The Receptor Kinases DRUS1 and DRUS2 Behave Distinctly in Osmotic Stress Tolerance by Modulating the Root System Architecture via Auxin Signaling
Abstract
1. Introduction
2. Results
2.1. drus1-1 and drus2 Mutants Displayed Distinct Tolerance to Osmotic Stress
2.2. drus2 but Not drus1-1 Seedlings Displayed a Strong Root System under OS
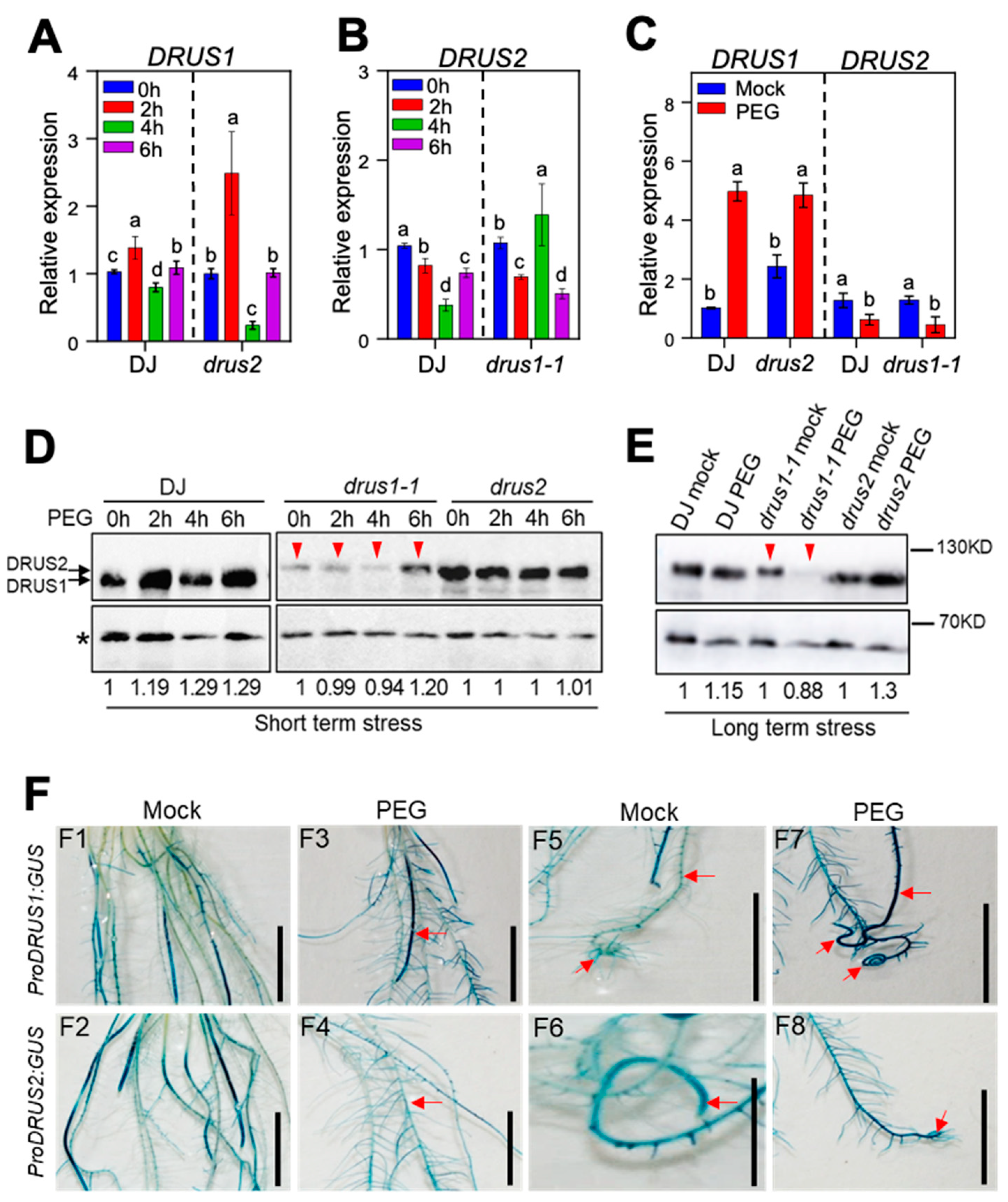
2.3. DRUS1 but Not DRUS2 Is Highly Induced in the Root by OS at Both the mRNA Level and the Protein Level
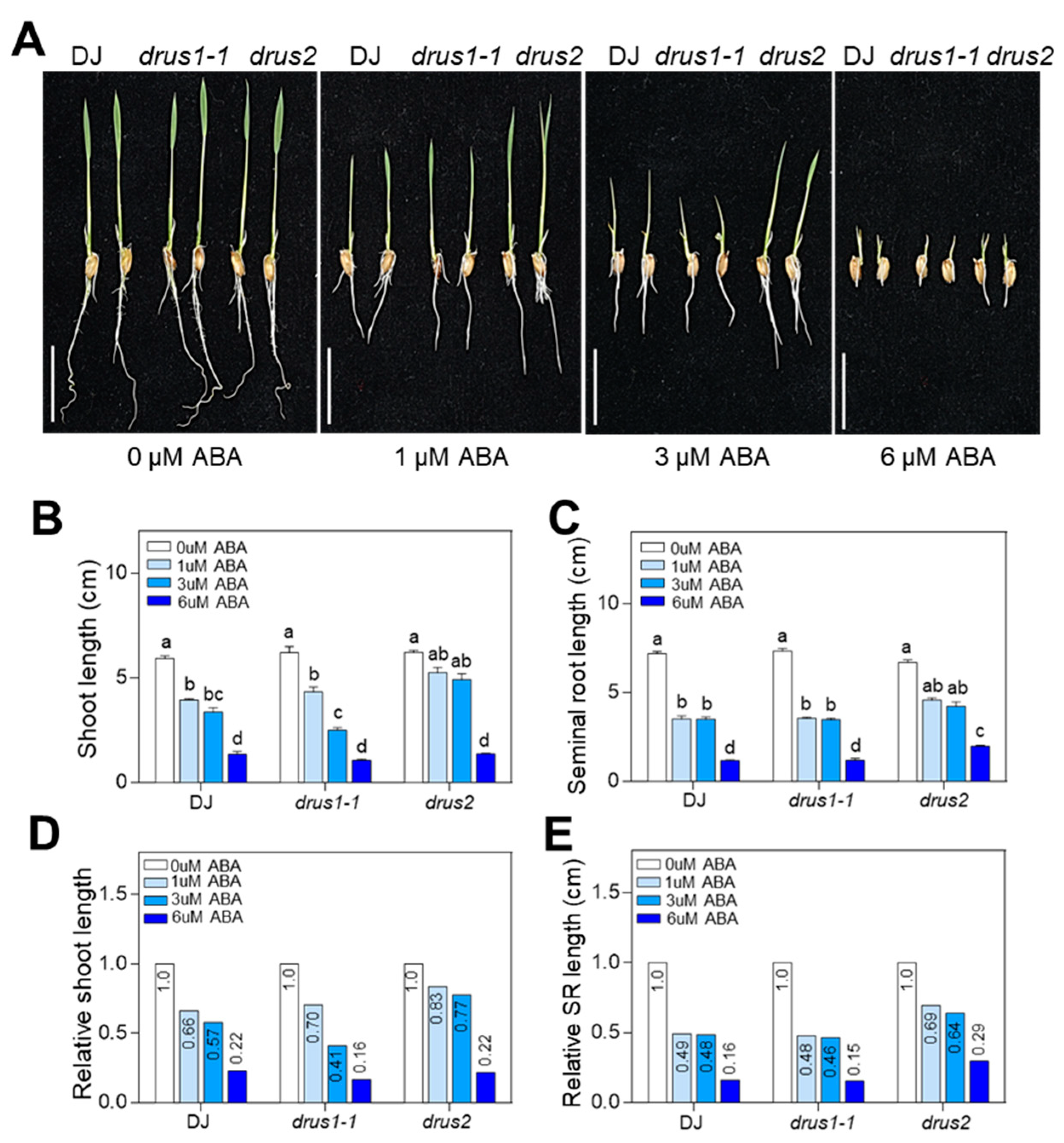
2.4. drus2 but Not drus1-1 Seedlings Are Insensitive to ABA Inhibition
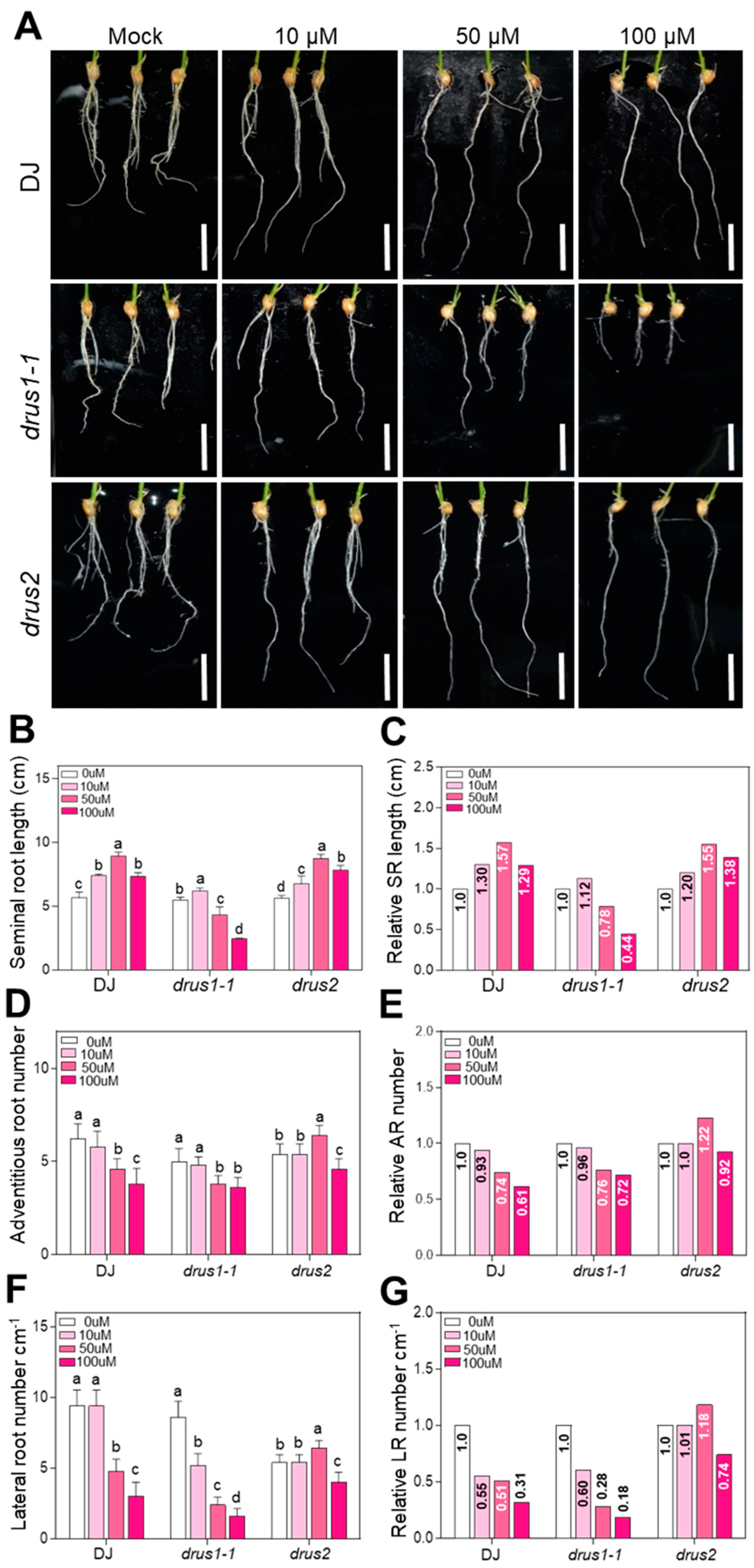
2.5. The Root Growth of drus1-1 Is Sensitive But drus2 Is Insensitive to Auxin Deprivation
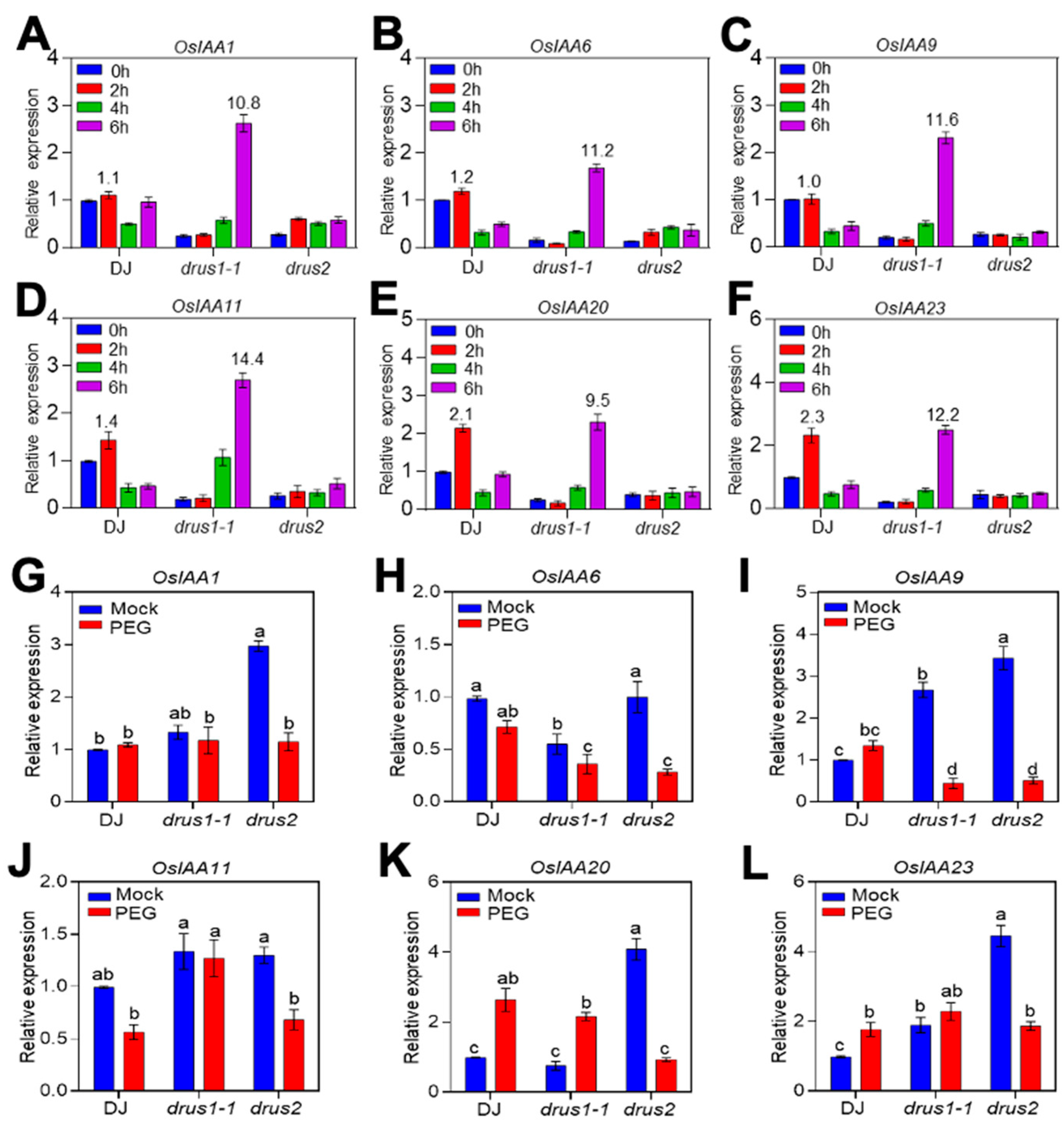
2.6. The OsIAAs Are Highly Induced in drus1-1 but Not in drus2 Roots
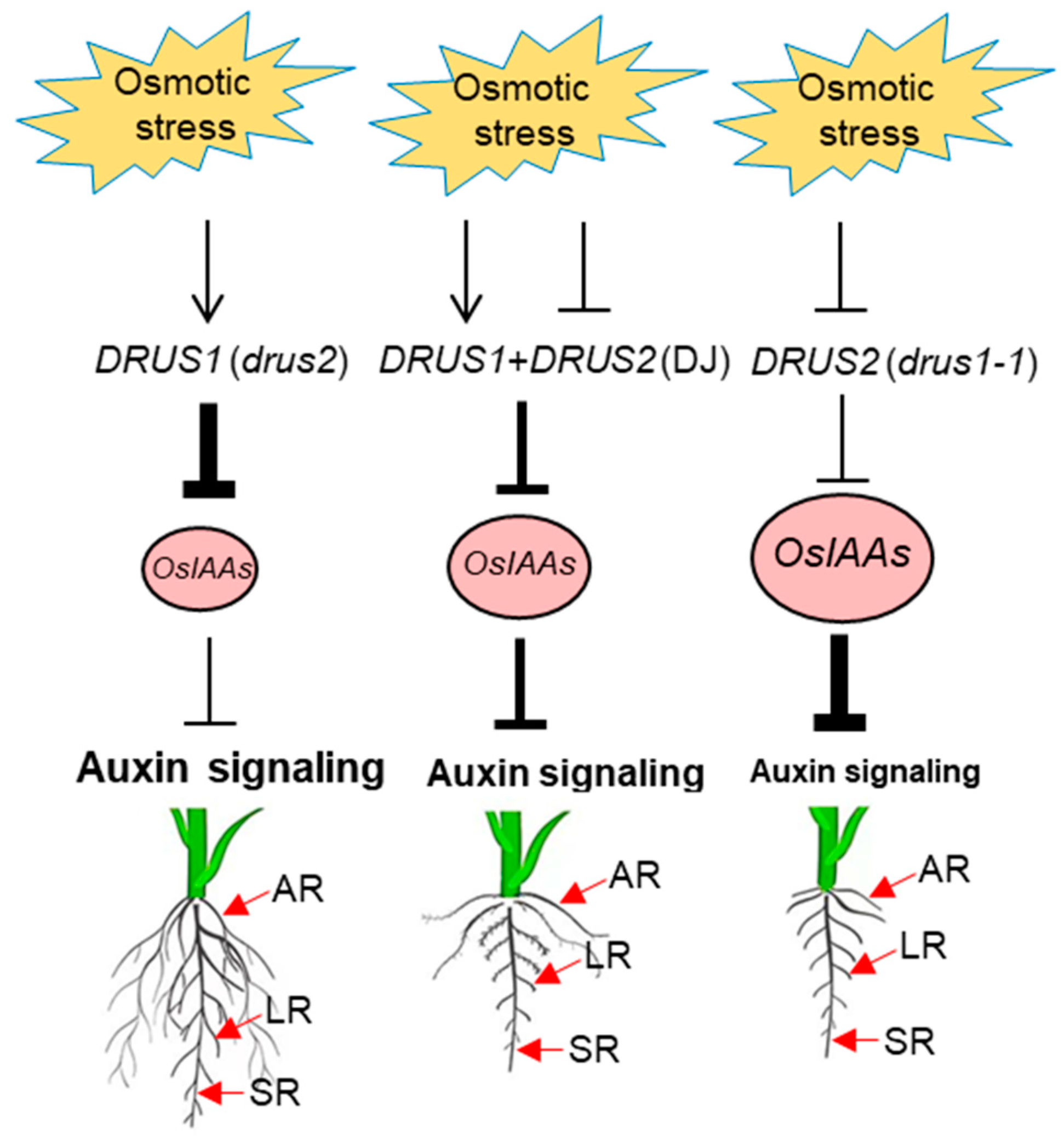
3. Discussion
3.1. DRUS1 Plays a Unique Role in the Osmotic Stress Tolerance of Rice Plants
3.2. DRUS1 Promotes Root Expansion under OS by Repressing OsIAAs and Activating Auxin Signaling
3.3. The Biological Significance of the Presence of DRUS1 and DRUS2 Homologues in Rice Genome
4. Materials and Methods
4.1. Plant Materials and Growth Conditions under Osmotic Stress
4.2. GUS Staining Assay
4.3. Immunoblot Analysis
4.4. Quantitative Reverse Transcription-PCR (qRT-PCR)
4.5. Auxin Content Measurement
4.6. Yucasin Treatment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hématy, K.; Höfte, H. Novel receptor kinases involved in growth regulation. Curr. Opin. Plant Biol. 2008, 11, 321–328. [Google Scholar] [CrossRef]
- Zhu, S.; Fu, Q.; Xu, F.; Zheng, H.; Yu, F. New paradigms in cell adaptation: Decades of discoveries on the Cr RLK1L receptor kinase signalling network. New Phytol. 2021, 232, 1168–1183. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Liu, M.-C.J.; Kita, D.; Jordan, S.S.; Yeh, F.-L.J.; Yvon, R.; Carpenter, H.; Federico, A.N.; Garcia-Valencia, L.E.; Eyles, S.J. FERONIA controls pectin-and nitric oxide-mediated male–female interaction. Nature 2020, 579, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Li, L.; Ye, H.; Yu, X.; Algreen, A.; Yin, Y. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2009, 106, 7648–7653. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Kita, D.; Li, C.; Cheung, A.Y.; Wu, H.-M. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA 2010, 107, 17821–17826. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xing, J.; Wang, L.; Liu, Y.; Qu, J.; Tan, Y.; Fu, X.; Lin, Q.; Deng, H.; Yu, F. Mutations of two FERONIA-like receptor genes enhance rice blast resistance without growth penalty. J. Exp. Bot. 2020, 71, 2112–2126. [Google Scholar] [CrossRef] [PubMed]
- Pu, C.-X.; Han, Y.-F.; Zhu, S.; Song, F.-Y.; Zhao, Y.; Wang, C.-Y.; Zhang, Y.-C.; Yang, Q.; Wang, J.; Bu, S.-L. The rice receptor-like kinases DWARF AND RUNTISH SPIKELET1 and 2 repress cell death and affect sugar utilization during reproductive development. Plant Cell 2017, 29, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-Y.; Liu, X.-X.; Xie, Y.; Lin, X.-Y.; Hu, Z.-J.; Wang, H.; Wang, L.-F.; Dang, W.-Q.; Zhang, L.-L.; Zhu, Y. Identification of FERONIA-like receptor genes involved in rice-Magnaporthe oryzae interaction. Phytopathol. Res. 2020, 2, 14. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, C.; Kuang, B.; Wei, L.; Yan, L.; Wang, T. Receptor-like kinase RUPO interacts with potassium transporters to regulate pollen tube growth and integrity in rice. PLoS Genet. 2016, 12, e1006085. [Google Scholar] [CrossRef]
- Wang, L.; Wang, D.; Yang, Z.; Jiang, S.; Qu, J.; He, W.; Liu, Z.; Xing, J.; Ma, Y.; Lin, Q. Roles of FERONIA-like receptor genes in regulating grain size and quality in rice. Sci. China Life Sci. 2021, 64, 294–310. [Google Scholar] [CrossRef]
- He, W.; Li, W.; Luo, X.; Tang, Y.; Wang, L.; Yu, F.; Lin, Q. Rice FERONIA-LIKE RECEPTOR 3 and 14 affect grain quality by regulating redox homeostasis during endosperm development. J. Exp. Bot. 2023, 74, 3003–3018. [Google Scholar] [CrossRef]
- Kou, X.; Han, W.; Kang, J. Responses of root system architecture to water stress at multiple levels: A meta-analysis of trials under controlled conditions. Front. Plant Sci. 2022, 13, 1085409. [Google Scholar] [CrossRef]
- Hazman, M.; Brown, K.M. Progressive drought alters architectural and anatomical traits of rice roots. Rice 2018, 11, 62. [Google Scholar] [CrossRef]
- Uga, Y.; Okuno, K.; Yano, M. Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 2011, 62, 2485–2494. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chen, Y.; Xing, H.; Ke, W.; Shi, Y.; Sui, Z.; Xu, R.; Gao, L.; Guo, G.; Li, J. Positional cloning and characterization reveal the role of a miRNA precursor gene ZmLRT in the regulation of lateral root number and drought tolerance in maize. J. Integr. Plant Biol. 2023, 65, 772–790. [Google Scholar] [CrossRef]
- Ranjan, A.; Sinha, R.; Singla-Pareek, S.L.; Pareek, A.; Singh, A.K. Shaping the root system architecture in plants for adaptation to drought stress. Physiol. Plant. 2022, 174, e13651. [Google Scholar] [CrossRef] [PubMed]
- Mehra, P.; Pandey, B.K.; Melebari, D.; Banda, J.; Leftley, N.; Couvreur, V.; Rowe, J.; Anfang, M.; De Gernier, H.; Morris, E. Hydraulic flux–responsive hormone redistribution determines root branching. Science 2022, 378, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Wang, J.; Zhou, J.; Qiao, J.; Li, Y.; Quan, R.; Huang, R. Abscisic acid promotes auxin biosynthesis to inhibit primary root elongation in rice. Plant Physiol. 2023, 191, 1953–1967. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Do, P.T.; Vo, K.T.X.; Van Le, A.T.; Tran, T.A.; Chu, H.H.; Jeon, J.-S.; To, T.M.H. The Plasmodesmal Protein Osger4 is Involved in Auxin Mediated Crown Root Development in Rice. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Xu, K.; Lou, Q.; Wang, D.; Li, T.; Chen, S.; Li, T.; Luo, L.; Chen, L. Overexpression of a novel small auxin-up RNA gene, OsSAUR11, enhances rice deep rootedness. BMC Plant Biol. 2023, 23, 319. [Google Scholar] [CrossRef]
- Kurepa, J.; Smalle, J.A. Auxin/cytokinin antagonistic control of the shoot/root growth ratio and its relevance for adaptation to drought and nutrient deficiency stresses. Int. J. Mol. Sci. 2022, 23, 1933. [Google Scholar] [CrossRef]
- Kudoyarova, G.R.; Dodd, I.C.; Veselov, D.S.; Rothwell, S.A.; Yu. Veselov, S. Common and specific responses to availability of mineral nutrients and water. J. Exp. Bot. 2015, 66, 2133–2144. [Google Scholar] [CrossRef]
- Salehin, M.; Bagchi, R.; Estelle, M. SCFTIR1/AFB-based auxin perception: Mechanism and role in plant growth and development. Plant Cell 2015, 27, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Hohl, M.; Schopfer, P. Water relations of growing maize coleoptiles: Comparison between mannitol and polyethylene glycol 6000 as external osmotica for adjusting turgor pressure. Plant Physiol. 1991, 95, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Shirazi, M.; Khan, M.; Arif, M. Effects of peg induced water stress on growth and physiological responses of rice genotypes at seedling stage. Pak. J. Bot. 2019, 51, 2013–2021. [Google Scholar]
- Li, X.; Zhang, L. SA and PEG-induced priming for water stress tolerance in rice seedling. In Information Technology and Agricultural Engineering; Springer: Berlin/Heidelberg, Germany, 2012; pp. 881–887. [Google Scholar]
- Liu, S.-C.; Jin, J.-Q.; Ma, J.-Q.; Yao, M.-Z.; Ma, C.-L.; Li, C.-F.; Ding, Z.-T.; Chen, L. Transcriptomic analysis of tea plant responding to drought stress and recovery. PLoS ONE 2016, 11, e0147306. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, Y.; Gu, P.; Zou, F.; Meng, L.; Song, W.; Yang, Y.; Wang, S.; Zhang, Y. Auxin is involved in lateral root formation induced by drought stress in tobacco seedlings. J. Plant Growth Regul. 2018, 37, 539–549. [Google Scholar] [CrossRef]
- Tsugafune, S.; Mashiguchi, K.; Fukui, K.; Takebayashi, Y.; Nishimura, T.; Sakai, T.; Shimada, Y.; Kasahara, H.; Koshiba, T.; Hayashi, K.-I. Yucasin DF, a potent and persistent inhibitor of auxin biosynthesis in plants. Sci. Rep. 2017, 7, 13992. [Google Scholar] [CrossRef] [PubMed]
- Dien, D.C.; Mochizuki, T.; Yamakawa, T. Effect of various drought stresses and subsequent recovery on proline, total soluble sugar and starch metabolisms in Rice (Oryza sativa L.) varieties. Plant Prod. Sci. 2019, 22, 530–545. [Google Scholar] [CrossRef]
- Lee, H.W.; Cho, C.; Pandey, S.K.; Park, Y.; Kim, M.-J.; Kim, J. LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 2019, 19, 46. [Google Scholar] [CrossRef]
- Quan, R.; Hu, S.; Zhang, Z.; Zhang, H.; Zhang, Z.; Huang, R. Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol. J. 2010, 8, 476–488. [Google Scholar] [CrossRef]
- Salvi, P.; Manna, M.; Kaur, H.; Thakur, T.; Gandass, N.; Bhatt, D.; Muthamilarasan, M. Phytohormone signaling and crosstalk in regulating drought stress response in plants. Plant Cell Rep. 2021, 40, 1305–1329. [Google Scholar] [CrossRef]
- Zhang, Q.; Yuan, W.; Wang, Q.; Cao, Y.; Xu, F.; Dodd, I.C.; Xu, W. ABA regulation of root growth during soil drying and recovery can involve auxin response. Plant Cell Environ. 2022, 45, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, R.; Xing, J.; Yan, L.; Wang, R.; Zhao, Y. The YUCCA-auxin-WOX11 module controls crown root development in rice. Front. Plant Sci. 2018, 9, 523. [Google Scholar] [CrossRef]
- Chen, L.; Tong, J.; Xiao, L.; Ruan, Y.; Liu, J.; Zeng, M.; Huang, H.; Wang, J.-W.; Xu, L. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 4273–4284. [Google Scholar] [CrossRef]
- Hentrich, M.; Sánchez-Parra, B.; Pérez Alonso, M.-M.; Carrasco Loba, V.; Carrillo, L.; Vicente-Carbajosa, J.; Medina, J.; Pollmann, S. YUCCA8 and YUCCA9 overexpression reveals a link between auxin signaling and lignification through the induction of ethylene biosynthesis. Plant Signal. Behav. 2013, 8, e26363. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.L.; Cheng, Z.J.; Zhang, X.S. Endogenous auxin biosynthesis and de novo root organogenesis. J. Exp. Bot. 2016, 67, 4011–4013. [Google Scholar] [CrossRef] [PubMed]
- Woo, Y.-M.; Park, H.-J.; Su’udi, M.; Yang, J.-I.; Park, J.-J.; Back, K.; Park, Y.-M.; An, G. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 2007, 65, 125–136. [Google Scholar] [CrossRef]
- Shen, C.; Wang, S.; Bai, Y.; Wu, Y.; Zhang, S.; Chen, M.; Guilfoyle, T.J.; Wu, P.; Qi, Y. Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L.). J. Exp. Bot. 2010, 61, 3971–3981. [Google Scholar] [CrossRef]
- Orosa-Puente, B.; Leftley, N.; Von Wangenheim, D.; Banda, J.; Srivastava, A.K.; Hill, K.; Truskina, J.; Bhosale, R.; Morris, E.; Srivastava, M. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 2018, 362, 1407–1410. [Google Scholar] [CrossRef]
- Song, Y.; Wang, L.; Xiong, L. Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 2009, 229, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.L. The gene OsIAA9 encoding auxin/indole-3-acetic acid proteins is a negative regulator of auxin-regulated root growth in rice. Biol. Plant 2019, 63, 210–218. [Google Scholar] [CrossRef]
- Song, Y.; Xu, Z.-F. Ectopic overexpression of an auxin/indole-3-acetic acid (Aux/IAA) gene OsIAA4 in rice induces morphological changes and reduces responsiveness to auxin. Int. J. Mol. Sci. 2013, 14, 13645–13656. [Google Scholar] [CrossRef]
- Zhu, Z.-X.; Liu, Y.; Liu, S.-J.; Mao, C.-Z.; Wu, Y.-R.; Wu, P. A gain-of-function mutation in OsIAA11 affects lateral root development in rice. Mol. Plant 2012, 5, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Kitomi, Y.; Inahashi, H.; Takehisa, H.; Sato, Y.; Inukai, Y. OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice. Plant Sci. 2012, 190, 116–122. [Google Scholar] [CrossRef]
- Ni, J.; Wang, G.; Zhu, Z.; Zhang, H.; Wu, Y.; Wu, P. OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant J. 2011, 68, 433–442. [Google Scholar]
- Jung, H.; Lee, D.-K.; Do Choi, Y.; Kim, J.-K. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 2015, 236, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Yang, X.; Lu, J.; Song, F.; Sun, J.; Wang, C.; Lian, J.; Zhao, L.; Zhao, B. OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci. 2021, 308, 110903. [Google Scholar] [CrossRef] [PubMed]
- Aloni, R.; Aloni, E.; Langhans, M.; Ullrich, C.I. Role of cytokinin and auxin in shaping root architecture: Regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 2006, 97, 883–893. [Google Scholar] [CrossRef]
- Li, C.; Yeh, F.-L.; Cheung, A.Y.; Duan, Q.; Kita, D.; Liu, M.-C.; Maman, J.; Luu, E.J.; Wu, B.W.; Gates, L. Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. elife 2015, 4, e06587. [Google Scholar] [CrossRef]
- Nguyen, Q.-N.; Lee, Y.-S.; Cho, L.-H.; Jeong, H.-J.; An, G.; Jung, K.-H. Genome-wide identification and analysis of Catharanthus roseus RLK1-like kinases in rice. Planta 2015, 241, 603–613. [Google Scholar] [CrossRef]
- Jing, X.-Q.; Shi, P.-T.; Zhang, R.; Zhou, M.-R.; Shalmani, A.; Wang, G.-F.; Liu, W.-T.; Li, W.-Q.; Chen, K.-M. Rice kinase OsMRLK63 contributes to drought tolerance by regulating reactive oxygen species production. Plant Physiol. 2023. [Google Scholar] [CrossRef]
- Feng, L.; Gao, Z.; Xiao, G.; Huang, R.; Zhang, H. Leucine-rich repeat receptor-like kinase FON1 regulates drought stress and seed germination by activating the expression of ABA-responsive genes in rice. Plant Mol. Biol. Report. 2014, 32, 1158–1168. [Google Scholar] [CrossRef]
- Ouyang, S.Q.; Liu, Y.F.; Liu, P.; Lei, G.; He, S.J.; Ma, B.; Zhang, W.K.; Zhang, J.S.; Chen, S.Y. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 2010, 62, 316–329. [Google Scholar] [CrossRef]
- Wu, F.; Sheng, P.; Tan, J.; Chen, X.; Lu, G.; Ma, W.; Heng, Y.; Lin, Q.; Zhu, S.; Wang, J. Plasma membrane receptor-like kinase leaf panicle 2 acts downstream of the DROUGHT AND SALT TOLERANCE transcription factor to regulate drought sensitivity in rice. J. Exp. Bot. 2015, 66, 271–281. [Google Scholar] [CrossRef]
- Chen, L.-J.; Wuriyanghan, H.; Zhang, Y.-Q.; Duan, K.-X.; Chen, H.-W.; Li, Q.-T.; Lu, X.; He, S.-J.; Ma, B.; Zhang, W.-K. An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol. 2013, 163, 1752–1765. [Google Scholar] [CrossRef]
- Pan, J.; Li, Z.; Wang, Q.; Yang, L.; Yao, F.; Liu, W. An S-domain receptor-like kinase, OsESG1, regulates early crown root development and drought resistance in rice. Plant Sci. 2020, 290, 110318. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.-H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.-H.; Mayer, K.F.X.; Li, W.-H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Malamy, J.E.; Benfey, P.N. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 1997, 124, 33–44. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Latif, A.; Yang, C.-G.; Zhang, L.-X.; Yang, X.-Y.; Liu, X.-Y.; Ai, L.-F.; Noman, A.; Pu, C.-X.; Sun, Y. The Receptor Kinases DRUS1 and DRUS2 Behave Distinctly in Osmotic Stress Tolerance by Modulating the Root System Architecture via Auxin Signaling. Plants 2024, 13, 860. https://doi.org/10.3390/plants13060860
Latif A, Yang C-G, Zhang L-X, Yang X-Y, Liu X-Y, Ai L-F, Noman A, Pu C-X, Sun Y. The Receptor Kinases DRUS1 and DRUS2 Behave Distinctly in Osmotic Stress Tolerance by Modulating the Root System Architecture via Auxin Signaling. Plants. 2024; 13(6):860. https://doi.org/10.3390/plants13060860
Chicago/Turabian StyleLatif, Ammara, Chen-Guang Yang, Lan-Xin Zhang, Xin-Yu Yang, Xin-Ye Liu, Lian-Feng Ai, Ali Noman, Cui-Xia Pu, and Ying Sun. 2024. "The Receptor Kinases DRUS1 and DRUS2 Behave Distinctly in Osmotic Stress Tolerance by Modulating the Root System Architecture via Auxin Signaling" Plants 13, no. 6: 860. https://doi.org/10.3390/plants13060860
APA StyleLatif, A., Yang, C.-G., Zhang, L.-X., Yang, X.-Y., Liu, X.-Y., Ai, L.-F., Noman, A., Pu, C.-X., & Sun, Y. (2024). The Receptor Kinases DRUS1 and DRUS2 Behave Distinctly in Osmotic Stress Tolerance by Modulating the Root System Architecture via Auxin Signaling. Plants, 13(6), 860. https://doi.org/10.3390/plants13060860






