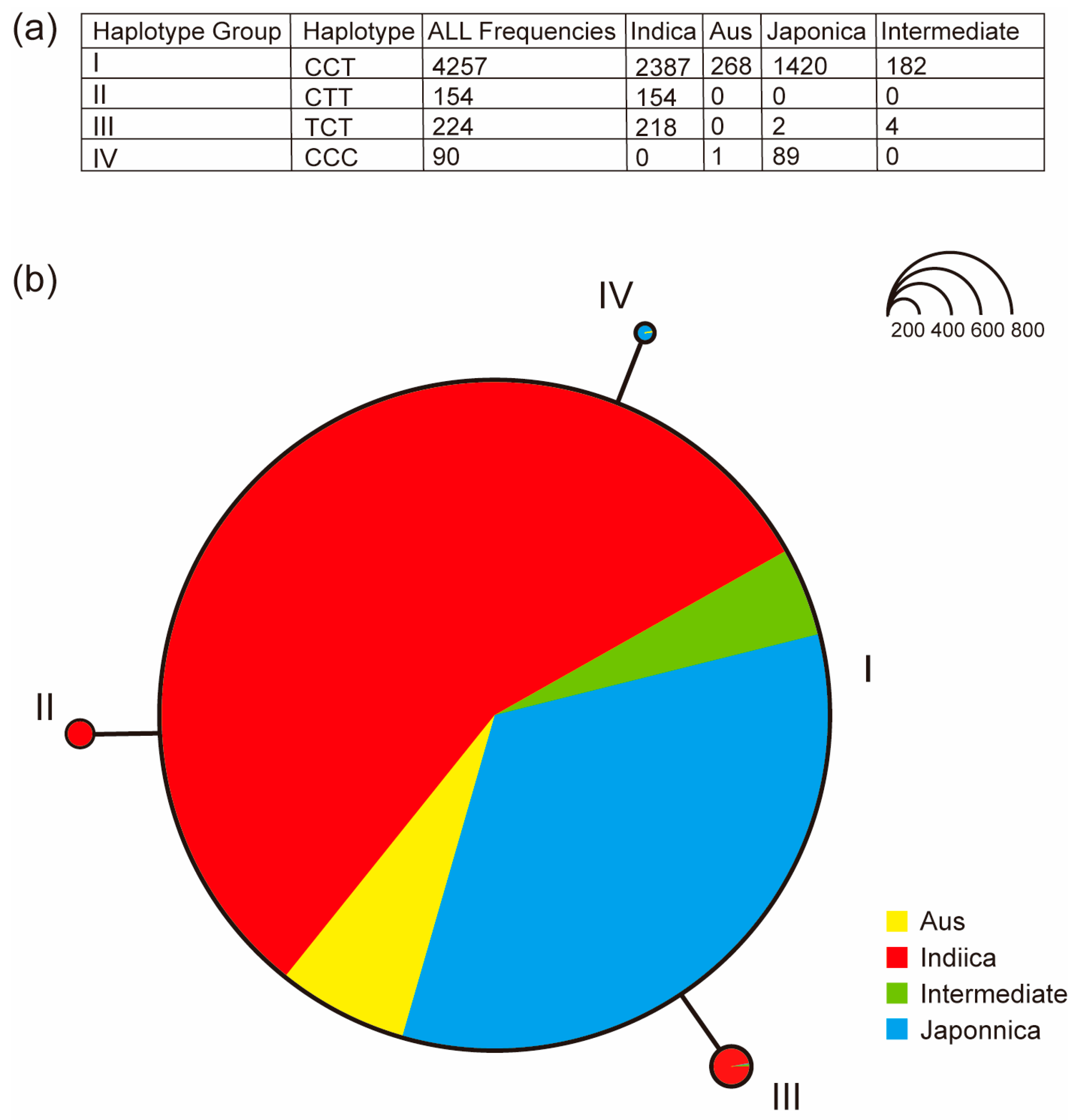
2.7. Haplotype Analysis of OsUGT88C3
2.8. Subcellular Localization Analysis of OsUGT88C3
2.9. OsUGT88C3 Is Highly Expressed in Leaves, Sheaths, Pistils, and Embryos
3. Discussion
3.1. Multiple UGTs Participate in the Synthesis of Anthocyanins in Rice Leaves
3.2. OsUGT88C3 Is Responsible for Malvidin Galactosylation
3.3. OsUGT88C3 Is a Unique Galactosyltransferase
3.4. The Catalytic Activity of OsUGT88C3 Is Conserved in Rice Germplasm
4. Materials and Methods
4.1. Plant Materials
4.2. Total Anthocyanin Content Measurement
4.3. Metabolic Sample Preparation and Profiling
4.4. RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction
4.5. RNA Sequencing and Data Analysis
4.6. KEGG Analysis
4.7. Recombinant Protein Expression and In Vitro Enzyme Assay
4.8. Phylogenetic Analysis and Multiple Sequence Alignment
4.9. Haplotype Analysis
4.10. Subcellular Localization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Bradshaw, H.D.; Schemske, D.W. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 2003, 426, 176–178. [Google Scholar] [CrossRef]
- Sarma, A.D.; Sharma, R. Anthocyanin-DNA copigmentation complex: Mutual protection against oxidative damage. Phytochemistry 1999, 52, 1313–1318. [Google Scholar] [CrossRef]
- Christie, P.J.; Alfenito, M.R.; Walbot, V. Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: Enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 1994, 194, 541–549. [Google Scholar] [CrossRef]
- Castellarin, S.D.; Pfeiffer, A.; Sivilotti, P.; Degan, M.; Peterlunger, E.; Gaspero, G.D. Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ. 2007, 30, 1381–1399. [Google Scholar] [CrossRef]
- Lorenc-Kukuła, K.; Jafra, S.; Oszmiański, J.; Szopa, J. Ectopic Expression of Anthocyanin 5-O-Glucosyltransferase in Potato Tuber Causes Increased Resistance to Bacteria. J. Agric. Food Chem. 2005, 53, 272–281. [Google Scholar] [CrossRef]
- Nistor, M.; Pop, R.; Daescu, A.; Pintea, A.; Socaciu, C.; Rugina, D. Anthocyanins as Key Phytochemicals Acting for the Prevention of Metabolic Diseases: An Overview. Molecules 2022, 27, 4254. [Google Scholar] [CrossRef]
- Wang, S.; Alseekh, S.; Fernie, A.R.; Luo, J. The Structure and Function of Major Plant Metabolite Modifications. Mol. Plant 2019, 12, 899–919. [Google Scholar] [CrossRef]
- Smeriglio, A.; Barreca, D.; Bellocco, E.; Trombetta, D. Chemistry, Pharmacology and Health Benefits of Anthocyanins. Phytother. Res. 2016, 30, 1265–1286. [Google Scholar] [CrossRef]
- Mackon, E.; Jeazet Dongho Epse Mackon, G.C.; Ma, Y.; Haneef Kashif, M.; Ali, N.; Usman, B.; Liu, P. Recent Insights into Anthocyanin Pigmentation, Synthesis, Trafficking, and Regulatory Mechanisms in Rice (Oryza sativa L.) Caryopsis. Biomolecules 2021, 11, 394. [Google Scholar] [CrossRef]
- Xia, D.; Zhou, H.; Wang, Y.; Li, P.; Fu, P.; Wu, B.; He, Y. How rice organs are colored: The genetic basis of anthocyanin biosynthesis in rice. Crop J. 2021, 9, 598–608. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Xia, X.; Zhang, Z.; He, J.; Nong, B.; Luo, T.; Feng, R.; Wu, Y.; Pan, Y.; et al. OsTTG1, a WD40 repeat gene, regulates anthocyanin biosynthesis in rice. Plant J. 2021, 107, 198–214. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, H.; Zhu, H.; Huang, C.; Liu, C.; Chang, Y.; Kong, Z.; Zhou, Z.; Wang, G.; Lin, Y.; et al. Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 2019, 223, 705–721. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef]
- Nagashima, S.; Okamoto, A.; Suzuki, H.; Asada, Y.; Kondo, T.; Yoshikawa, T. Anthocyanin galactosyltransferase from Aralia cordata, cDNA cloning and characterization. Plant Biotechnol. 2004, 21, 191–195. [Google Scholar] [CrossRef]
- Xu, Z.S.; Ma, J.; Wang, F.; Ma, H.Y.; Wang, Q.X.; Xiong, A.S. Identification and characterization of DcUCGalT1, a galactosyltransferase responsible for anthocyanin galactosylation in purple carrot (Daucus carota L.) taproots. Sci. Rep. 2016, 6, 27356. [Google Scholar] [CrossRef]
- Feng, K.; Xu, Z.S.; Liu, J.X.; Li, J.W.; Wang, F.; Xiong, A.S. Isolation, purification, and characterization of AgUCGalT1, a galactosyltransferase involved in anthocyanin galactosylation in purple celery (Apium graveolens L.). Planta 2018, 247, 1363–1375. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, B.; Qi, Y.; Liu, C.; Liu, Z.; Ren, X. Biochemical and functional characterization of AcUFGT3a, a galactosyltransferase involved in anthocyanin biosynthesis in the red-fleshed kiwifruit (Actinidia chinensis). Physiol. Plant. 2018, 162, 409–426. [Google Scholar] [CrossRef]
- Peng, M.; Shahzad, R.; Gul, A.; Subthain, H.; Shen, S.; Lei, L.; Zheng, Z.; Zhou, J.; Lu, D.; Wang, S.; et al. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat. Commun. 2017, 8, 1975. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, F.; Deng, Y.; Sun, L.; Mao, M.; Chen, R.; Qiang, Q.; Zhou, J.; Long, T.; Zhao, X.; et al. Integrated Metabolomics and Transcriptomics Analyses Reveal the Metabolic Differences and Molecular Basis of Nutritional Quality in Landraces and Cultivated Rice. Metabolites 2022, 12, 384. [Google Scholar] [CrossRef]
- Zhong, R.; Cui, D.; Phillips, D.R.; Ye, Z.-H. A Novel Rice Xylosyltransferase Catalyzes the Addition of 2-O-Xylosyl Side Chains onto the Xylan Backbone. Plant Cell Physiol. 2018, 59, 554–565. [Google Scholar] [CrossRef]
- Shi, Y.; Phan, H.; Liu, Y.; Cao, S.; Zhang, Z.; Chu, C.; Schläppi, M.R. Glycosyltransferase OsUGT90A1 helps protect the plasma membrane during chilling stress in rice. J. Exp. Bot. 2020, 71, 2723–2739. [Google Scholar] [CrossRef]
- He, X.; Huang, R.; Liu, L.; Li, Y.; Wang, W.; Xu, Q.; Yu, Y.; Zhou, T. CsUGT78A15 catalyzes the anthocyanidin 3-O-galactoside biosynthesis in tea plants. Plant Physiol. Biochem. 2021, 166, 738–749. [Google Scholar] [CrossRef]
- Ford, C.M.; Boss, P.K.; Høj, P.B. Cloning and characterization of Vitis vinifera UDP-glucose:flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J. Biol. Chem. 1998, 273, 9224–9233. [Google Scholar] [CrossRef]
- Li, X.J.; Zhang, J.Q.; Wu, Z.C.; Lai, B.; Huang, X.M.; Qin, Y.H.; Wang, H.C.; Hu, G.B. Functional characterization of a glucosyltransferase gene, LcUFGT1, involved in the formation of cyanidin glucoside in the pericarp of Litchi chinensis. Physiol. Plant. 2016, 156, 139–149. [Google Scholar] [CrossRef]
- Zhu, Q.; Yu, S.; Zeng, D.; Liu, H.; Wang, H.; Yang, Z.; Xie, X.; Shen, R.; Tan, J.; Li, H.; et al. Development of “Purple Endosperm Rice” by Engineering Anthocyanin Biosynthesis in the Endosperm with a High-Efficiency Transgene Stacking System. Mol. Plant 2017, 10, 918–929. [Google Scholar] [CrossRef]
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J.; Awazuhara, M.; Inoue, E.; Takahashi, H.; Goodenowe, D.B.; Kitayama, M.; et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005, 42, 218–235. [Google Scholar] [CrossRef]
- Yamazaki, M.; Gong, Z.; Fukuchi-Mizutani, M.; Fukui, Y.; Tanaka, Y.; Kusumi, T.; Saito, K. Molecular cloning and biochemical characterization of a novel anthocyanin 5-O-glucosyltransferase by mRNA differential display for plant forms regarding anthocyanin. J. Biol. Chem. 1999, 274, 7405–7411. [Google Scholar] [CrossRef]
- Ono, E.; Ruike, M.; Iwashita, T.; Nomoto, K.; Fukui, Y. Co-pigmentation and flavonoid glycosyltransferases in blue Veronica persica flowers. Phytochemistry 2010, 71, 726–735. [Google Scholar] [CrossRef]
- Morita, Y.; Hoshino, A.; Kikuchi, Y.; Okuhara, H.; Ono, E.; Tanaka, Y.; Fukui, Y.; Saito, N.; Nitasaka, E.; Noguchi, H.; et al. Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O-glucoside-2″-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J. 2005, 42, 353–363. [Google Scholar]
- Yonekura-Sakakibara, K.; Fukushima, A.; Nakabayashi, R.; Hanada, K.; Matsuda, F.; Sugawara, S.; Inoue, E.; Kuromori, T.; Ito, T.; Shinozaki, K.; et al. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 2012, 69, 154–167. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Nakabayashi, R.; Sugawara, S.; Tohge, T.; Ito, T.; Koyanagi, M.; Kitajima, M.; Takayama, H.; Saito, K. A flavonoid 3-O-glucoside:2”-O-glucosyltransferase responsible for terminal modification of pollen-specific flavonols in Arabidopsis thaliana. Plant J. 2014, 79, 769–782. [Google Scholar] [CrossRef]
- Zhao, H.; Yao, W.; Ouyang, Y.; Yang, W.; Wang, G.; Lian, X.; Xing, Y.; Chen, L.; Xie, W. RiceVarMap: A comprehensive database of rice genomic variations. Nucleic Acids Res. 2015, 43, D1018–D1022. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Batistic, O.; Waadt, R.; Steinhorst, L.; Held, K.; Kudla, J. CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 2010, 61, 211–222. [Google Scholar] [CrossRef]
- Zhu, Q.; Wang, B.; Tan, J.; Liu, T.; Li, L.; Liu, Y.G. Plant Synthetic Metabolic Engineering for Enhancing Crop Nutritional Quality. Plant Commun. 2020, 1, 100017. [Google Scholar] [CrossRef]
- He, Y.; Sun, S.; Zhao, J.; Huang, Z.; Peng, L.; Huang, C.; Tang, Z.; Huang, Q.; Wang, Z. UDP-glucosyltransferase OsUGT75A promotes submergence tolerance during rice seed germination. Nat. Commun. 2023, 14, 2296. [Google Scholar] [CrossRef]
- Itkin, M.; Rogachev, I.; Alkan, N.; Rosenberg, T.; Malitsky, S.; Masini, L.; Meir, S.; Iijima, Y.; Aoki, K.; de Vos, R.; et al. GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell 2011, 23, 4507–4525. [Google Scholar] [CrossRef]
- McCue, K.F.; Shepherd, L.V.T.; Allena, P.V.; Maccree, M.M.; Rockhold, D.R.; Corsini, D.L.; Davies, H.V.; Belknap, W.R. Metabolic compensation of steroidal glycoalkaloid biosynthesis in transgenic potato tubers: Using reverse genetics to confirm the in vivo enzyme function of a steroidal alkaloid galactosyltransferase. Plant Sci. 2005, 168, 267–273. [Google Scholar] [CrossRef]
- Ren, Z.; Ji, X.; Jiao, Z.; Luo, Y.; Zhang, G.-Q.; Tao, S.; Lei, Z.; Zhang, J.; Wang, Y.; Liu, Z.-J.; et al. Functional analysis of a novel C-glycosyltransferase in the orchid Dendrobium catenatum. Hortic. Res. 2020, 7, 111. [Google Scholar] [CrossRef]
- Souza, L.P.d.; Scossa, F.; Proost, S.; Bitocchi, E.; Papa, R.; Tohge, T.; Fernie, A.R. Multi-tissue integration of transcriptomic and specialized metabolite profiling provides tools for assessing the common bean (Phaseolus vulgaris) metabolome. Plant J. 2019, 97, 1132–1153. [Google Scholar] [CrossRef]
- Noguchi, A.; Horikawa, M.; Fukui, Y.; Fukuchi-Mizutani, M.; Iuchi-Okada, A.; Ishiguro, M.; Kiso, Y.; Nakayama, T.; Ono, E. Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in Lamiales. Plant Cell 2009, 21, 1556–1572. [Google Scholar] [CrossRef]
- Kim, B.G.; Jung, N.R.; Joe, E.J.; Hur, H.G.; Lim, Y.; Chong, Y.; Ahn, J.H. Bacterial synthesis of a flavonoid deoxyaminosugar conjugate in Escherichia coli expressing a glycosyltransferase of Arabidopsis thaliana. ChemBioChem 2010, 11, 2389–2392. [Google Scholar] [CrossRef]
- Itkin, M.; Heinig, U.; Tzfadia, O.; Bhide, A.J.; Shinde, B.; Cardenas, P.D.; Bocobza, S.E.; Unger, T.; Malitsky, S.; Finkers, R.; et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 2013, 341, 175–179. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Z.; Gu, Y. Caryopsis Development and Anthocyanidin Accumulation of Colored Rice. Chin. J. Rice Sci./Zhongguo Shuidao Kexue 2011, 25, 392–398. [Google Scholar]
- Yang, J.; Chen, R.; Wang, C.; Li, C.; Ye, W.; Zhang, Z.; Wang, S. A widely targeted metabolite modificomics strategy for modified metabolites identification in tomato. J. Integr. Plant Biol. 2024, 00, 1–14. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Stat. Method) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Zhu, F.; Cai, Y.-Z.; Bao, J.; Corke, H. Effect of γ-irradiation on phenolic compounds in rice grain. Food Chem. 2010, 120, 74–77. [Google Scholar] [CrossRef]




| Locus | Gene Symbol | Tissue a | Annotation b | Function c | Reference |
|---|---|---|---|---|---|
| LOC_Os04g24110 | OsUGT707A3 | HN-L3/HN-L1, S2-L3/S2-L1 | UDP-glucoronosyl and UDP-glucosyl transferase domain containing protein, expressed | Glucosylates 3-OH of kaempferol | [21] |
| LOC_Os05g45200 | OsGT4, OsCGT | Not detected | UDP-glucoronosyl and UDP-glucosyl transferase domain containing protein, expressed | Glucosylates cyanidin, malvidin, peonidin, and procyanidin A1 | [22] |
| LOC_Os07g32620 | OsUGT88C3 | HN-L3/HN-L1, S2-L3/S2-L1 | Anthocyanidin 5,3-O-glucosyltransferase, putative, expressed | No activity on flavones, flavonols, and flavanones | [21] |
| LOC_Os02g37690 | OsGT1 | HN-L3/HN-L1, S2-L3/S2-L1 | UDP-glucoronosyl and UDP-glucosyl transferase domain containing protein, expressed | Catalyzes cyanidin as a substrate to form cyanidin 3-O-glucoside | [22] |
| LOC_Os06g49300 | OsXYXT1 | Not detected | Glycosyltransferase protein, putative, expressed | Catalyzes the addition of 2-O-xylosyl side chains onto the xylan backbone | [23] |
| LOC_Os07g32630 | OsGT2 | Not detected | UDP-glucoronosyl and UDP-glucosyl transferase, putative, expressed | Glucosylates cyanidin, malvidin, and procyanidin B2 | [22] |
| LOC_Os01g53370 | OsGT5 | HN-L3/HN-L1, S2-L3/S2-L1 | Anthocyanidin 5,3-O-glucosyltransferase, putative, expressed | Glucosylates cyanidin, peonidin, and procyanidin A1 | [22] |
| LOC_Os06g18010 | OsUGT706E2 | HN-L3/HN-L2, S2-L3/S2-L1 | Anthocyanidin 5,3-O-glucosyltransferase, putative, expressed | No activity on flavones, flavonols, and flavanones | [21] |
| LOC_Os07g32020 | OsUGT90A1 | HN-L3/HN-L1, HN-L3/HN-L2, S2-L3/S2-L1S2-3/S2-1 | Anthocyanin 3-O-beta-glucosyltransferase, putative, expressed | Helps protect plasma membranes during chilling stress in rice | [24] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Fu, S.; Cao, Z.; Liu, H.; Huang, S.; Li, C.; Zhang, Z.; Yang, H.; Wang, S.; Luo, J.; et al. OsUGT88C3 Encodes a UDP-Glycosyltransferase Responsible for Biosynthesis of Malvidin 3-O-Galactoside in Rice. Plants 2024, 13, 697. https://doi.org/10.3390/plants13050697
Zhao S, Fu S, Cao Z, Liu H, Huang S, Li C, Zhang Z, Yang H, Wang S, Luo J, et al. OsUGT88C3 Encodes a UDP-Glycosyltransferase Responsible for Biosynthesis of Malvidin 3-O-Galactoside in Rice. Plants. 2024; 13(5):697. https://doi.org/10.3390/plants13050697
Chicago/Turabian StyleZhao, Sihan, Shuying Fu, Zhenfeng Cao, Hao Liu, Sishu Huang, Chun Li, Zhonghui Zhang, Hongbo Yang, Shouchuang Wang, Jie Luo, and et al. 2024. "OsUGT88C3 Encodes a UDP-Glycosyltransferase Responsible for Biosynthesis of Malvidin 3-O-Galactoside in Rice" Plants 13, no. 5: 697. https://doi.org/10.3390/plants13050697
APA StyleZhao, S., Fu, S., Cao, Z., Liu, H., Huang, S., Li, C., Zhang, Z., Yang, H., Wang, S., Luo, J., & Long, T. (2024). OsUGT88C3 Encodes a UDP-Glycosyltransferase Responsible for Biosynthesis of Malvidin 3-O-Galactoside in Rice. Plants, 13(5), 697. https://doi.org/10.3390/plants13050697






