Physiological and Proteomic Analyses of mtn1 Mutant Reveal Key Players in Centipedegrass Tiller Development
Abstract
1. Introduction
2. Results
2.1. Phenotype of mtn1 Mutant
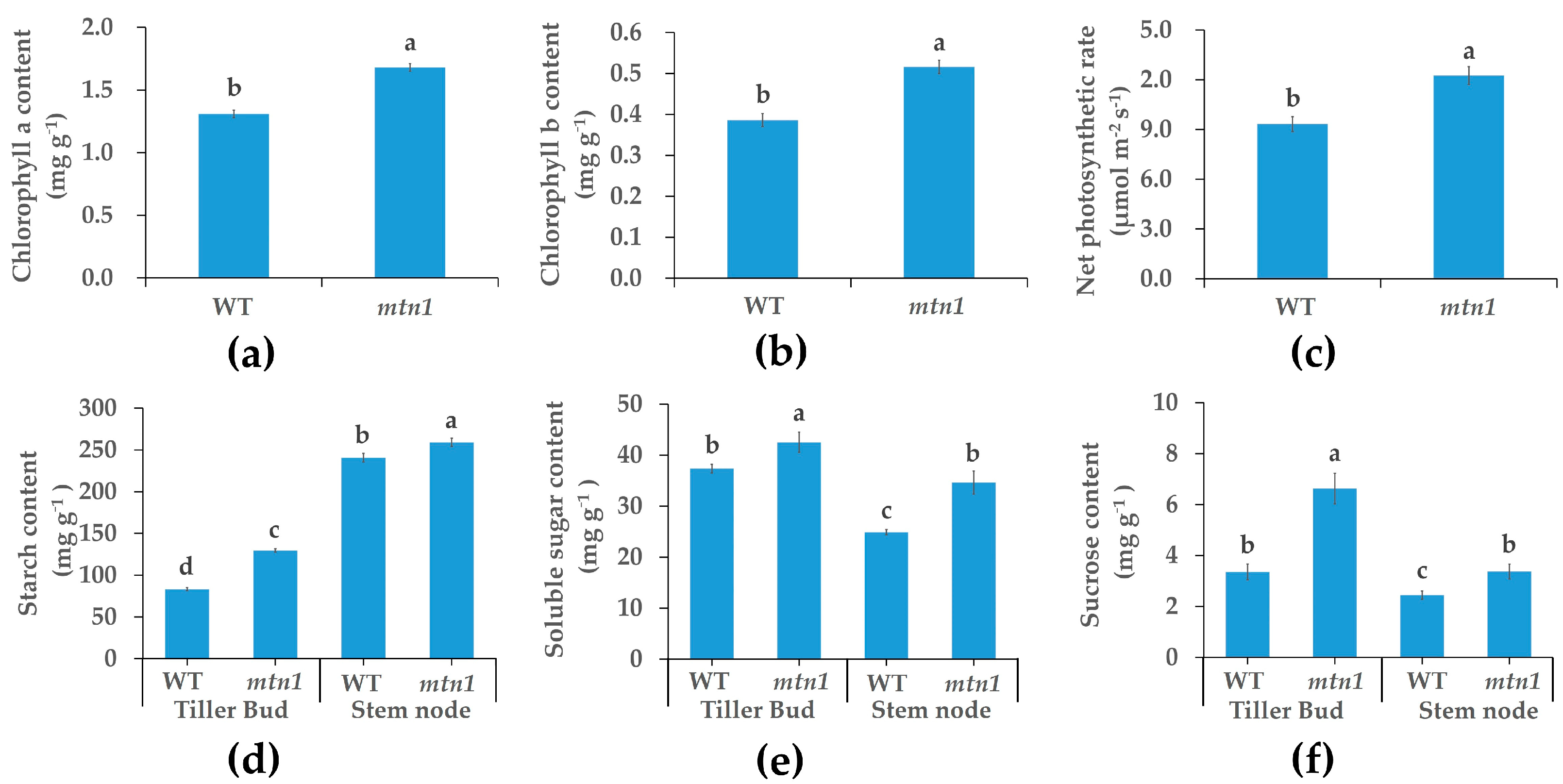
2.2. Physiological Characteristics
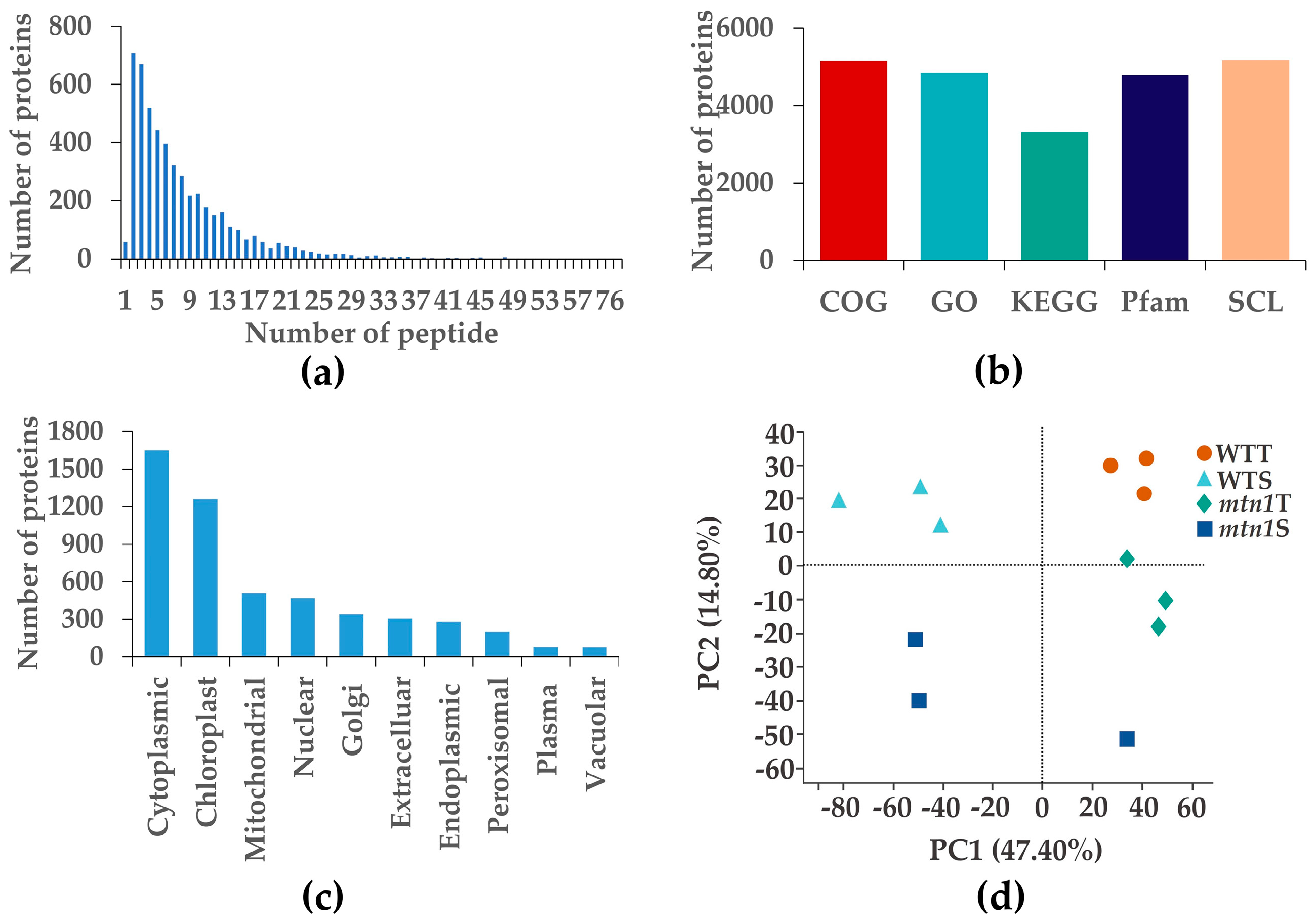
2.3. Protein Data Quality
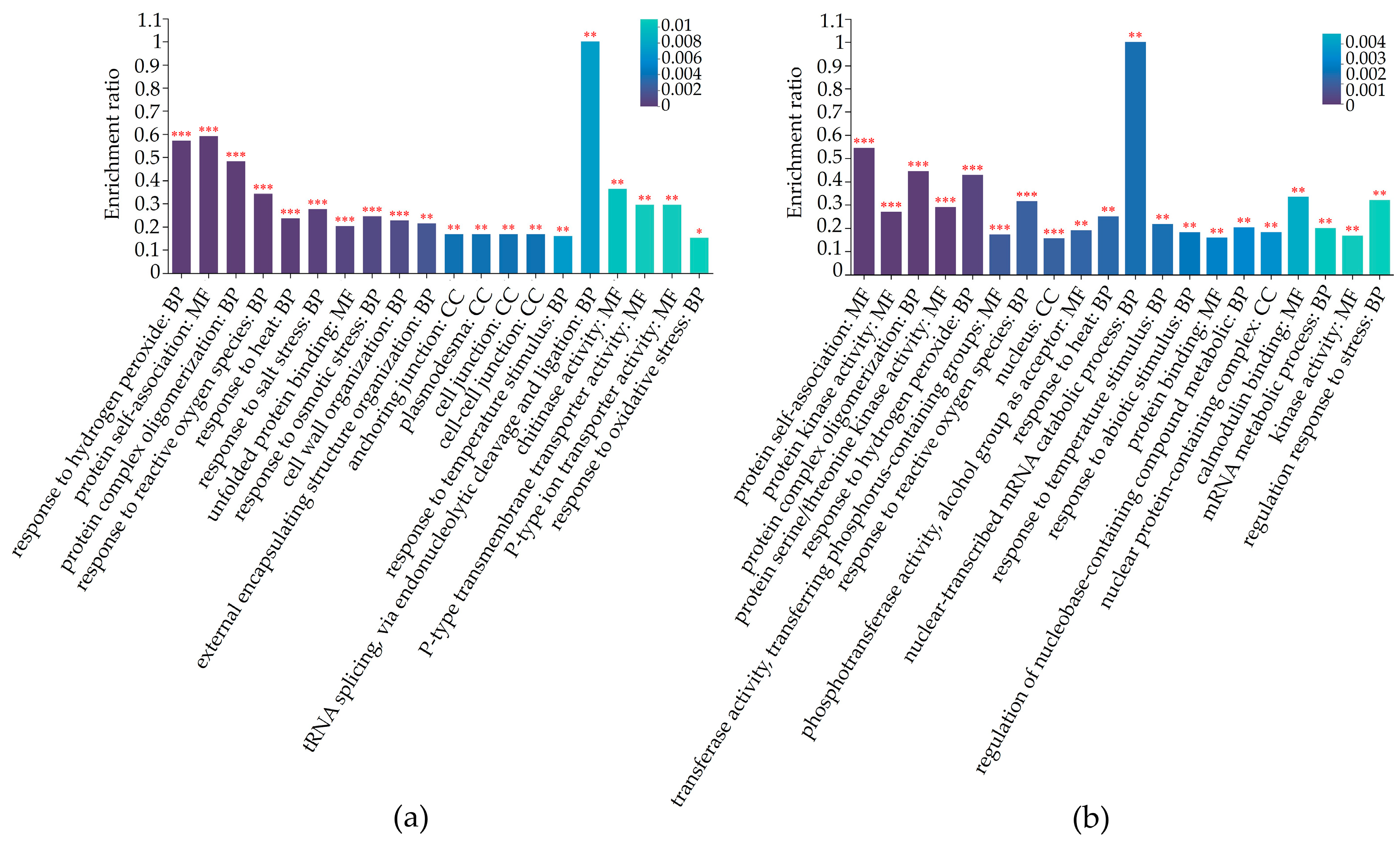
2.4. Differential Expression Proteins Analysis
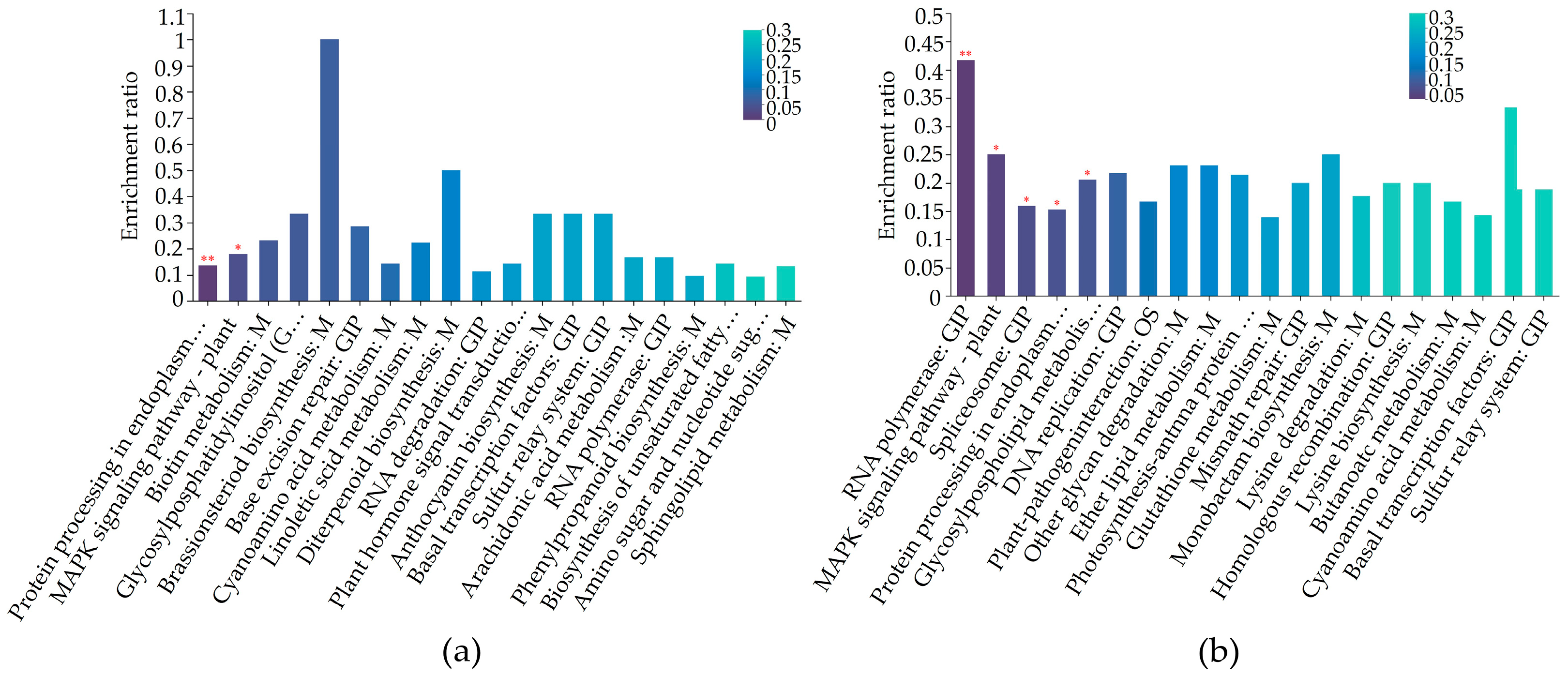
2.5. Differentially Regulated Biochemical Pathways of mtn1 Mutant and WT
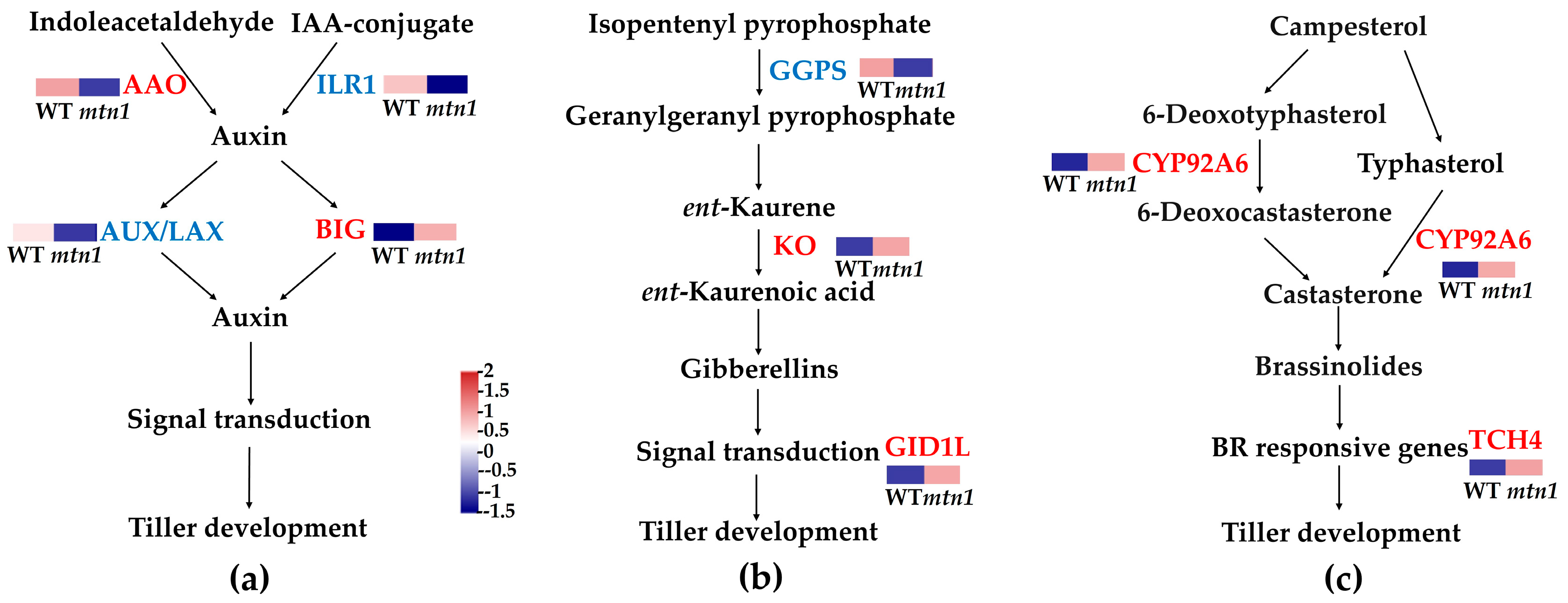
2.6. DEPs Involved in Plant Hormone Signal Transduction
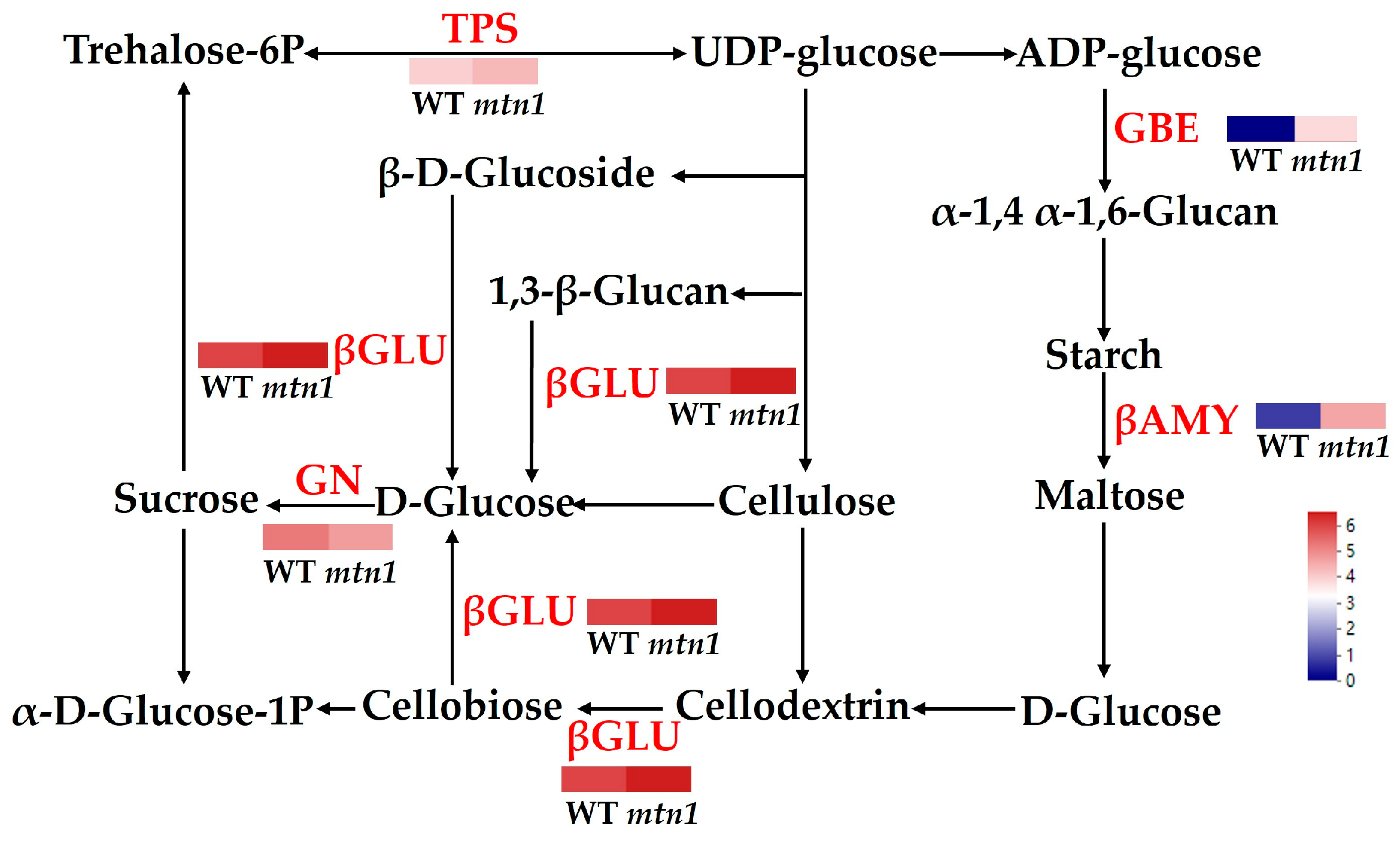
2.7. DAPs Involved in Starch and Sucrose Metabolism
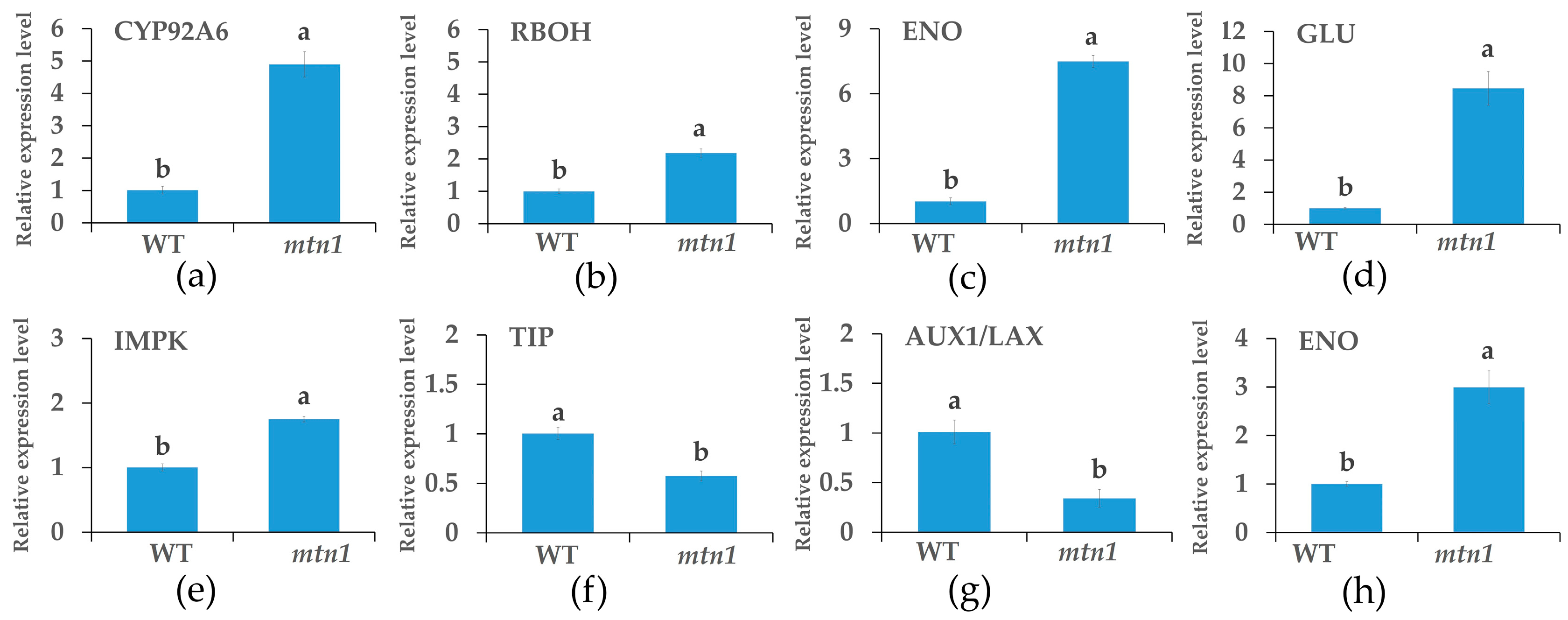
2.8. qRT-PCR
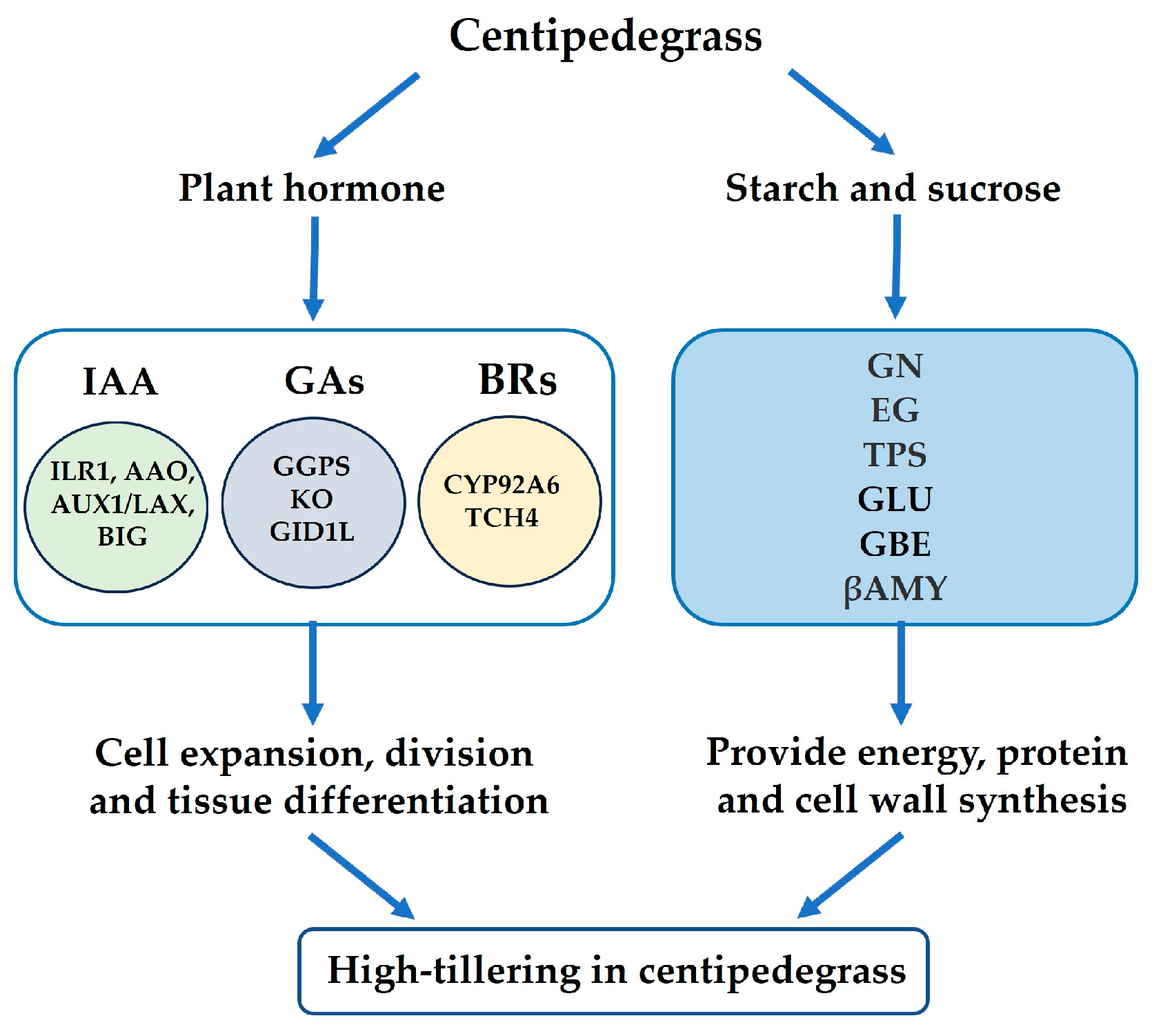
2.9. Summary Analysis of the Physiological Parameters and Proteomes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Experimental Design
4.2. Physiological Parameters’ Determination
4.3. Protein Extraction and Trypsin Digestion
4.4. Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS) Analysis
4.5. Protein Identification and Bioinformatics Analysis
4.6. qRT-PCR
4.7. Data Statistics and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, J.; Hanna, W.; Elsner, E. Morphological and seed set characteristics of centipedegrass accessions collected in China. Econ. Bot. 2003, 57, 380–388. [Google Scholar] [CrossRef]
- Li, J.; Guo, H.; Zong, J.; Chen, J.; Li, D.; Liu, J. Genetic diversity in centipedegrass [Eremochloa ophiuroides (Munro) Hack.]. Hortic. Res. 2020, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Abllenda, J.A.; Navarro, C.; Prat, S. Flowering and tuberization: A tale of two nightshades. Trends Plant Sci. 2014, 19, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Abllenda, J.A.; Navarro, C.; Prat, S. From the model to the crop: Genes controlling tuber formation in potato. Curr. Opin. Biotechnol. 2011, 22, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Plunkert, M.; Luo, X.; Liu, Z. Developmental regulation of stolon and rhizome. Curr. Opin. Plant Biol. 2021, 59, 101970. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Smith, S.M.; Li, J. Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Yang, J.; Cui, H.; Li, Z.; Jia, F.; Chen, J.; Li, X. Effects of plant density on tillering in the weed grass Aegilops tauschii and its phytohormonal regulation. Plant Physiol. Bioch. 2020, 157, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Chaoqing Ding, C.; Zhang, G.; Hu, J.; Zhu, L.; Zeng, D.; Qian, Q.; Ren, D. Genetic and environmental control of rice tillering. Crop J. 2023, 11, 1287–1302. [Google Scholar] [CrossRef]
- Li, H.; Sun, H.; Jiang, J.; Sun, X.; Tan, L.; Sun, C. TAC4 controls tiller angle by regulating the endogenous auxin content and distribution in rice. Plant Biotechnol. J. 2021, 19, 64–73. [Google Scholar] [CrossRef]
- Zhao, Y.; Zha, M.; Xu, C.; Hou, F.; Wang, Y. Proteomic analysis revealed the antagonistic effect of decapitation and strigolactones on the tillering control in rice. Plants 2021, 21, 91. [Google Scholar] [CrossRef]
- Li, L.; Xie, C.; Zong, J.; Guo, H.; Li, D.; Liu, J. Physiological and comparative transcriptome analyses of the high-tillering mutant mtn1 reveal regulatory mechanisms in the tillering of centipedegrass (Eremochloa ophiuroides (Munro) Hack.). Int. J. Mol. Sci. 2022, 23, 11580. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Sun, F.; Wang, Y.; Shi, L.; Liu, S.; Xi, Y. The PIN1 family gene PvPIN1 is involved in auxin-dependent root emergence and tillering in switchgrass. Gene. Mol. Biol. 2016, 39, 62–72. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tabuchi, H.; Zhang, Y.; Hattori, S.; Omae, M.; Shimizu-Sato, S.; Oikawa, T.; Qian, Q.; Nishimura, M.; Kitano, H.; Xie, H.; et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 2011, 23, 3276–3287. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Gao, C.; Chen, M.S.; Pan, B.Z.; Ye, K.; Xu, Z.F. Gibberellin promotes shoot branching in the perennial woody plant Jatropha curcas. Plant Cell Physiol. 2015, 56, 1655–1666. [Google Scholar] [CrossRef] [PubMed]
- Tenreira, T.; Lange, M.J.P.; Lange, T.; Bres, C.; Labadie, M.; Monfort, A.; Hernould, M.; Rothan, C.; Denoyes, B. A Specific gibberellin 20-oxidase dictates the flowering-runnering decision in diploid strawberry. Plant Cell 2017, 29, 2168–2182. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, L.; Ge, Y.; Wang, J.; Yu, J.; Yang, Z.; Huang, B. Gibberellic acid inhibition of tillering in tall fescue involving crosstalks with cytokinins and transcriptional regulation of genes controlling axillary bud outgrowth. Plant Sci. 2019, 287, 110168. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Feng, G.; Yang, Z.; Liu, Q.; Nie, G.; Li, D.; Huang, T.; Huang, L.; Zhang, X. Transcriptome Analysis reveals the potential molecular mechanisms of tiller bud development in orchardgrass. Int. J. Mol. Sci. 2023, 24, 15762. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.F.; Yang, S.Y.; Chen, K.T.; Hsing, Y.I.; Zeevaart, J.A.D.; Chen, L.J.; Yu, S.M. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef]
- Xu, H.; Liu, Q.; Yao, T.; Fu, X. Shedding light on integrative GA signaling. Curr. Opin. Plant Biol. 2014, 21, 89–95. [Google Scholar] [CrossRef]
- Wu, K.; Xu, H.; Gao, X.; Fu, X. New insights into gibberellin signaling in regulating plant growth-metabolic coordination. Curr. Opin. Plant Biol. 2021, 63, 102074. [Google Scholar] [CrossRef]
- Yang, C.J.; Zhang, C.; Lu, Y.N.; Jin, J.Q.; Wang, X.L. The mechanisms of brassinosteroids’ action: From signal transduction to plant development. Mol. Plant 2011, 4, 588–600. [Google Scholar] [CrossRef]
- Oklestkova, J.; Rárová, L.; Kvasnica, M.; Strnad, M. Brassinosteroids: Synthesis and biological activities. Phytochem. Rev. 2015, 14, 1053–1072. [Google Scholar] [CrossRef]
- Tong, H.; Chu, C. Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci. 2018, 23, 1016–1028. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Ji, Y.; Hu, J.; Guo, R.; Sun, S.; Wang, X. Strigolactones and brassinosteroids antagonistically regulate the stability of the D53–OsBZR1 complex to determine FC1 expression in rice tillering. Mol. Plant 2020, 13, 586–597. [Google Scholar] [CrossRef]
- Barbier, F.; Péron, T.; Lecerf, M.; Perez-Garcia, M.D.; Barrière, Q.; Rolčík, J.; Boutet-Mercey, S.; Citerne, S.; Lemoine, R.; Porcheron, B. Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J. Exp. Bot. 2015, 66, 2569–2582. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.B.; Barbier, F.F.; Zhao, J.; Zafar, S.A.; Uzair, M.; Sun, Y.; Fang, J.; Perez-Garcia, M.D.; Bertheloot, J.; Sakr, S.; et al. Sucrose promotes D53 accumulation and tillering in rice. New Phytol. 2022, 234, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Fichtner, F.; Barbier, F.F.; Feil, R.; Watanabe, M.; Annunziata, M.G.; Chabikwa, T.G.; Höfgen, R.; Stitt, M.; Beveridge, C.A.; Lunn, J.E. Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). Plant J. 2017, 92, 611–623. [Google Scholar] [CrossRef]
- Fichtner, F.; Barbier, F.F.; Annunziata, M.G.; Feil, R.; Olas, J.J.; Mueller-Roeber, B.; Stitt, M.; Beveridge, C.A.; Lunn, J.E. Regulation of shoot branching in Arabidopsis by trehalose 6-phosphate. New Phytol. 2021, 229, 2135–2151. [Google Scholar] [CrossRef]
- Li, M.; Li, H.; Zhu, Q.; Liu, D.; Li, Z.; Chen, H.; Luo, J.; Gong, P.; Ismail, A.M.; Zhang, Z. Knockout of the sugar transporter OsSTP15 enhances grain yield by improving tiller number due to increased sugar content in the shoot base of rice (Oryza sativa L.). New Phytol. 2024, 241, 1250–1265. [Google Scholar] [CrossRef]
- Bertheloot, J.; Barbier, F.; Boudon, F.; Perez-Garcia, M.D.; Péron, T.; Citerne, S.; Dun, E.; Beveridge, C.; Godin, C.; Sakr, S. Sugar availability suppresses the auxin-induced strigolactone pathway to promote bud outgrowth. New Phytol. 2020, 225, 866–879. [Google Scholar] [CrossRef]
- Shi, J.; Wang, N.; Zhou, H.; Xu, Q.; Yan, G. Transcriptome analyses provide insights into the homeostatic regulation of axillary buds in upland cotton (G. hirsutum L.). BMC Plant Bio. 2020, 20, 228. [Google Scholar] [CrossRef]
- He, R.; Ni, Y.; Li, J.; Jiao, Z.; Zhu, X.; Jiang, Y.; Li, Q.; Niu, J. Quantitative changes in the transcription of phytohormone-related genes: Some transcription factors are major causes of the wheat mutant dmc not tillering. Int. J. Mol. Sci. 2018, 19, 1324. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ni, B.; Zeng, Y.; He, C.; Zhang, J. Transcriptomic analysis reveals hormonal control of branching in Salix matsudana. Forests 2020, 11, 287. [Google Scholar] [CrossRef]
- Li, Y. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Bio. 2018, 69, 417–435. [Google Scholar]
- Hayashi, K.I.; Arai, K.; Aoi, Y.; Tanaka, Y.; Hira, H.; Guo, R.; Hu, Y.; Ge, C.; Zhao, Y.; Kasahara, H.; et al. The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun. 2021, 12, 6752. [Google Scholar] [CrossRef] [PubMed]
- Winer, L.; Riov, J.; Goren, R. Catabolism of indole-3-acetic acid in citrus leaves: Identification and characterization of indole-3-aldehyde oxidase. Physiol. Plant. 1993, 89, 220–226. [Google Scholar] [CrossRef]
- Swarup, R.; Bhosale, R. Developmental roles of AUX1/LAX auxin influx carriers in plants. Front. Plant Sci. 2019, 10, 1306. [Google Scholar] [CrossRef]
- Tong, H.; Chu, C. Coordinating gibberellin and brassinosteroid signaling beyond green revolution. J. Genet. Genom. 2023, 50, 459–461. [Google Scholar] [CrossRef]
- Castro-Camba, R.; Sánchez, C.; Vidal, N.; Vielba, J.M. Plant development and crop yield: The role of gibberellins. Plants 2022, 11, 2650. [Google Scholar] [CrossRef]
- LaMantia, J.M.; Chandra, A.; Huff, D.R. Development of ent-kaurene oxidase-based conserved intron spanning primers for species identification in the genus poa (Poaceae; Bluegrass). Agronomy 2018, 8, 58. [Google Scholar] [CrossRef]
- Mauriat, M.; Sandberg, L.G.; Moritz, T. Proper gibberellin localization in vascular tissue is required to control auxin-dependent leaf development and bud outgrowth in hybrid aspen. Plant J. 2011, 67, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Agharkar, M.; Lomba, P.; Altpeter, F.; Zhang, H.; Kenworthy, K.; Lange, T. Stable expression of AtGA2ox1 in a low-input turfgrass (Paspalum notatum Flugge) reduces bioactive gibberellin levels and improves turf quality under field conditions. Plant Biotechnol. J. 2007, 5, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Wang, S.; Song, W.; Zhang, J.; Wang, Y.; Liu, Q.; Yu, J.; Ye, Y.; Li, S.; Chen, J.; et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 2020, 367, eaaz2046. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zheng, H.; Lin, L.; Wang, F.; Sui, N. Roles of brassinosteroids in plant growth and abiotic stress response. Plant Growth Regul. 2021, 93, 29–38. [Google Scholar] [CrossRef]
- Zhang, C.; Bai, M.; Chong, K. Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 2014, 33, 683–696. [Google Scholar] [CrossRef] [PubMed]
- González-Grandío, E.; Poza-Carrión, C.; Sorzano, C.O.; Cubas, P. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 2013, 25, 834–850. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Trieu, A.; Radhakrishnan, P.; Kwok, S.F.; Harris, S.; Zhang, K.; Wang, J.; Wan, J.; Zhai, H.; Takatsuto, S.; et al. Brassinosteroids regulate grain filling in rice. Plant Cell 2008, 20, 2130–2145. [Google Scholar] [CrossRef]
- Iliev, E.A.; Wei, X.; Polisensky, D.H.; Torisky, R.S.; Braam, C.J. Transcriptional and posttranscriptional regulation of Arabidopsis TCH4 expression by diverse stimuli roles of cis regions and brassinosteroids. Plant Physiol. 2002, 130, 770–783. [Google Scholar] [CrossRef]
- Wang, M.; Pérez-Garcia, M.D.; Davière, J.M.; Barbier, F.; Ogé, L.; Gentilhomme, J.; Voisine, L.; Péron, T.; Launay-Avon, A.; Clément, G.; et al. Axillary bud outgrowth in rose is controlled by sugar metabolic and signaling pathways. J. Exp. Bot. 2021, 72, 3044–3060. [Google Scholar] [CrossRef]
- Shim, S.H.; Mahong, B.; Lee, S.K.; Kongdin, M.; Lee, C.; Kim, Y.J.; Qu, G.; Zhang, D.; Ketudat Cairns, J.R.; Jeon, J.S. Rice β-glucosidase Os12BGlu38 is required for synthesis of intine cell wall and pollen fertility. J. Exp. Bot. 2022, 73, 784–800. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, J.; Li, D.; Chen, J.; Guo, A.; Guo, H.; Zong, J.; Liu, J. A Method for Creating Zoysia Mutants by Oxygen Cold Plasma Mutagenesis. CN112825767B, 31 January 2023. [Google Scholar]
- Zhang, S.; Hu, F.; Li, H.; Li, X. Influence of earthworm mucus and amino acids on tomato seedling growth and cadmium accumulation. Environ. Pollut. 2009, 157, 2737–2742. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, K.; Kuwabara, Y.; Motobayashi, T. Photosynthetic light-use efficiency of rice leaves under fluctuating incident light. Agrosys. Geosci. Environ. 2020, 3, e20030. [Google Scholar] [CrossRef]
- Ma, Z.; Chen, S.; Wang, Z.; Liu, J.; Zhang, B. Proteome analysis of bermudagrass stolons and rhizomes provides new insights into the adaptation of plant stems to aboveground and underground growth. J. Proteom. 2021, 241, 104245. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zi, H.; Wang, R.; Liu, J.; Wang, H.; Chen, R.; Li, L.; Guo, H.; Chen, J.; Li, J.; et al. A high-quality chromosome-scale assembly of the centipedegrass [Eremochloa ophiuroides (Munro) Hack.] genome provides insights into chromosomal structural evolution and prostrate growth habit. Hortic. Res. 2021, 8, 201. [Google Scholar] [CrossRef] [PubMed]
- Nikita, P.; Koch, H.; Koch, S.; Lubeck, M.; Heilig, R.; Brehmer, S.; Fischer, R.; Cox, J. MaxQuant Software for Ion mobility enhanced shotgun proteomics. Mol. Cell. Proteom. 2020, 19, 1058–1069. [Google Scholar]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S.; AmiGO, H. Web presence working, AmiGO: Online access to ontology and annotation data. Bioinformatics 2009, 25, 288–289. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, C.; Chen, R.; Sun, Q.; Hao, D.; Zong, J.; Guo, H.; Liu, J.; Li, L. Physiological and Proteomic Analyses of mtn1 Mutant Reveal Key Players in Centipedegrass Tiller Development. Plants 2024, 13, 1028. https://doi.org/10.3390/plants13071028
Xie C, Chen R, Sun Q, Hao D, Zong J, Guo H, Liu J, Li L. Physiological and Proteomic Analyses of mtn1 Mutant Reveal Key Players in Centipedegrass Tiller Development. Plants. 2024; 13(7):1028. https://doi.org/10.3390/plants13071028
Chicago/Turabian StyleXie, Chenming, Rongrong Chen, Qixue Sun, Dongli Hao, Junqin Zong, Hailin Guo, Jianxiu Liu, and Ling Li. 2024. "Physiological and Proteomic Analyses of mtn1 Mutant Reveal Key Players in Centipedegrass Tiller Development" Plants 13, no. 7: 1028. https://doi.org/10.3390/plants13071028
APA StyleXie, C., Chen, R., Sun, Q., Hao, D., Zong, J., Guo, H., Liu, J., & Li, L. (2024). Physiological and Proteomic Analyses of mtn1 Mutant Reveal Key Players in Centipedegrass Tiller Development. Plants, 13(7), 1028. https://doi.org/10.3390/plants13071028




