Soil, Plant, and Microorganism Interactions Drive Secondary Succession in Alpine Grassland Restoration
Abstract
1. Introduction
2. Results
2.1. Plant Communities
2.2. Soil Properties and Microbial Activity
2.3. Soil Microbial Community Composition
2.4. Relationship between Soil Properties, Plant Communities, and Microbial Populations
3. Discussion
3.1. Effect of Secondary Succession on Plant and Soil Properties
3.2. Effect of Secondary Succession on Soil Microorganisms
3.3. Relationships among Plants, Soil Properties, and Microbial Succession
4. Materials and Methods
4.1. Study Sites
4.2. Experimental Design and Sampling
4.3. Soil Chemical Analysis
4.4. Microbial Biomass and Enzymatic Activities
4.5. Soil DNA Extraction and Sequencing
4.6. Bioinformatics Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, J.; Li, X.L. Degradation of frigid swampy meadows on the Qinghai–Tibet Plateau. Prog. Phys. Geogr. 2016, 40, 794–810. [Google Scholar] [CrossRef]
- Li, X.L.; Perry, G.L.W.; Brierley, G.; Sun, H.Q.; Li, C.H.; Lu, G.X. Quantitative assessment of degradation classifications for degraded alpine meadows (heitutan), Sanjiangyuan, Western China. Land Degrad. Dev. 2012, 25, 417–427. [Google Scholar] [CrossRef]
- Li, X.L.; Xue, Z.P.; Gao, J. Dynamic changes of plateau wetlands in Madou county, the Yellow River source zone of China: 1990–2013. Wetlands 2016, 36, 299–310. [Google Scholar] [CrossRef]
- Li, X.L.; Xue, Z.P.; Gao, J. Environmental influence on vegetation properties of frigid wetlands on the Qinghai-Tibet Plateau, Western China. Wetlands 2016, 36, 807–819. [Google Scholar] [CrossRef]
- Gao, J.; Li, X.L. A knowledge-based approach to mapping degraded meadows on the Qinghai–Tibet Plateau, China. Int. J. Remote Sens. 2017, 38, 6147–6163. [Google Scholar] [CrossRef]
- Fierer, N.; Wood, S.A.; Bueno de Mesquita, C.P. How microbes can, and cannot, be used to assess soil health. Soil Biol. Biochem. 2021, 153, 108111. [Google Scholar] [CrossRef]
- Coban, O.; De Deyn, G.B.; van der Ploeg, M. Soil microbiota as game-changers in restoration of degraded lands. Science 2022, 375, abe0725. [Google Scholar] [CrossRef]
- Van der Putten, W.H.; Bardgett, R.D.; Bever, J.D.; Bezemer, T.M.; Casper, B.B.; Fukami, T.; Kardol, P.; Klironomos, J.N.; Kulmatiski, A.; Schweitzer, J.A.; et al. Plant–soil feedbacks: The past, the present and future challenges. J. Ecol. 2013, 101, 265–276. [Google Scholar] [CrossRef]
- Cui, Y.X.; Fang, L.C.; Guo, X.B.; Wang, X.; Zhang, Y.J.; Li, P.F.; Zhang, X.C. Ecoenzymatic stoichiometry and microbial nutrient limitation in rhizosphere soil in the arid area of the northern Loess Plateau, China. Soil Biol. Biochem. 2018, 116, 11–21. [Google Scholar] [CrossRef]
- Cui, Y.X.; Wang, X.; Zhang, X.C.; Ju, W.L.; Duan, C.J.; Guo, X.B.; Wang, Y.Q.; Fang, L.C. Soil moisture mediates microbial carbon and phosphorus metabolism during vegetation succession in a semiarid region. Soil Biol. Biochem. 2020, 147, 107814. [Google Scholar] [CrossRef]
- Haichar, F.e.Z.; Marol, C.; Berge, O.; Rangel-Castro, J.I.; Prosser, J.I.; Balesdent, J.; Heulin, T.; Achouak, W. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008, 2, 1221–1230. [Google Scholar] [CrossRef]
- Kardol, P.; De Deyn, G.B.; Laliberté, E.; Mariotte, P.; Hawkes, C.V.; van der Putten, W. Biotic plant–soil feedbacks across temporal scales. J. Ecol. 2013, 101, 309–315. [Google Scholar] [CrossRef]
- Bitas, V.; Kim, H.-S.; Bennett, J.W.; Kang, S. Sniffing on microbes: Diverse roles of microbial volatile organic compounds in plant health. Mol. Plant. Microbe Interact. 2013, 26, 835–843. [Google Scholar] [CrossRef]
- Wang, J.; Liu, G.B.; Zhang, C.; Wang, G.L.; Fang, L.C.; Cui, Y.X. Higher temporal turnover of soil fungi than bacteria during long-term secondary succession in a semiarid abandoned farmland. Soil Till. Res. 2019, 194, 104305. [Google Scholar] [CrossRef]
- Zhong, Y.Q.W.; Yan, W.M.; Wang, R.W.; Wang, W.; Shangguan, Z.P. Decreased occurrence of carbon cycle functions in microbial communities along with long-term secondary succession. Soil Biol. Biochem. 2018, 123, 207–217. [Google Scholar] [CrossRef]
- Sun, S.; Li, S.; Avera, B.N.; Strahm, B.D.; Badgley, B.D.; Löffler, F.E. Soil bacterial and fungal communities show distinct recovery patterns during forest ecosystem restoration. Appl. Environ. Microbiol. 2017, 83, e00966-17. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, G.B.; Song, Z.; Wang, J.; Guo, L. Interactions of soil bacteria and fungi with plants during long-term grazing exclusion in semiarid grasslands. Soil Biol. Biochem. 2018, 124, 47–58. [Google Scholar] [CrossRef]
- Schmidt, S.K.; Nemergut, D.R.; Darcy, J.L.; Lynch, R. Do bacterial and fungal communities assemble differently during primary succession. Mol. Ecol. 2014, 23, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Bååth, E. Fungal biomass production and turnover in soil estimated using the acetate-in-ergosterol technique. Soil Biol. Biochem. 2007, 39, 2173–2177. [Google Scholar] [CrossRef]
- Cui, Y.X.; Fang, L.C.; Guo, X.B.; Han, F.; Ju, W.L.; Ye, L.P.; Wang, X.; Tan, W.F.; Zhang, X.C. Natural grassland as the optimal pattern of vegetation restoration in arid and semi-arid regions: Evidence from nutrient limitation of soil microbes. Sci. Total Environ. 2019, 648, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Panchal, P.; Preece, C.; Peñuelas, J.; Giri, J. Soil carbon sequestration by root exudates. Trends Plant Sci. 2022, 27, 749–757. [Google Scholar] [CrossRef]
- Lozano, Y.M.; Hortal, S.; Armas, C.; Pugnaire, F.I. Interactions among soil, plants, and microorganisms drive secondary succession in a dry environment. Soil Biol. Biochem. 2014, 78, 298–306. [Google Scholar] [CrossRef]
- Scott, A.J.; Morgan, J.W. Recovery of soil and vegetation in semi-arid Australian old fields. J. Arid Environ. 2012, 76, 61–71. [Google Scholar] [CrossRef]
- Dana, E.D.; Mota, J.F. Vegetation and soil recovery on gypsum outcrops in semi-arid Spain. J. Arid Environ. 2006, 65, 444–459. [Google Scholar] [CrossRef]
- Cheng, J.M.; Jing, G.H.; Wei, L.; Jing, Z.B. Long-term grazing exclusion effects on vegetation characteristics, soil properties and bacterial communities in the semi-arid grasslands of China. Ecol. Eng. 2016, 97, 170–178. [Google Scholar] [CrossRef]
- Baddeley, J.A.; Edwards, A.C.; Watson, C.A. Changes in soil C and N stocks and C:N stoichiometry 21 years after land use change on an arable mineral topsoil. Geoderma 2017, 303, 19–26. [Google Scholar] [CrossRef]
- Zeng, Q.C.; Liu, Y.; Fang, Y.; Ma, R.T.; Lal, R.; An, S.S.; Huang, Y.M. Impact of vegetation restoration on plants and soil C:N:P stoichiometry on the Yunwu Mountain Reserve of China. Ecol. Eng. 2017, 109, 92–100. [Google Scholar] [CrossRef]
- Yang, Y.h.; Luo, Y.Q. Carbon: Nitrogen stoichiometry in forest ecosystems during stand development. Glob. Ecol. Biogeogr. 2011, 20, 354–361. [Google Scholar] [CrossRef]
- Han, C.L.; Zhou, W.D.; Gu, Y.J.; Wang, J.Q.; Zhou, Y.F.; Xue, Y.Y.; Shi, Z.G.; Siddique, K.H.M. Effects of tillage regime on soil aggregate-associated carbon, enzyme activity, and microbial community structure in a semiarid agroecosystem. Plant Soil 2024. [Google Scholar] [CrossRef]
- Mitchell, R.J.; Hester, A.J.; Campbell, C.D.; Chapman, S.J.; Cameron, C.M.; Hewison, R.L.; Potts, J.M. Explaining the variation in the soil microbial community: Do vegetation composition and soil chemistry explain the same or different parts of the microbial variation? Plant Soil 2011, 351, 355–362. [Google Scholar] [CrossRef]
- Jangid, K.; Williams, M.A.; Franzluebbers, A.J.; Schmidt, T.M.; Coleman, D.C.; Whitman, W.B. Land-use history has a stronger impact on soil microbial community composition than aboveground vegetation and soil properties. Soil Biol. Biochem. 2011, 43, 2184–2193. [Google Scholar] [CrossRef]
- Kuramae, E.; Gamper, H.; van Veen, J.; Kowalchuk, G. Soil and plant factors driving the community of soil-borne microorganisms across chronosequences of secondary succession of chalk grasslands with a neutral pH. FEMS Microbiol. Ecol. 2011, 77, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, C.; Jiang, L.; Luo, Y. Trends in soil microbial communities during secondary succession. Soil Biol. Biochem. 2017, 115, 92–99. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Banning, N.C.; Gleeson, D.B.; Grigg, A.H.; Grant, C.D.; Andersen, G.L.; Brodie, E.L.; Murphy, D.V. Soil microbial community successional patterns during forest ecosystem restoration. Appl. Environ. Microbiol. 2011, 77, 6158–6164. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.; Blaalid, R.; Vik, U.; Carlsen, T.; Kauserud, H.; Eidesen, P.B. Primary succession of Bistorta vivipara (L.) Delabre (Polygonaceae) root-associated fungi mirrors plant succession in two glacial chronosequences. Environ. Microbiol. 2015, 17, 2777–2790. [Google Scholar] [CrossRef] [PubMed]
- Fry, E.L.; Manning, P.; Macdonald, C.; Hasegawa, S.; De Palma, A.; Power, S.A.; Singh, B.K. Shifts in microbial communities do not explain the response of grassland ecosystem function to plant functional composition and rainfall change. Soil Biol. Biochem. 2016, 92, 199–210. [Google Scholar] [CrossRef]
- Zeng, Q.C.; An, S.S.; Liu, Y. Soil bacterial community response to vegetation succession after fencing in the grassland of China. Sci. Total Environ. 2017, 609, 2–10. [Google Scholar] [CrossRef]
- Millard, P.; Singh, B.K. Does grassland vegetation drive soil microbial diversity? Nutr. Cycl. Agroecosyst. 2010, 88, 147–158. [Google Scholar] [CrossRef]
- Pugnaire, F.I.; Armas, C.; Valladares, F. Soil as a mediator in plant-plant interactions in a semi-arid community. J. Veg. Sci. 2004, 15, 85–92. [Google Scholar] [CrossRef]
- Thakur, M.P.; Milcu, A.; Manning, P.; Niklaus, P.A.; Roscher, C.; Power, S.; Reich, P.B.; Scheu, S.; Tilman, D.; Ai, F.; et al. Plant diversity drives soil microbial biomass carbon in grasslands irrespective of global environmental change factors. Glob. Chang. Biol. 2015, 21, 4076–4085. [Google Scholar] [CrossRef]
- Steinauer, K.; Tilman, D.; Wragg, P.D.; Cesarz, S.; Cowles, J.M.; Pritsch, K.; Reich, P.B.; Weisser, W.W.; Eisenhauer, N. Plant diversity effects on soil microbial functions and enzymes are stronger than warming in a grassland experiment. Ecology 2015, 96, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Tscherko, D.; Hammesfahr, U.; Zeltner, G.; Kandeler, E.; Böcker, R. Plant succession and rhizosphere microbial communities in a recently deglaciated alpine terrain. Basic Appl. Ecol. 2005, 6, 367–383. [Google Scholar] [CrossRef]
- Boudsocq, S.; Niboyet, A.; Lata, J.C.; Raynaud, X.; Loeuille, N.; Mathieu, J.; Blouin, M.; Abbadie, L.; Barot, S. Plant preference for ammonium versus nitrate: A neglected determinant of ecosystem functioning? Am. Nat. 2012, 180, 60–69. [Google Scholar] [CrossRef]
- Fontaine, S.; Henault, C.; Aamor, A.; Bdioui, N.; Bloor, J.M.G.; Maire, V.; Mary, B.; Revaillot, S.; Maron, P.A. Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect. Soil Biol. Biochem. 2011, 43, 86–96. [Google Scholar] [CrossRef]
- Santiago, T.; Pablo, L.P.; Olga, S.C.; Veronica, G.; Marina, G.-P. Soil microbial communities respond to an environmental gradient of grazing intensity in south Patagonia Argentina. J. Arid Environ. 2021, 184, 104300. [Google Scholar] [CrossRef]
- El Zahar Haichar, F.; Achouak, W.; Christen, R.; Heulin, T.; Marol, C.; Marais, M.F.; Mougel, C.; Ranjard, L.; Balesdent, J.; Berge, O. Identification of cellulolytic bacteria in soil by stable isotope probing. Environ. Microbiol. 2007, 9, 625–634. [Google Scholar] [CrossRef]
- Poll, C.; Marhan, S.; Ingwersen, J.; Kandeler, E. Dynamics of litter carbon turnover and microbial abundance in a rye detritusphere. Soil Biol. Biochem. 2008, 40, 1306–1321. [Google Scholar] [CrossRef]
- Shi, J.Y.; Gong, J.R.; Li, X.B.; Zhang, Z.H.; Zhang, W.Y.; Li, Y.; Song, L.Y.; Zhang, S.Q.; Dong, J.; Baoyin, T.-t. Plant–microbial linkages regulate soil organic carbon dynamics under phosphorus application in a typical temperate grassland in northern China. Agric. Ecosyst. Environ. 2022, 335, 108006. [Google Scholar] [CrossRef]
- Yuan, Z.Q.; Yu, K.L.; Epstein, H.; Fang, C.; Li, J.T.; Liu, Q.Q.; Liu, X.W.; Gao, W.J.; Li, F.M. Effects of legume species introduction on vegetation and soil nutrient development on abandoned croplands in a semi-arid environment on the Loess Plateau, China. Sci. Total Environ. 2016, 541, 692–700. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Hill, B.H.; Follstad Shah, J.J. Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature 2009, 462, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Sinsabaugh, R.L. Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 2010, 42, 391–404. [Google Scholar] [CrossRef]
- Bell, C.W.; Fricks, B.E.; Rocca, J.D.; Steinweg, J.M.; McMahon, S.K.; Wallenstein, M.D. High-throughput fluorometric measurement of potential soil extracellular enzyme activities. JOVE J. Vis. Exp. 2013, 81, e50961. [Google Scholar]

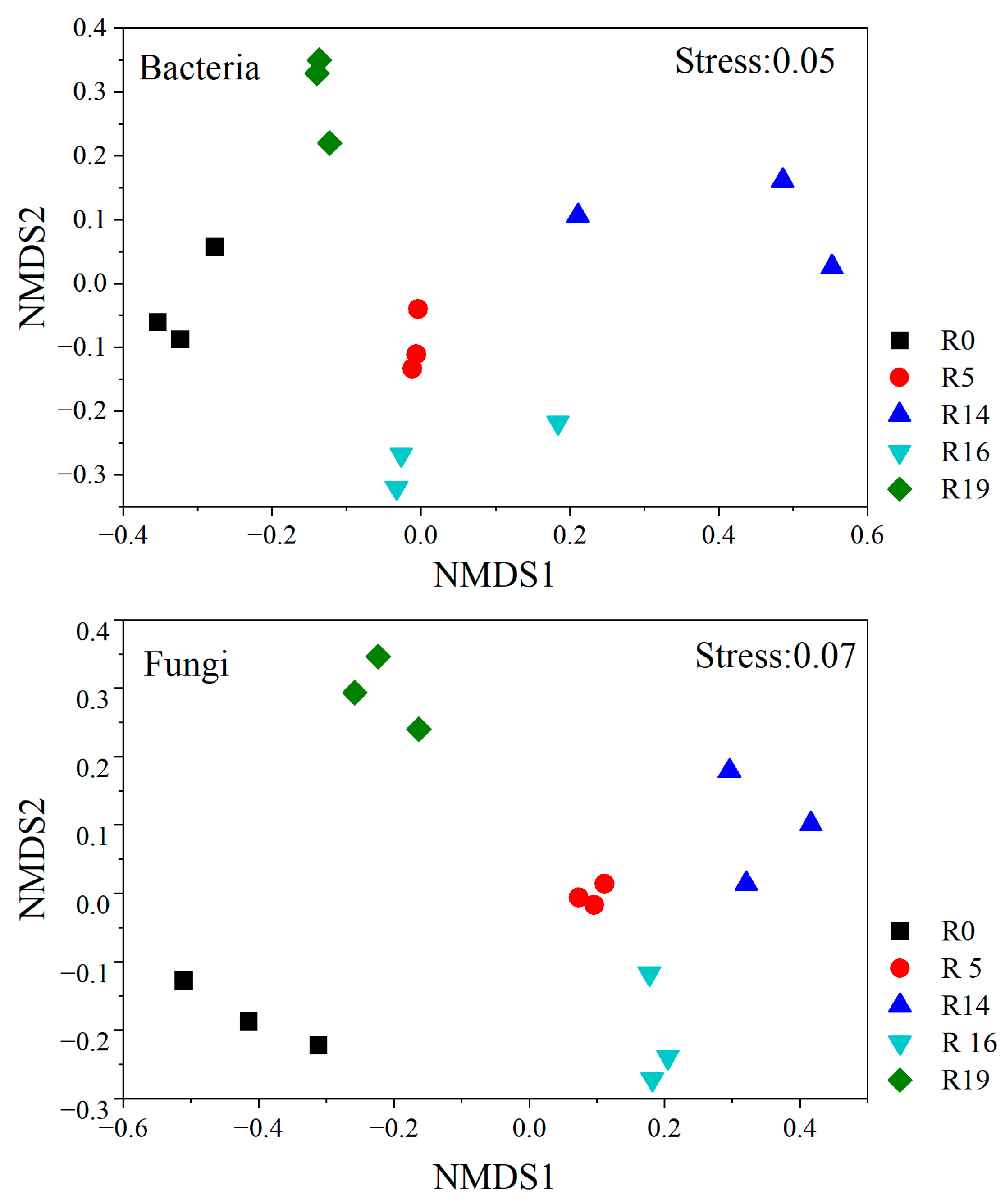
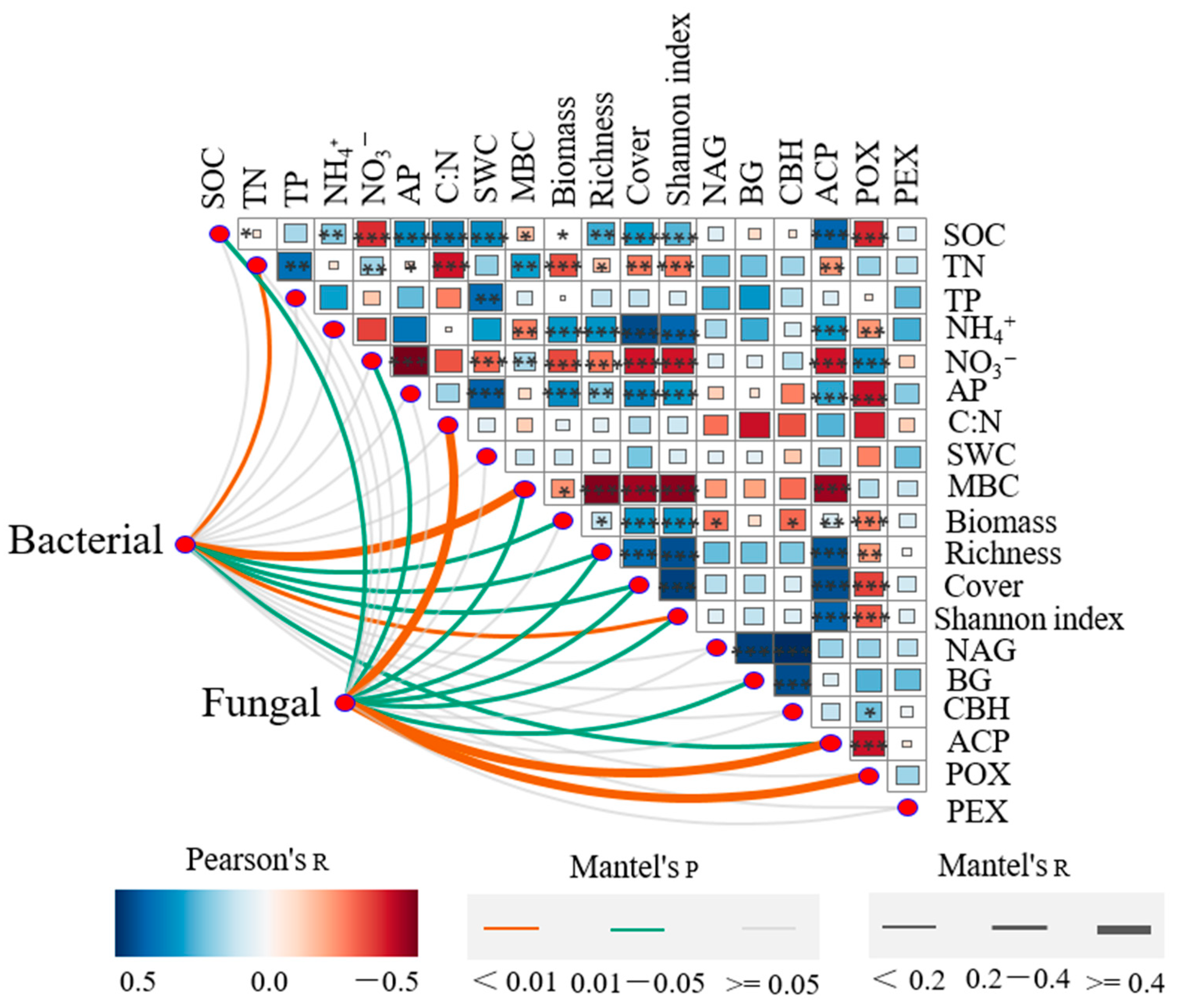

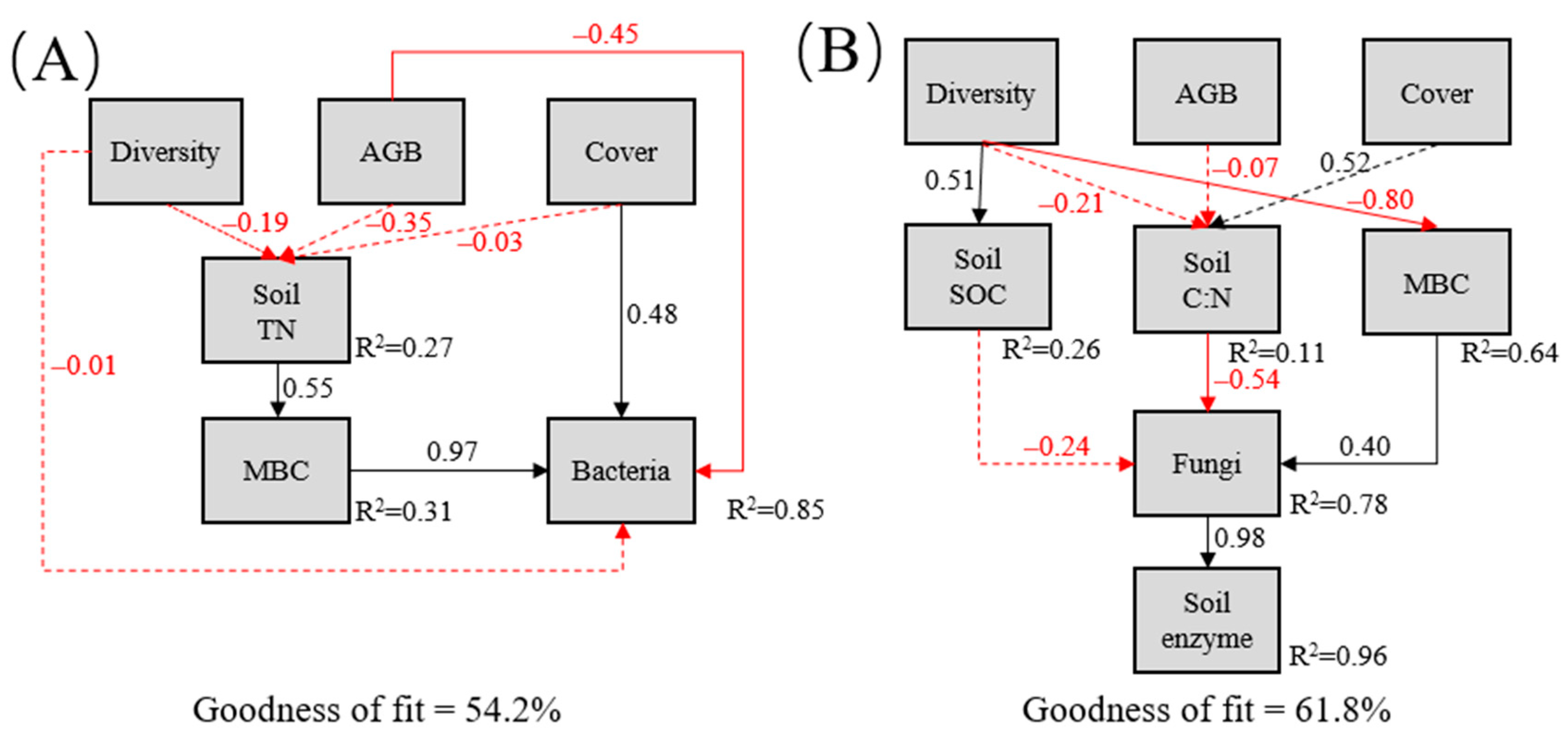
| Parameters | R0 | R5 | R14 | R16 | R19 | F | p |
|---|---|---|---|---|---|---|---|
| Aboveground biomass (kg ha−1) | 1098 ± 74.8 c | 1476 ± 29.8 b | 2253 ± 52.8 a | 1016 ± 16.1 c | 1437 ± 76.9 d | 77.57 | <0.001 |
| Cover (%) | 67.7 ± 2.60 c | 90.7 ± 2.60 ab | 96.3 ± 2.03 a | 87.0 ± 1.53 b | 96.3 ± 2.33 a | 27.49 | <0.001 |
| Richness | 10.3 ± 1.33 b | 19.7 ± 1.20 a | 18.3 ± 1.33 a | 20.0 ± 1.15 a | 20.3 ± 1.20 a | 11.37 | 0.001 |
| Shannon Index | 2.06 ± 0.05 c | 2.61 ± 0.05 ab | 2.77 ± 0.05 a | 2.55 ± 0.07 b | 2.69 ± 0.09 ab | 18.02 | 0.0001 |
| Parameters | R0 | R5 | R14 | R16 | R19 | F | p |
|---|---|---|---|---|---|---|---|
| SOC (g kg−1) | 32.4 ± 0.28 c | 32.8 ± 0.40 c | 35.2 ± 2.26 a | 36.8 ± 0.29 b | 42.6 ± 0.38 a | 15.0 | <0.001 |
| TN (g kg−1) | 3.12 ± 0.08 b | 3.65 ± 0.18 a | 3.05 ± 0.07 b | 3.22 ± 0.08 b | 3.68 ± 0.05 a | 8.93 | 0.002 |
| TP (g kg−1) | 0.40 ± 0.01 b | 0.43 ± 0.01 a | 0.39 ± 0.01 b | 0.38 ± 0.00 b | 0.44 ± 0.00 a | 10.4 | 0.001 |
| NO3−-N (mg kg−1) | 6.65 ± 1.11 a | 5.94 ± 0.06 a | 3.62 ± 0.11 b | 5.61 ± 0.02 a | 3.42 ± 0.38 b | 7.46 | 0.005 |
| NH4+-N (mg kg−1) | 5.98 ± 0.47 b | 9.89 ± 0.62 a | 9.31 ± 0.06 a | 7.02 ± 0.49 b | 10.2 ± 0.19 a | 19.9 | <0.001 |
| AP (mg kg−1) | 2.65 ± 0.07 cd | 2.94 ± 0.03 c | 3.83 ± 0.07 b | 2.56 ± 0.18 d | 4.24 ± 0.14 a | 43.8 | <0.001 |
| pH | 8.05 ± 0.01 a | 7.72 ± 0.03 b | 6.65 ± 0.09 c | 7.99 ± 0.05 a | 7.86 ± 0.12 ab | 68.6 | <0.001 |
| C:N ratio | 10.4 ± 0.27 ab | 9.07 ± 0.43 b | 11.6 ± 0.89 a | 11.5 ± 0.37 a | 11.6 ± 0.14 a | 4.92 | 0.019 |
| MBC (mg kg−1) | 744 ± 12.7 a | 468 ± 16.2 c | 383 ± 5.02 e | 430 ± 0.52 d | 552 ± 5.90 b | 208 | <0.001 |
| MBN (mg kg−1) | 85.1 ± 3.64 a | 72.5 ± 0.96 b | 68.2 ± 1.32 b | 67.0 ± 0.48 b | 70.8 ± 3.31 b | 9.65 | 0.002 |
| MBP (mg kg−1) | 8.63 ± 1.85 d | 30.2 ± 2.33 b | 39.5 ± 0.84 a | 18.9 ± 0.78 c | 8.85 ± 1.88 d | 66.6 | <0.001 |
| SMC (%) | 25.0 ± 0.55 b | 25.7 ± 0.64 b | 25.7 ± 0.24 b | 22.0 ± 0.98 c | 32.7 ± 1.42 a | 20.6 | <0.001 |
| NAG (μmol g−1 h−1) | 60.7 ± 3.87 c | 210 ± 23.6 a | 145 ± 5.88 b | 32.5 ± 9.88 c | 146 ± 17.2 b | 26.0 | <0.001 |
| BG (μmol g−1 h−1) | 184 ± 0.79 d | 1010 ± 62.0 a | 364 ± 17.5 c | 197 ± 24.1 d | 479 ± 6.16 b | 120 | <0.001 |
| CBH (μmol g−1 h−1) | 83.2 ± 2.15 d | 163 ± 10.4 a | 138 ± 10.50 b | 71.5 ± 1.10 d | 108 ± 4.23 c | 29.6 | <0.001 |
| ACP (μmol g−1 h−1) | 412 ± 2.05 d | 630 ± 28.8 c | 695 ± 15.6 b | 779 ± 15.2 a | 830 ± 1.94 a | 101 | <0.001 |
| POX (nmol g−1 h−1) | 262 ± 11.1 a | 280 ± 0.94 a | 224 ± 9.13 b | 206 ± 11.5 b | 199 ± 7.57 b | 15.6 | <0.001 |
| PEX (nmol g−1 h−1) | 85.0 ± 1.40 ab | 96.0 ± 1.87 a | 70.6 ± 5.39 b | 83.8 ± 9.40 ab | 92.5 ± 1.70 a | 3.81 | 0.039 |
| Parameters | R0 | R5 | R14 | R16 | R19 | F | p | |
|---|---|---|---|---|---|---|---|---|
| Observed OTUs | Bacteria | 2822 ± 9.23 a | 2622 ± 49.0 b | 2533 ± 16.6 b | 2598 ± 31.1 b | 2754 ± 38.8 a | 14.1 | 0.0004 |
| Fungi | 837 ± 14.4 a | 850 ± 19.6 a | 679 ± 9.50 b | 672 ± 21.9 b | 673 ± 9.50 b | 34.3 | <0.0001 | |
| Chao1 | Bacteria | 2902 ± 8.13 a | 2715 ± 14.2 c | 2543 ± 4.79 e | 2685 ± 1.38 d | 2764 ± 8.18 b | 236 | <0.0001 |
| Fungi | 838 ± 14.0 a | 851 ± 19.5 a | 679 ± 9.90 b | 673 ± 21.8 b | 674 ± 9.70 b | 34.7 | <0.0001 | |
| ACE | Bacteria | 2895 ± 10.9 a | 2712 ± 12.2 c | 2541 ± 4.68 e | 2673 ± 1.99 d | 2762 ± 7.12 b | 242 | <0.0001 |
| Fungi | 839 ± 14.1 a | 852 ± 19.6 a | 680 ± 9.80 b | 675 ± 21.9 b | 675 ± 9.70 b | 34.1 | <0.0001 | |
| Shannon index | Bacteria | 7.29 ± 0.006 ab | 7.29 ± 0.021 a | 7.21 ± 0.006 c | 7.24 ± 0.021 bc | 7.30 ± 0.015 a | 6.73 | 0.0068 |
| Fungi | 5.09 ± 0.17 ab | 5.33 ± 0.11 a | 4.85 ± 0.11 b | 4.75 ± 0.14 bc | 4.41 ± 0.05 c | 8.37 | 0.0031 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, C.; Liang, D.; Zhou, W.; Xu, Q.; Xiang, M.; Gu, Y.; Siddique, K.H.M. Soil, Plant, and Microorganism Interactions Drive Secondary Succession in Alpine Grassland Restoration. Plants 2024, 13, 780. https://doi.org/10.3390/plants13060780
Han C, Liang D, Zhou W, Xu Q, Xiang M, Gu Y, Siddique KHM. Soil, Plant, and Microorganism Interactions Drive Secondary Succession in Alpine Grassland Restoration. Plants. 2024; 13(6):780. https://doi.org/10.3390/plants13060780
Chicago/Turabian StyleHan, Chenglong, Defei Liang, Weidi Zhou, Qiuyun Xu, Mingxue Xiang, Yanjie Gu, and Kadambot H. M. Siddique. 2024. "Soil, Plant, and Microorganism Interactions Drive Secondary Succession in Alpine Grassland Restoration" Plants 13, no. 6: 780. https://doi.org/10.3390/plants13060780
APA StyleHan, C., Liang, D., Zhou, W., Xu, Q., Xiang, M., Gu, Y., & Siddique, K. H. M. (2024). Soil, Plant, and Microorganism Interactions Drive Secondary Succession in Alpine Grassland Restoration. Plants, 13(6), 780. https://doi.org/10.3390/plants13060780







