Transcriptome-Based Weighted Gene Co-Expression Network Analysis Reveals the Photosynthesis Pathway and Hub Genes Involved in Promoting Tiller Growth under Repeated Drought–Rewatering Cycles in Perennial Ryegrass
Abstract
1. Introduction
2. Results
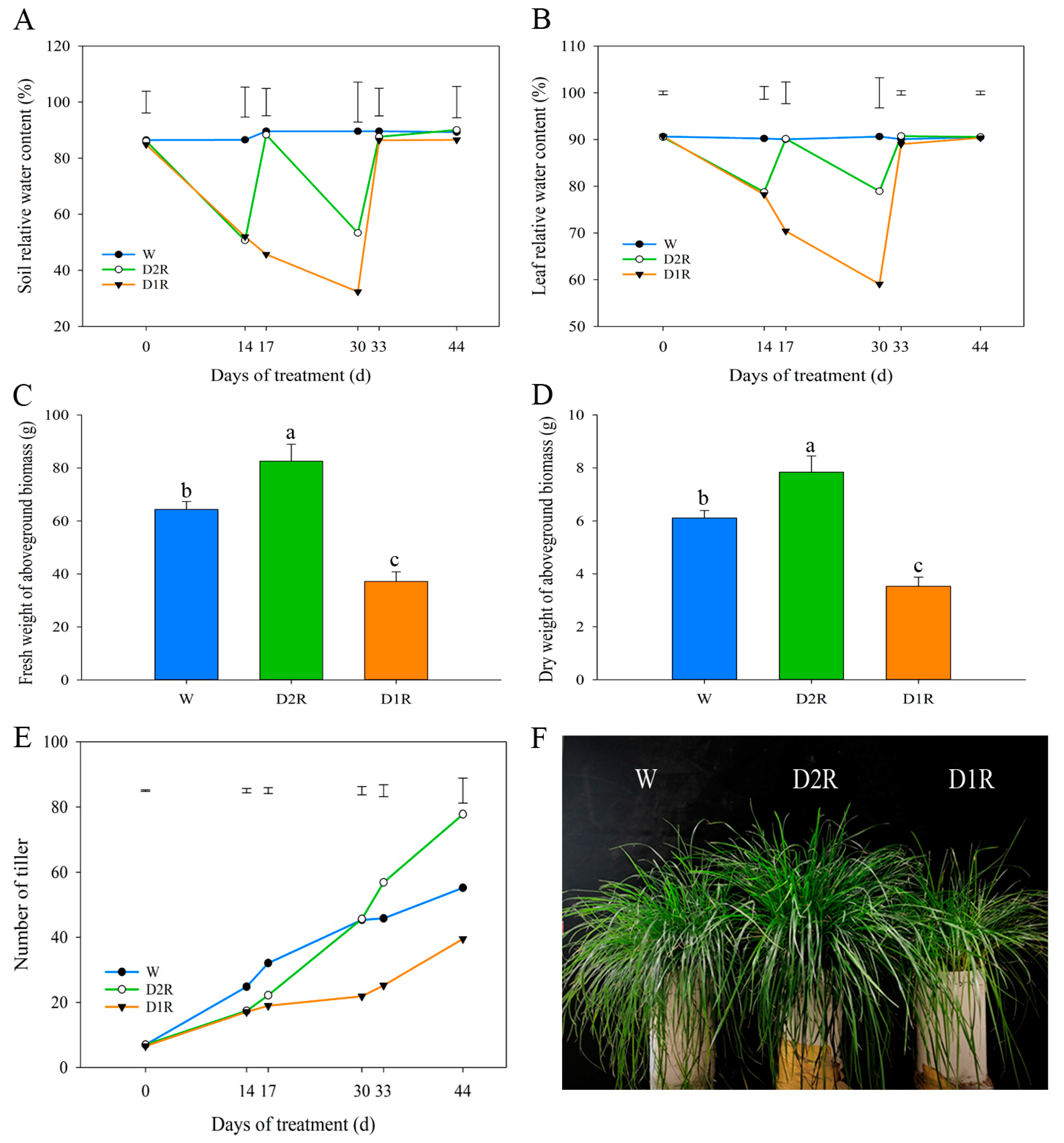
2.1. Effects of Drought–Rewatering Treatment on the Perennial Ryegrass Phenotype
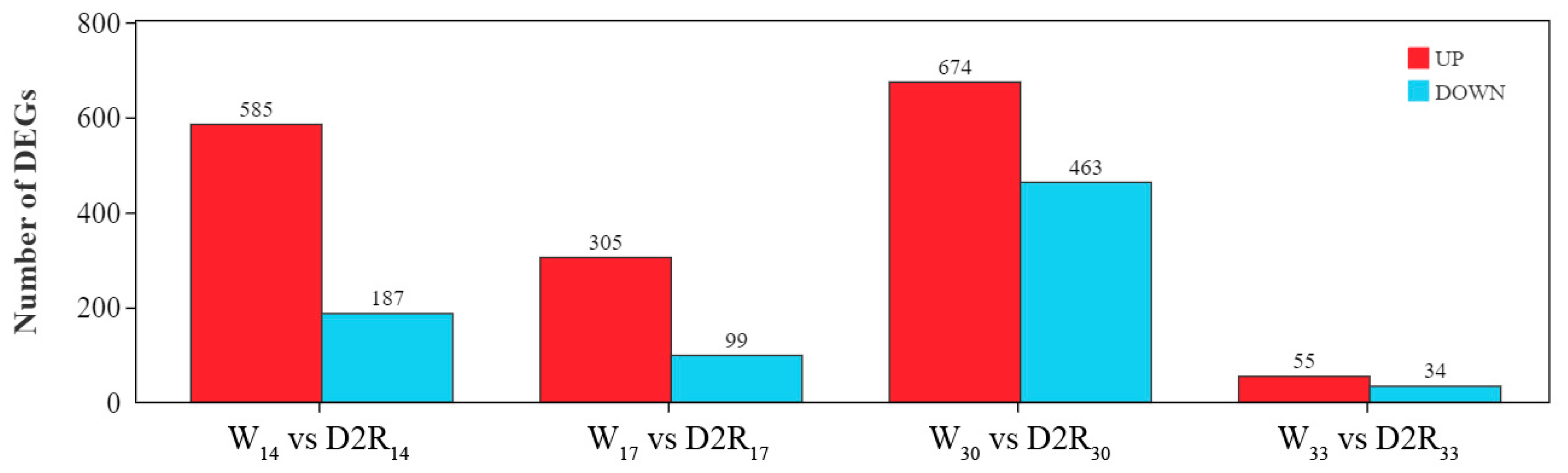
2.2. Identification of DEGs in Different Treatments
2.3. WGCNA to Identify DEGs Associated with Tiller Number
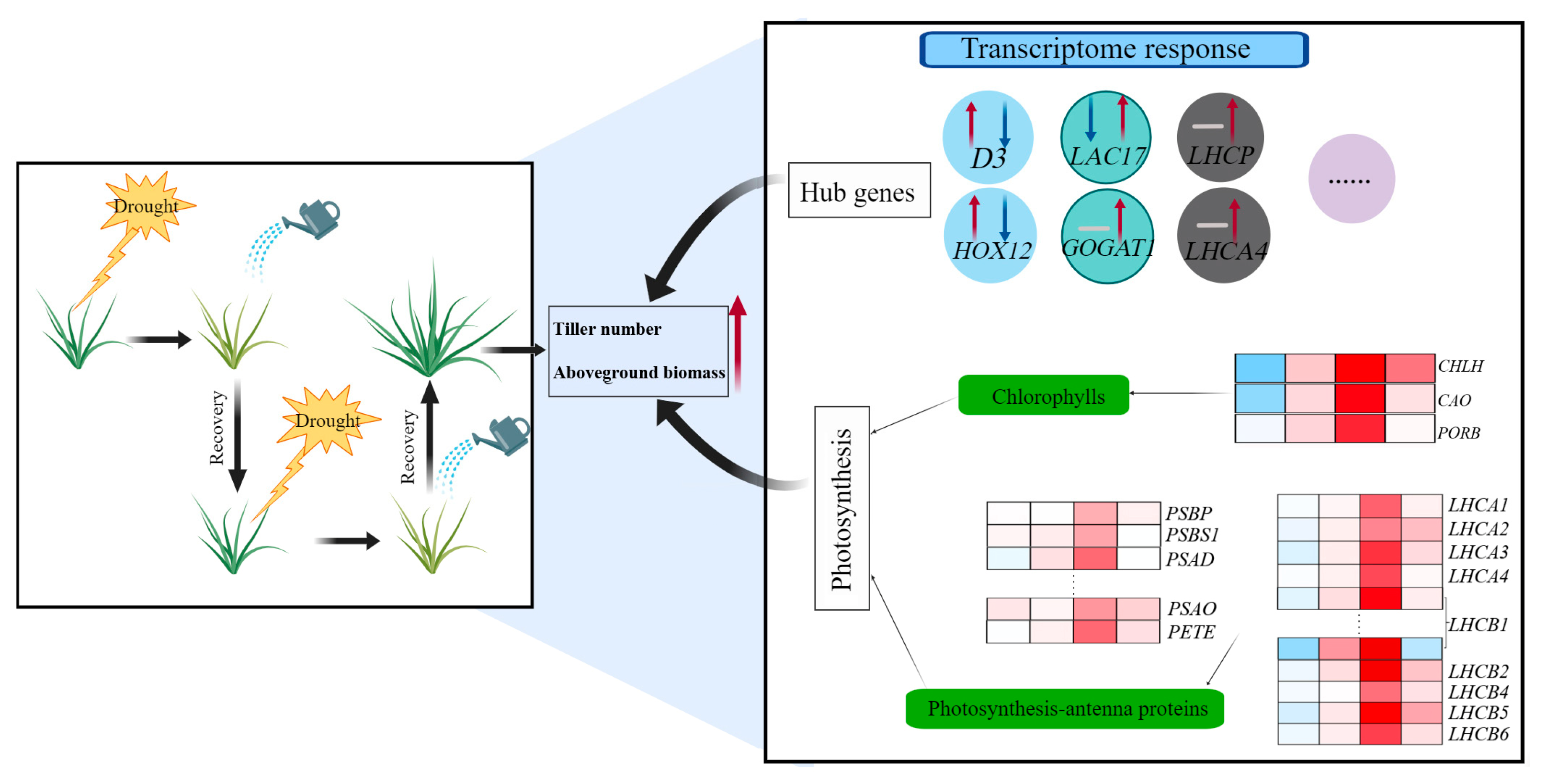
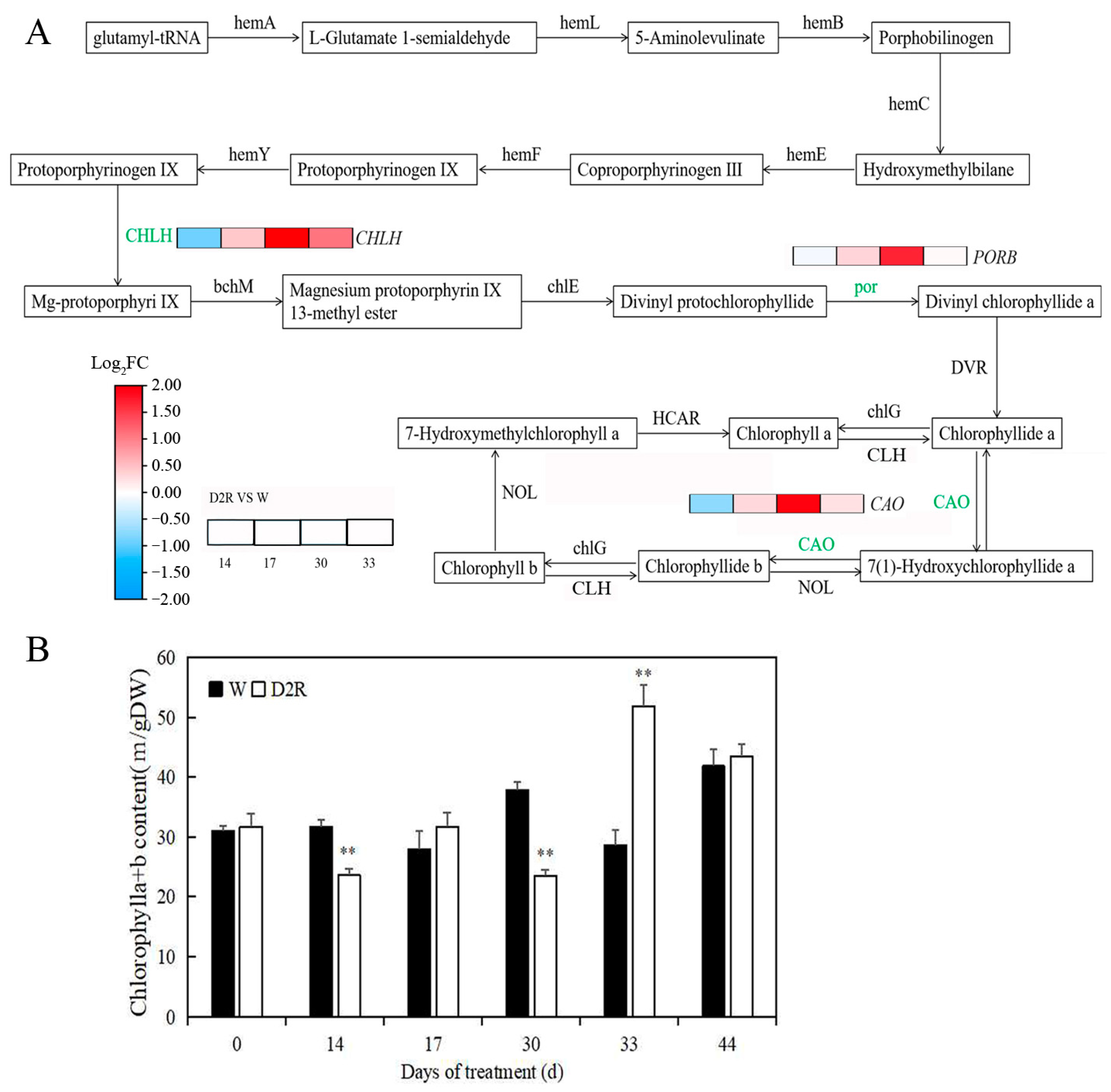
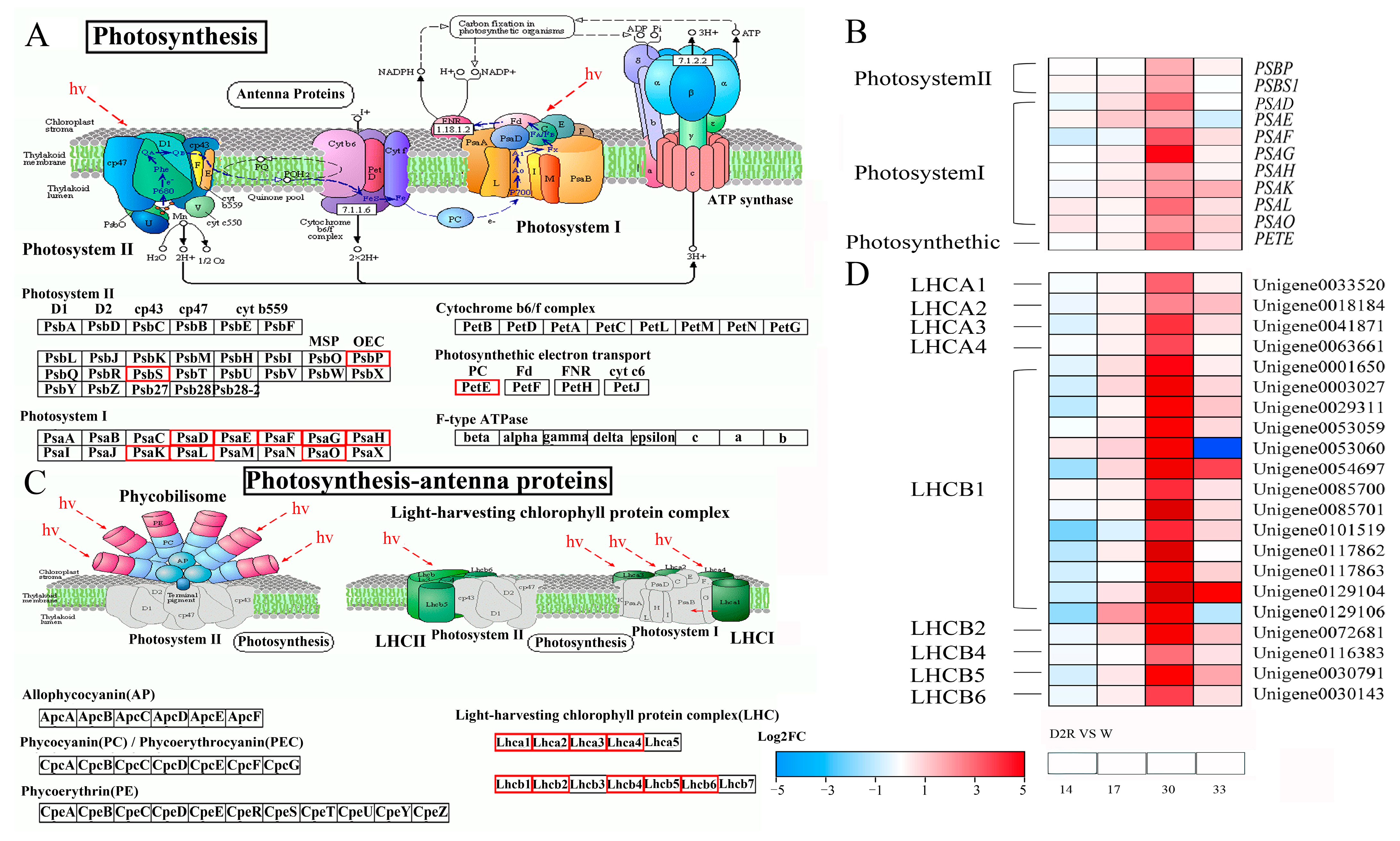
2.4. Photosynthesis Played Significant Roles in Regulating Tiller Development under Two Cycles of Drought–Rewatering Treatment
2.5. Validation of RNA-Seq Expression Data by qRT-PCR
3. Discussion
3.1. Moderate Drought Conditions and Rewatering Cycles Could Lead to More Vigorous Growth
3.2. Transcriptional Changes in Hub Genes Were Associated with Enhanced Tiller Development under Repeated Drought–Rewatering Treatment
3.3. Enhanced Expression of Photosynthesis and Photosynthesis-Antenna Genes Contributed to Increased Tiller Growth of Perennial Ryegrass under Repeated Drought–Rewatering
4. Material and Methods
4.1. Plant Material and Growth Conditions
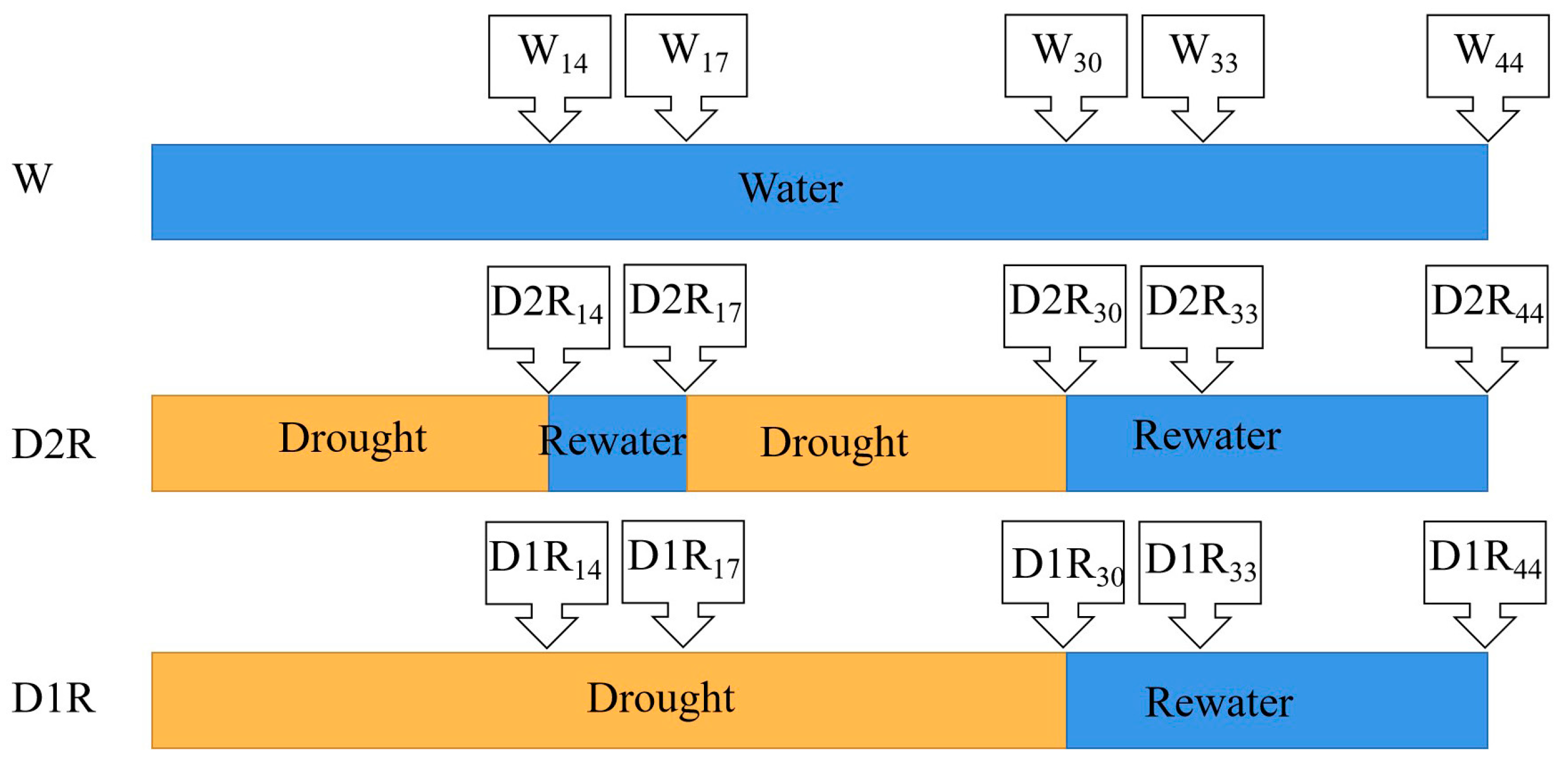
4.2. Treatments and Experimental Design
4.3. Determination of Soil Relative Water Content and Leaf Relative Water Content
4.4. Measurements of Chlorophyll Content
4.5. Total RNA Isolation, Library Preparation, and Transcriptome Sequencing
4.6. Differential Expressed Genes (DEGs), Weighted Gene Co-Expression Network Analysis (WGCNA), Gene Ontology (GO), and Pathway Enrichment Analysis
4.7. Validation and Expression Analysis by qRT-PCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hussain, S.; Hussain, S.; Qadir, T.; Khaliq, A.; Ashraf, U.; Parveen, A.; Saqib, M.; Rafiq, M. Drought stress in plants: An overview on implications, tolerance mechanisms and agronomic mitigation strategies. Plant Sci. Today 2019, 6, 389–402. [Google Scholar] [CrossRef]
- Bostock, R.M. Signal crosstalk and induced resistance: Straddling the line between cost and benefit. Annu. Rev. Phytopathol. 2005, 43, 545–580. [Google Scholar] [CrossRef]
- Busso, C.A.; Richards, J.H. Drought and clipping effects on tiller demography and growth of two tussock grasses in Utah. J. Arid Environ. 1995, 29, 239–251. [Google Scholar] [CrossRef]
- Huang, B.; Dacota, M.; Jiang, Y. Research advances in mechanisms of turfgrass tolerance to abiotic stresses: From physiology to molecular biology: Stress biology of specialty crops. Crit. Rev. Plant Sci. 2014, 33, 141–189. [Google Scholar] [CrossRef]
- Slatyer, R.O. Plant-Water Relationships. J. Appl. Ecol. 1968, 1, 261–262. [Google Scholar] [CrossRef]
- Schulze, W.X.; Altenbuchinger, M.; He, M.; Kränzlein, M.; Zörb, C. Proteome profiling of repeated drought stress reveals genotype-specific responses and memory effects in maize. Plant Physiol. Bioch. 2021, 159, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Avramova, Z. Defence-related priming and responses to recurring drought: Two manifestations of plant transcriptional memory mediated by the ABA and JA signalling pathways. Plant Cell Environ. 2019, 42, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, e740. [Google Scholar] [CrossRef] [PubMed]
- Avramova, V.; AbdElgawad, H.; Zhang, Z.; Fotschki, B.; Casadevall, R.; Vergauwen, L.; Knapen, D.; Taleisnik, E.; Guisez, Y.; Asard, H.; et al. Drought induces distinct growth response, protection, and recovery mechanisms in the maize leaf growth zone. Plant Physiol. 2015, 169, 1382–1396. [Google Scholar] [CrossRef] [PubMed]
- Crisp, P.A.; Ganguly, D.; Eichten, S.R.; Borevitz, J.O.; Pogson, B.J. Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2016, 2, e1501340. [Google Scholar] [CrossRef]
- Wedeking, R.; Maucourt, M.; Deborde, C.; Moing, A.; Gibon, Y.; Goldbach, H.E.; Wimmer, M.A. 1H-NMR metabolomic profiling reveals a distinct metabolic recovery response in shoots and roots of temporarily drought-stressed sugar beets. PLoS ONE 2018, 13, e196102. [Google Scholar] [CrossRef]
- Ding, Y.; Virlouvet, L.; Liu, N.; Riethoven, J.; Fromm, M.; Avramova, Z. Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biol. 2014, 14, 141. [Google Scholar] [CrossRef]
- Ben Abdallah, M.; Methenni, K.; Nouairi, I.; Zarrouk, M.; Youssef, N.B. Drought priming improves subsequent more severe drought in a drought-sensitive cultivar of olive cv. Chétoui. Sci. Hortic. 2017, 221, 43–52. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, X.; Zhuang, L.; Gao, Y.; Huang, B. Abscisic acid mediation of drought priming-enhanced heat tolerance in tall fescue (Festuca arundinacea) and Arabidopsis. Physiol. Plantarum 2019, 167, 488–501. [Google Scholar] [CrossRef]
- Brito, C.; Dinis, L.; Ferreira, H.; Moutinho-Pereira, J.; Correia, C.M. Foliar pre-treatment with abscisic acid enhances olive tree drought adaptability. Plants 2020, 9, 341. [Google Scholar] [CrossRef]
- Nosalewicz, A.; Siecińska, J.; Kondracka, K.; Nosalewicz, M. The functioning of Festuca arundinacea and Lolium perenne under drought is improved to a different extend by the previous exposure to water deficit. Environ. Exp. Bot. 2018, 156, 271–278. [Google Scholar] [CrossRef]
- Marcos, F.C.C.; Silveira, N.M.; Marchiori, P.E.R.; Machado, E.C.; Souza, G.M.; Landell, M.G.A.; Ribeiro, R.V. Drought tolerance of sugarcane propagules is improved when origin material faces water deficit. PLoS ONE 2018, 13, e206716. [Google Scholar] [CrossRef] [PubMed]
- Mantoan, L.P.B.; Corrêa, C.V.; Rainho, C.A.; de Almeida, L.F.R. Rapid dehydration induces long-term water deficit memory in sorghum seedlings: Advantages and consequences. Environ. Exp. Bot. 2020, 180, 104252. [Google Scholar] [CrossRef]
- Chieppa, J.; Power, S.A.; Nielsen, U.N.; Tissue, D.T. Plant functional traits affect competitive vigor of pasture grasses during drought and following recovery. Ecosphere 2022, 13, e4156. [Google Scholar] [CrossRef]
- Xu, Z.; Zhou, G.; Shimizu, H. Are plant growth and photosynthesis limited by pre-drought following rewatering in grass? J. Exp. Bot. 2009, 60, 3737–3749. [Google Scholar] [CrossRef]
- Flexas, J.; Bota, J.; Cifre, J.; Mariano Escalona, J.; Galmes, J.; Gulias, J.; Lefi, E.; Florinda Martinez-Canells, S.; Teresa Moreno, M.; Ribas-Carbo, M.; et al. Understanding down-regulation of photosynthesis under water stress: Future prospects and searching for physiological tools for irrigation management. Ann. Appl. Biol. 2004, 144, 273–283. [Google Scholar] [CrossRef]
- Shi, J.; Yasuor, H.; Yermiyahu, U.; Zuo, Q.; Ben-Gal, A. Dynamic responses of wheat to drought and nitrogen stresses during re-watering cycles. Agr. Water Manag. 2014, 146, 163–172. [Google Scholar] [CrossRef]
- Fry, J.; Huang, B. Applied Turfgrass Science and Physiology; Wiley: Hoboken, NJ, USA, 2004; p. 130. [Google Scholar]
- Cattani, D.J.; Clark, K.W. Influence of wear-stress on turfgrass growth components and visual density ratings. Can. J. Plant Sci. 1991, 1, 305–308. [Google Scholar] [CrossRef][Green Version]
- Baalbaki, R.; Hajj Hassan, N.; Zurayk, R. Aegilops species from semiarid areas of lebanon: Variation in quantitative attributes under water stress. Crop Sci. 2006, 46, 799–806. [Google Scholar] [CrossRef]
- Moeller, C.; Evers, J.B.; Rebetzke, G. Canopy architectural and physiological characterization of near-isogenic wheat lines differing in the tiller inhibition gene tin. Front. Plant Sci. 2014, 5, e617. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Su, J.; Luo, X.; Meng, J.; Zhang, H.; Li, P.; Sun, Y.; Song, J.; Peng, X.; Yu, C. Nitrogen limits zinc-mediated stimulation of tillering in rice by modifying phytohormone balance under low-temperature stress. Food Energy Secur. 2022, 11, e359. [Google Scholar] [CrossRef]
- Kane, K.H. Effects of endophyte infection on drought stress tolerance of Lolium perenne accessions from the Mediterranean region. Environ. Exp. Bot. 2011, 71, 337–344. [Google Scholar] [CrossRef]
- Borrajo, C.I.; Sánchez-Moreiras, A.M.; Reigosa, M.J. Morpho-physiological responses of tall wheatgrass populations to different levels of water stress. PLoS ONE 2018, 13, e209281. [Google Scholar] [CrossRef]
- Walter, J.; Nagy, L.; Hein, R.; Rascher, U.; Beierkuhnlein, C.; Willner, E.; Jentsch, A. Do plants remember drought? Hints towards a drought-memory in grasses. Environ. Exp. Bot. 2011, 71, 34–40. [Google Scholar] [CrossRef]
- Cyriac, D.; Hofmann, R.W.; Stewart, A.; Sathish, P.; Winefield, C.S.; Moot, D.J. Intraspecific differences in long-term drought tolerance in perennial ryegrass. PLoS ONE 2018, 13, e194977. [Google Scholar] [CrossRef]
- Svobodová, I.; Míša, P. Effect of drought stress on the formation of yield elements in spring barley and the potential of stress expression reduction by foliar application of fertilizers and growth stimulator. Plant Soil Environ. 2004, 50, 439–446. [Google Scholar] [CrossRef]
- Zhuang, L.; Wang, J.; Huang, B. Drought inhibition of tillering in Festuca arundinacea associated with axillary bud development and strigolactone signaling. Environ. Exp. Bot. 2017, 142, 15–23. [Google Scholar] [CrossRef]
- Mastalerczuk, G.; Borawska-Jarmułowicz, B. Physiological and morphometric response of forage grass species and their biomass distribution depending on the term and frequency of water deficiency. Agronomy 2021, 11, 2471. [Google Scholar] [CrossRef]
- Ding, Y.; Liu, N.; Virlouvet, L.; Riethoven, J.J.; Fromm, M.; Avramova, Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, e229. [Google Scholar] [CrossRef]
- Ma, X.; Yu, J.; Zhuang, L.; Shi, Y.; Meyer, W.; Huang, B. Differential regulatory pathways associated with drought-inhibition and post-drought recuperation of rhizome development in perennial grass. Ann Bot. 2020, 126, 481–497. [Google Scholar] [CrossRef]
- Xu, J.; Ding, C.; Ding, Y.; Tang, S.; Zha, M.; Luo, B.; Wang, S. A proteomic approach to analyze differential regulation of proteins during bud outgrowth under apical dominance based on the auxin transport canalization model in rice (Oryza sativa L.). J. Plant Growth Regul. 2015, 34, 122–136. [Google Scholar] [CrossRef]
- Ishikawa, S.; Maekawa, M.; Arite, T.; Onishi, K.; Takamure, I.; Kyozuka, J. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 2005, 46, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, T.; Wang, M.; Liu, Y.; Yuan, S.; Gao, Y.; Yin, L.; Sun, W.; Peng, L.; Zhang, W.; et al. DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching. Plant Cell Physiol. 2014, 55, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Lv, T.; Shen, H.; Luong, P.; Wang, J.; Wang, Z.; Huang, Z.; Xiao, L.; Engineer, C.; Kim, T.H.; et al. Regulation of drought tolerance by the F-Box protein MAX2 in Arabidopsis. Plant Physiol. 2014, 164, 424–439. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hu, Q.; Yan, J.; Sun, K.; Liang, Y.; Jia, M.; Meng, X.; Fang, S.; Wang, Y.; Jing, Y.; et al. z-Carotene isomerase suppresses tillering in rice through the coordinated biosynthesis of strigolactone and abscisic acid. Mol. Plant 2020, 13, 1784–1801. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Zhao, C.; Zhao, M.; Han, Y.; Li, S. Lignin synthesis, affected by sucrose in lotus (Nelumbo nucifera) seedlings, was involved in regulation of root formation in the Arabidopsis thanliana. Int. J. Mol. Sci. 2022, 23, 2250. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, C.; Zhu, M.; Yu, Y.; Zhang, Y.; Wei, Z. Generation and characterization of transgenic poplar plants overexpressing a cotton laccase gene. Plant Cell Tissue Organ Cult. 2008, 93, 303–310. [Google Scholar] [CrossRef]
- Berthet, S.; Demont-Caulet, N.; Pollet, B.; Bidzinski, P.; Cézard, L.; Le Bris, P.; Borrega, N.; Hervé, J.; Blondet, E.; Balzergue, S.; et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 2011, 23, 1124–1137. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Shi, H.; Yu, S.; Zhou, W.; Li, J.; Liu, S.; Deng, M.; Ma, J.; Wei, Y.; Zheng, Y.; et al. Comprehensive transcriptomics, proteomics, and metabolomics analyses of the mechanisms regulating tiller production in low-tillering wheat. Theor. Appl. Genet. 2019, 132, 2181–2193. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xiao, Y.; Zhang, X.; Huang, X.; Fan, G. Maize straw mulching with uniconazole application increases the tillering capacity and grain yield of dryland winter wheat (Triticum aestivum L.). Field Crops Res. 2022, 284, e108573. [Google Scholar] [CrossRef]
- Tamura, W.; Hidaka, Y.; Tabuchi, M.; Kojima, S.; Hayakawa, T.; Sato, T.; Obara, M.; Kojima, M.; Sakakibara, H.; Yamaya, T. Reverse genetics approach to characterize a function of NADH-glutamate synthase1 in rice plants. Amino Acids 2010, 39, 1003–1012. [Google Scholar] [CrossRef]
- Lee, S.; Marmagne, A.; Park, J.; Fabien, C.; Yim, Y.; Kim, S.J.; Kim, T.H.; Lim, P.O.; Masclaux Daubresse, C.; Nam, H.G. Concurrent activation of OsAMT1;2 and OsGOGAT1 in rice leads to enhanced nitrogen use efficiency under nitrogen limitation. Plant J. 2020, 103, 7–20. [Google Scholar] [CrossRef]
- Liu, Z.; Zhu, Y.; Dong, Y.; Tang, L.; Zheng, Y.; Xiao, J. Interspecies interaction for nitrogen use efficiency via up-regulated glutamine and glutamate synthase under wheat-faba bean intercropping. Field Crops Res. 2021, 274, 108324. [Google Scholar] [CrossRef]
- Pintó-Marijuan, M.; Cotado, A.; Fleta-Soriano, E.; Munné-Bosch, S. Drought stress memory in the photosynthetic mechanisms of an invasive CAM species, Aptenia cordifolia. Photosynth. Res. 2017, 131, 241–253. [Google Scholar] [CrossRef]
- Hu, H.; He, B.; Ma, L.; Chen, X.; Han, P.; Luo, Y.; Liu, Y.; Fei, X.; Wei, A. Physiological and transcriptome analyses reveal the photosynthetic response to drought stress in drought-sensitive (Fengjiao) and drought-tolerant (Hanjiao) Zanthoxylum bungeanum cultivars. Front. Plant Sci. 2022, 13, e968714. [Google Scholar] [CrossRef]
- Al-Oudat, M.; Baydoun, S.A.; Mohammad, A. Effects of enhanced UV-B on growth and yield of two Syrian crops wheat (Triticum durum var. Horani) and broad beans (Vicia faba) under field conditions. Environ. Exp. Bot. 1998, 40, 11–16. [Google Scholar] [CrossRef]
- Pandey, V.; Shukla, A. Acclimation and tolerance strategies of rice under drought stress. Rice sci. 2015, 22, 147–161. [Google Scholar] [CrossRef]
- Rochaix, J.D. Fine-tuning photosynthesis. Science 2013, 342, 50–51. [Google Scholar] [CrossRef] [PubMed]
- Ostria-Gallardo, E.; Larama, G.; Berríos, G.; Fallard, A.; Gutiérrez-Moraga, A.; Ensminger, I.; Manque, P.; Bascuñán-Godoy, L.; Bravo, L.A. Decoding gene networks modules that explain the recovery of Hymenoglossum cruentum Cav. after extreme desiccation. Front. Plant Sci. 2020, 11, e574. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Yi, J.; Yang, Y.; Lei, C.; Gong, M. Transcriptome response to drought, rehydration and re-dehydration in potato. Int. J. Mol. Sci. 2020, 21, 159. [Google Scholar] [CrossRef]
- Ganeteg, U.; Külheim, C.; Andersson, J.; Jansson, S. Is each light-harvesting complex protein important for plant fitness? Plant Physiol. 2004, 134, 502–509. [Google Scholar] [CrossRef]
- Aghdasi, M.; Fattahi, S.; Sadeghipour, H.R. Functional analysis of LHCB1 in Arabidopsis growth, development and photosynthetic capacity. In Applied Photosynthesis; Najafpou, M., Ed.; InTech: Rijeka, Croatia, 2012; pp. 389–404. [Google Scholar]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. In Circular: California Agricultural Experiment Station, 2nd ed.; College of Agriculture, University of California: Berkeley, CA, USA, 1950; Volume 347, p. 32. [Google Scholar]
- Wang, X.; Vignjevic, M.; Jiang, D.; Jacobsen, S.; Wollenweber, B. Improved tolerance to drought stress after anthesis due to priming before anthesis in wheat (Triticum aestivum L.) var. Vinjett. J. Exp. Bot. 2014, 65, 6441–6456. [Google Scholar] [CrossRef] [PubMed]
- Tait, M.A.; Hik, D.S. Is dimethylsulfoxide a reliable solvent for extracting chlorophyll under field conditions? Photosynth. Res. 2003, 78, 87–91. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, e550. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, e559. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 1, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yan, H.; Jiang, X.; Yin, G.; Zhang, X.; Qi, X.; Zhang, Y.; Yan, Y.; Ma, X.; Peng, Y. Identification of candidate reference genes in perennial ryegrass for quantitative RT-PCR under various abiotic stress conditions. PLoS ONE 2014, 9, e93724. [Google Scholar] [CrossRef] [PubMed]
- Panina, Y.; Germond, A.; David, B.G.; Watanabe, T.M. Pairwise efficiency: A new mathematical approach to qPCR data analysis increases the precision of the calibration curve assay. BMC Bioinform. 2019, 20, 295. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]

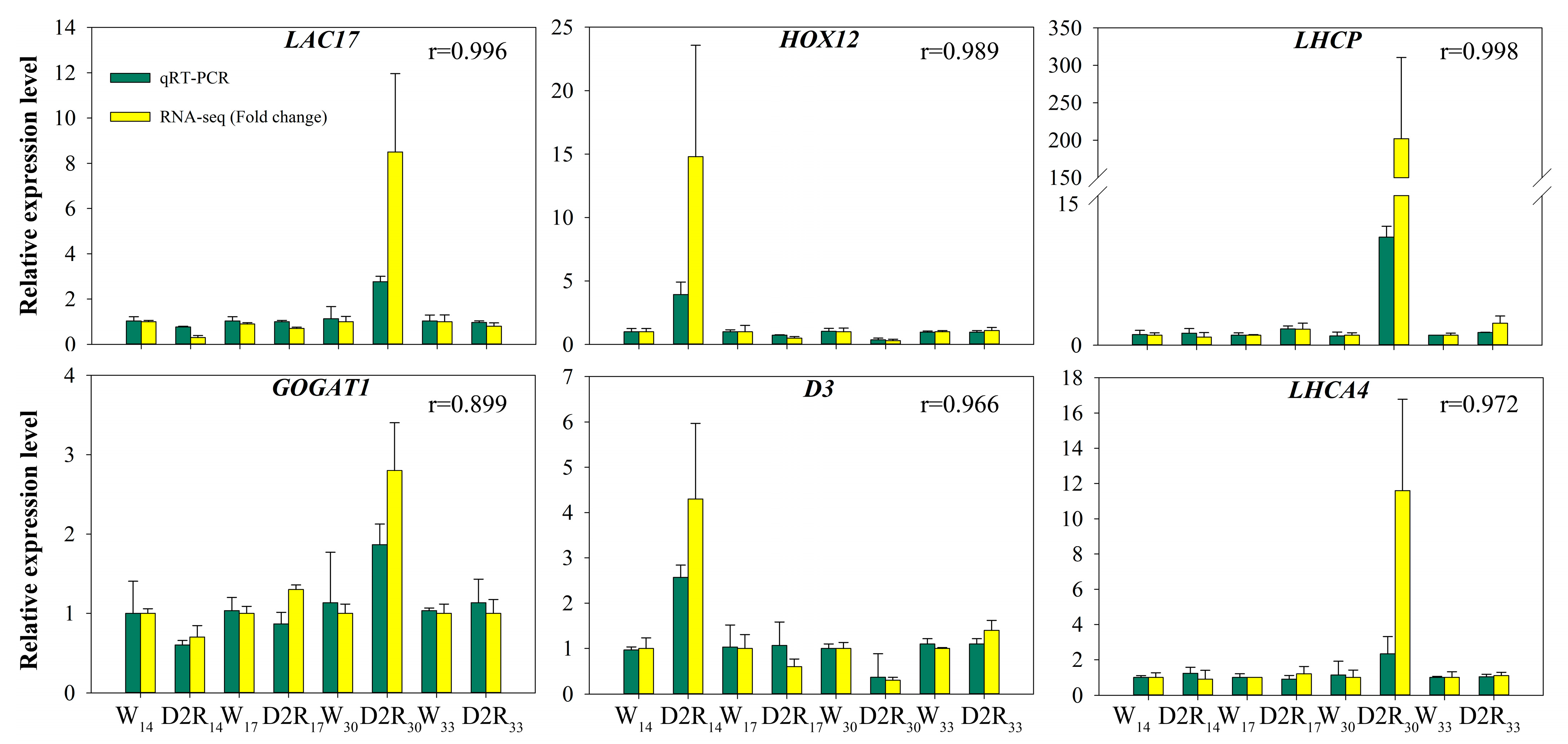
| Gene ID. | Gene Name | Annotation of Unigenes | kWithin |
|---|---|---|---|
| Turquoise | |||
| Unigene0061484 | LAC17 | XP_051186142.1 putative laccase-17 [Lolium perenne] | 74.3 |
| Unigene0109552 | GOGAT1 | XP_003566997.1 glutamate synthase 1 [NADH], chloroplastic [Brachypodium distachyon] | 70.8 |
| Blue | |||
| Unigene0122450 | HOX12 | XP_047088374.1 homeobox-leucine zipper protein HOX12-like [Lolium rigidum] | 63.3 |
| Unigene0047820 | D3 | XP_003564315.1 F-box/LRR-repeat MAX2 homolog [Brachypodium distachyon] | 58.8 |
| Black | |||
| Unigene0072681 | LHCP | KAE8804416.1 Chlorophyll a-b binding protein, chloroplastic [Hordeum vulgare] | 36.8 |
| Unigene0063661 | LHCA4 | XP_010235157.1 chlorophyll a-b binding protein 4, chloroplastic [Brachypodium distachyon] | 36.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Y.; Zhang, X.; Li, J.; Wang, R.; Chen, J.; Kong, L.; Li, X.; Yang, Z.; Zhuang, L. Transcriptome-Based Weighted Gene Co-Expression Network Analysis Reveals the Photosynthesis Pathway and Hub Genes Involved in Promoting Tiller Growth under Repeated Drought–Rewatering Cycles in Perennial Ryegrass. Plants 2024, 13, 854. https://doi.org/10.3390/plants13060854
Ding Y, Zhang X, Li J, Wang R, Chen J, Kong L, Li X, Yang Z, Zhuang L. Transcriptome-Based Weighted Gene Co-Expression Network Analysis Reveals the Photosynthesis Pathway and Hub Genes Involved in Promoting Tiller Growth under Repeated Drought–Rewatering Cycles in Perennial Ryegrass. Plants. 2024; 13(6):854. https://doi.org/10.3390/plants13060854
Chicago/Turabian StyleDing, Yunjia, Xiaxiang Zhang, Jialei Li, Ruying Wang, Jie Chen, Lingna Kong, Xin Li, Zhimin Yang, and Lili Zhuang. 2024. "Transcriptome-Based Weighted Gene Co-Expression Network Analysis Reveals the Photosynthesis Pathway and Hub Genes Involved in Promoting Tiller Growth under Repeated Drought–Rewatering Cycles in Perennial Ryegrass" Plants 13, no. 6: 854. https://doi.org/10.3390/plants13060854
APA StyleDing, Y., Zhang, X., Li, J., Wang, R., Chen, J., Kong, L., Li, X., Yang, Z., & Zhuang, L. (2024). Transcriptome-Based Weighted Gene Co-Expression Network Analysis Reveals the Photosynthesis Pathway and Hub Genes Involved in Promoting Tiller Growth under Repeated Drought–Rewatering Cycles in Perennial Ryegrass. Plants, 13(6), 854. https://doi.org/10.3390/plants13060854







