Diversity of the Maize Root Endosphere and Rhizosphere Microbiomes Modulated by the Inoculation with Pseudomonas fluorescens UM270 in a Milpa System
Abstract
1. Introduction
2. Results
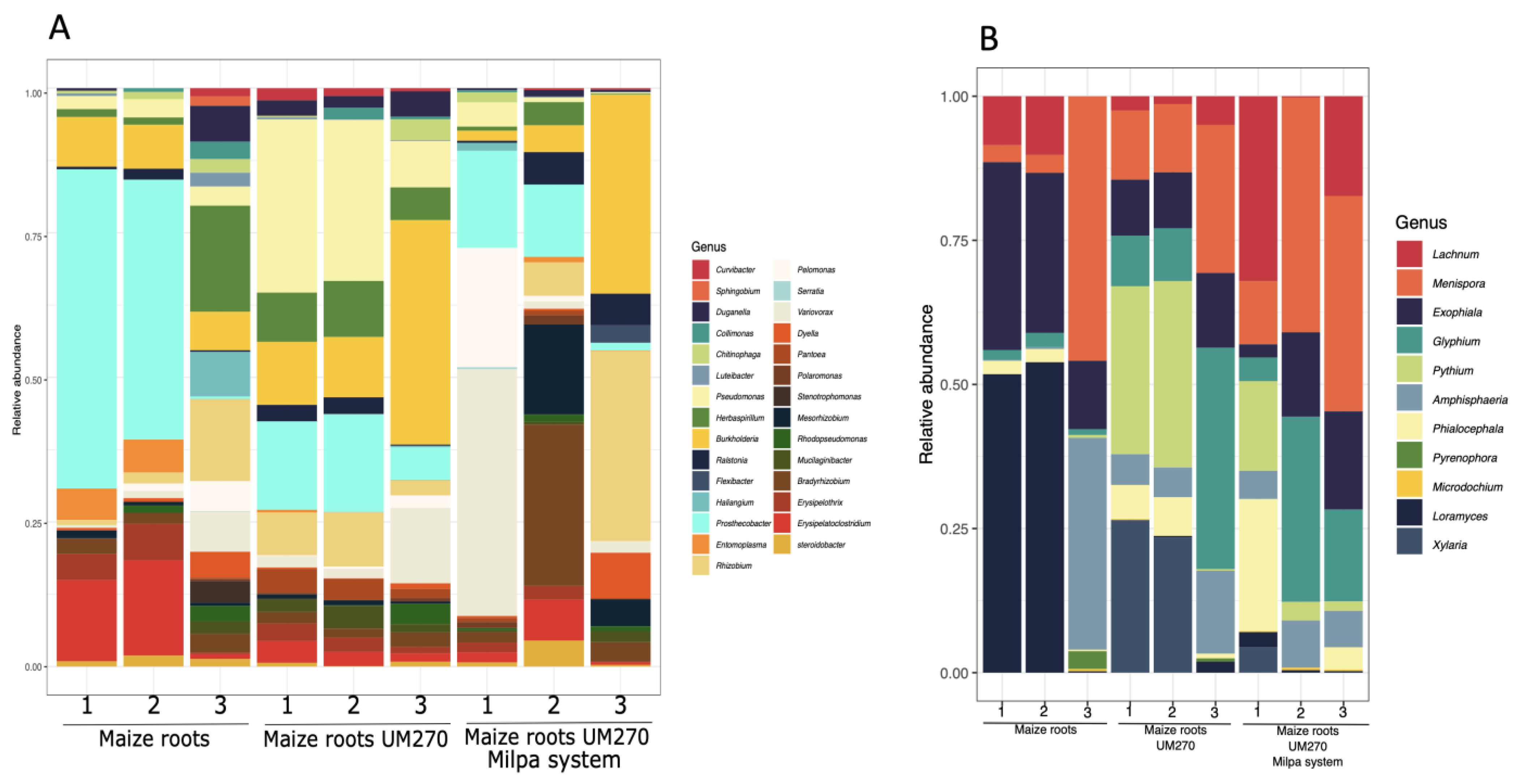
2.1. Endobiome Analysis of Maize Roots
2.2. Index Diversity Analysis
2.3. Endobiome Network Analysis
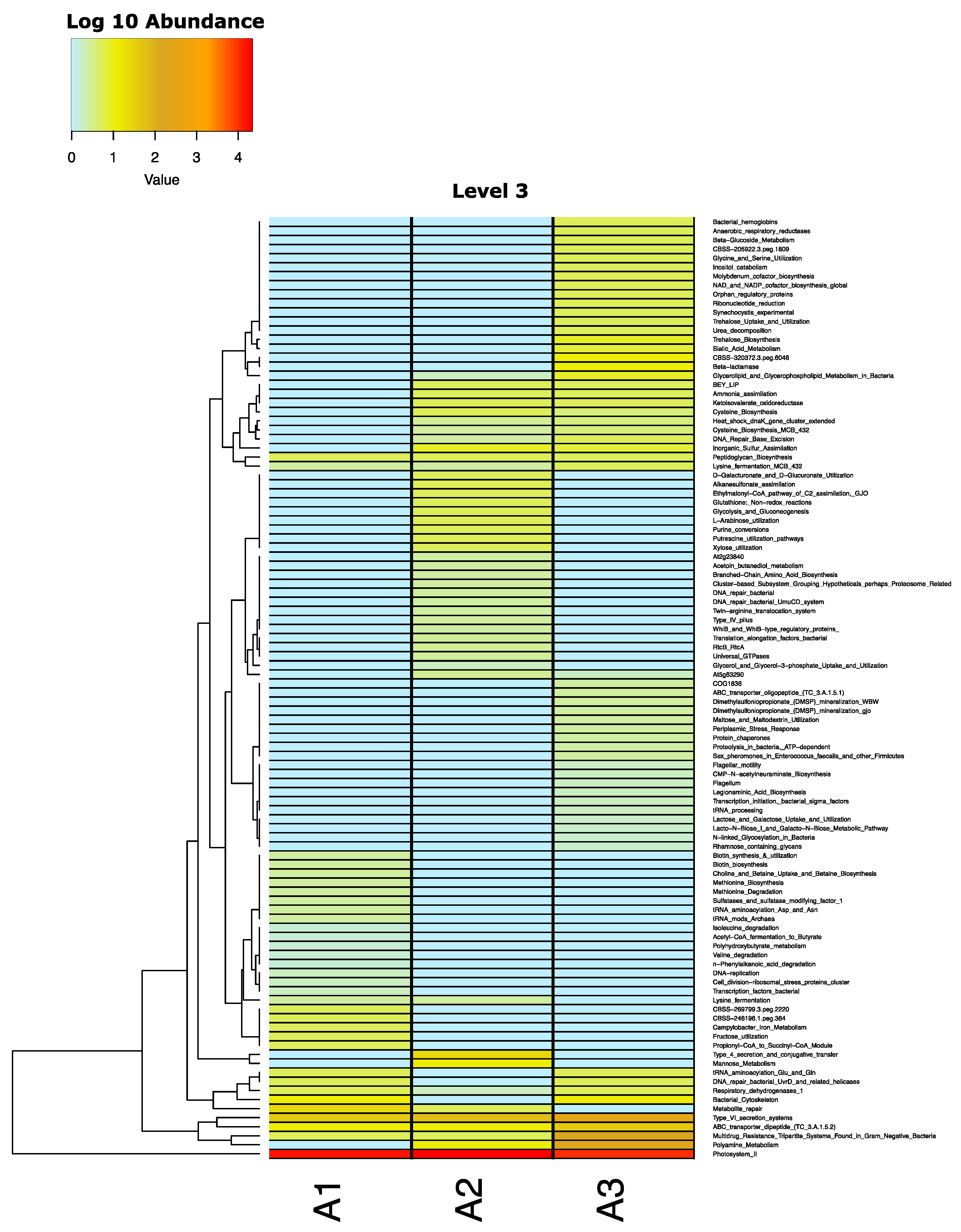
2.4. Metagenomics of the Rhizosphere
3. Discussion
4. Materials and Methods
4.1. Experimental Site
4.2. Biological Material
4.3. Inoculum Preparation
4.4. Seed Treatments
4.5. Establishment of the Experiment under Field Conditions
4.6. Experimental Design
4.7. Endophytic DNA Extraction and Illumina Sequencing
4.8. Data Processing
4.9. Analysis Alpha and Beta Diversities
4.10. Endobiome Network Analysis
4.11. Metagenomic DNA Isolation and Analysis of Soil Rhizosphere
4.12. Sequence Accession Numbers
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nigh, R.; Diemont, S.A.W. The Maya milpa: Fire and the legacy of living soil. Front. Ecol. Environ. 2013, 11, e45–e54. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, Z.; Liu, J.; Li, X.; Wang, X.; Dai, C.; Zhang, T.; Carrión, V.J.; Wei, Z.; Cao, F.; et al. Crop rotation and native microbiome inoculation restore soil capacity to suppress a root disease. Nat. Commun. 2023, 14, 8126. [Google Scholar] [CrossRef] [PubMed]
- Ebel, R.; Pozas, J.; Soria, F.; Cruz, J. Manejo orgánico de la milpa: Rendimiento de maíz, frijol y calabaza en monocultivo y policultivo Organic milpa: Yields of maize, beans, and squash in mono-and polycropping systems. Terra Latinoam. 2017, 35, 149–160. [Google Scholar] [CrossRef]
- Ureta, C.; González, E.J.; Espinosa, A.; Trueba, A.; Piñeyro-Nelson, A.; Álvarez-Buylla, E.R. Maize yield in Mexico under climate change. Agric. Syst. 2020, 177, 102697. [Google Scholar] [CrossRef]
- Ariza-Mejía, D.; Oyoque-Salcedo, G.; Angóa-Pérez, V.; Mena-Violante, H.G.; Álvarez-Bernal, D.; Torres-García, J.R. Diversity and Potential Function of the Bacterial Rhizobiome Associated to Physalis Ixocarpa Broth. in a Milpa System, in Michoacan, Mexico. Agronomy 2022, 12, 1780. [Google Scholar] [CrossRef]
- Erel, R.; Bérard, A.; Capowiez, L.; Doussan, C.; Arnal, D.; Souche, G.; Gavaland, A.; Fritz, C.; Visser, E.J.W.; Salvi, S.; et al. Soil type determines how root and rhizosphere traits relate to phosphorus acquisition in field-grown maize genotypes. Plant Soil 2017, 412, 115–132. [Google Scholar] [CrossRef]
- Rudolph-Mohr, N.; Tötzke, C.; Kardjilov, N.; Oswald, S.E. Mapping water, oxygen, and pH dynamics in the rhizosphere of young maize roots. J. Plant Nutr. Soil Sci. 2017, 180, 336–346. [Google Scholar] [CrossRef]
- Li, Y.; Qu, Z.; Xu, W.; Chen, W.; Hu, Y.; Wang, Z. Maize (Zea mays L.) genotypes induce the changes of rhizosphere microbial communities. Arch. Microbiol. 2022, 204, 321. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Ley, R.E. Exploring the maize rhizosphere microbiome in the field: A glimpse into a highly complex system. Commun. Integr. Biol. 2013, 6, e25177. [Google Scholar] [CrossRef]
- Correa-Galeote, D.; Bedmar, E.J.; Arone, G.J. Maize endophytic bacterial diversity as affected by soil cultivation history. Front. Microbiol. 2018, 9, 484. [Google Scholar] [CrossRef]
- Chen, Q.-L.; An, X.-L.; Zheng, B.-X.; Ma, Y.-B.; Su, J.-Q. Long-term organic fertilization increased antibiotic resistome in phyllosphere of maize. Sci. Total Environ. 2018, 645, 1230–1237. [Google Scholar] [CrossRef]
- Khatoon, Z.; Huang, S.; Rafique, M.; Fakhar, A.; Kamran, M.A.; Santoyo, G. Unlocking the potential of plant growth-promoting rhizobacteria on soil health and the sustainability of agricultural systems. J. Environ. Manag. 2020, 273, 111118. [Google Scholar] [CrossRef]
- Pereira, S.I.A.; Abreu, D.; Moreira, H.; Vega, A.; Castro, P.M.L. Plant growth-promoting rhizobacteria (PGPR) improve the growth and nutrient use efficiency in maize (Zea mays L.) under water deficit conditions. Heliyon 2020, 6, e05106. [Google Scholar] [CrossRef]
- Ferrarezi, J.A.; Carvalho-Estrada, P.d.A.; Batista, B.D.; Aniceto, R.M.; Tschoeke, B.A.P.; Andrade, P.A.d.M.; Lopes, B.d.M.; Bonatelli, M.L.; Odisi, E.J.; Azevedo, J.L.; et al. Effects of inoculation with plant growth-promoting rhizobacteria from the Brazilian Amazon on the bacterial community associated with maize in field. Appl. Soil Ecol. 2022, 170, 104297. [Google Scholar] [CrossRef]
- da Cunha, E.T.; Pedrolo, A.M.; Arisi, A.C.M. Thermal and salt stress effects on the survival of plant growth-promoting bacteria Azospirillum brasilense in inoculants for maize cultivation. J. Sci. Food Agric. 2024. [Google Scholar] [CrossRef]
- Trivedi, P.; Mattupalli, C.; Eversole, K.; Leach, J.E. Enabling sustainable agriculture through understanding and enhancement of microbiomes. New Phytol. 2021, 230, 2129–2147. [Google Scholar] [CrossRef]
- Liu, N.; Dong, L.; Deng, X.; Liu, D.; Liu, Y.; Li, M.; Hu, Y.; Yan, Y. Genome-wide identification, molecular evolution, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon L. BMC Plant Biol. 2018, 18, 336. [Google Scholar] [CrossRef]
- Santoyo, G. How plants recruit their microbiome? New insights into beneficial interactions. J. Adv. Res. 2021, 40, 45–58. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, X.; Shen, Z.; Zhu, C.; Jiao, Z.; Li, R.; Shen, Q. Pre-colonization of PGPR triggers rhizosphere microbiota succession associated with crop yield enhancement. Plant Soil 2019, 439, 553–567. [Google Scholar] [CrossRef]
- Real-Sosa, K.M.; Hernández-Calderón, E.; Flores-Cortez, I.; Valencia-Cantero, E. Bacteria-derived N,N-dimethylhexadecylamine modulates the endophytic microbiome of Medicago truncatula in vitro. Rhizosphere 2022, 21, 100470. [Google Scholar] [CrossRef]
- Li, B.; Li, Y.Y.; Wu, H.M.; Zhang, F.F.; Li, C.J.; Li, X.X.; Lambers, H.; Li, L. Root exudates drive interspecific facilitation by enhancing nodulation and N2 fixation. Proc. Natl. Acad. Sci. USA 2016, 113, 6496–6501. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, A.C.; Gutiérrez, R.T.; Santana, R.C.; Urrutia, A.R.; Fauvart, M.; Michiels, J.; Vanderleyden, J. Effects of co-inoculation of native Rhizobium and Pseudomonas strains on growth parameters and yield of two contrasting Phaseolus vulgaris L. genotypes under Cuban soil conditions. Eur. J. Soil Biol. 2014, 62, 105–112. [Google Scholar] [CrossRef]
- Coniglio, A.; Larama, G.; Molina, R.; Mora, V.; Torres, D.; Marin, A.; Avila, A.I.; Lede NoirCarlan, C.; Erijman, L.; Figuerola, E.L.; et al. Modulation of Maize Rhizosphere Microbiota Composition by Inoculation with Azospirillum argentinense Az39 (Formerly A. brasilense Az39). J. Soil Sci. Plant Nutr. 2022, 22, 3553–3567. [Google Scholar] [CrossRef]
- Ferreira, L.D.V.M.; De Carvalho, F.; Fonseca Colombo Andrade, J.; Padua Oliveira, D.; Vasconcelos De Madeiros, F.H.; De Souza Moreira, F.M. Co-inoculation of selected nodule endophytic rhizobacterial strains with Rhizobium tropici promotes plant growth and controls damping off in common bean. Pedosphere 2020, 30, 98–108. [Google Scholar] [CrossRef]
- Korir, H.; Mungai, N.W.; Thuita, M.; Hamba, Y.; Masso, C. Co-inoculation effect of rhizobia and plant growth promoting rhizobacteria on common bean growth in a low phosphorus soil. Front. Plant Sci. 2017, 8, 141. [Google Scholar] [CrossRef] [PubMed]
- Leite, R.d.A.; Martins, L.C.; Ferreira, L.V.d.S.F.; Barbosa, E.S.; Alves, B.J.R.; Zilli, J.E.; Araújo, A.P.; Jesus, E. da C. Co-inoculation of Rhizobium and Bradyrhizobium promotes growth and yield of common beans. Appl. Soil Ecol. 2022, 172, 104356. [Google Scholar] [CrossRef]
- Hidalgo Rodríguez, E.J.; Ramos Otiniano, C.C.; Lezama Asencio, P.B.; Chuna Mogollón, P.; Chaman Medina, E. Coinoculación de Rhizophagus irregularis y Rhizobium sp. en Phaseolus vulgaris L. var. canario (Fabaceae) “frijol canario”. Arnaldoa 2019, 26, 991–1006. [Google Scholar]
- Renaut, S.; Daoud, R.; Masse, J.; Vialle, A.; Hijri, M. Inoculation with Rhizophagus irregularis does not alter arbuscular mycorrhizal fungal community structure within the roots of corn, wheat, and soybean crops. Microorganisms 2020, 8, 83. [Google Scholar] [CrossRef]
- Chen, Q.; Deng, X.; Elzenga, J.T.M.; van Elsas, J.D. Effect of soil bacteriomes on mycorrhizal colonization by Rhizophagus irregularis—Interactive effects on maize (Zea mays L.) growth under salt stress. Biol. Fertil. Soils 2022, 58, 515–525. [Google Scholar] [CrossRef]
- Romero-Munar, A.; Aroca, R.; Zamarreño, A.M.; García-Mina, J.M.; Perez-Hernández, N.; Ruiz-Lozano, J.M. Dual Inoculation with Rhizophagus irregularis and Bacillus megaterium Improves Maize Tolerance to Combined Drought and High Temperature Stress by Enhancing Root Hydraulics, Photosynthesis and Hormonal Responses. Int. J. Mol. Sci. 2023, 24, 5193. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Li, T.; Shen, M.; Yang, Z.L.; Zhao, Z.W. Evidence for a Dark Septate Endophyte (Exophiala Pisciphila, H93) Enhancing Phosphorus Absorption by Maize Seedlings. Plant Soil 2020, 452, 249–266. [Google Scholar] [CrossRef]
- Lorenzo, L.E.; Messuti, M.I. Glyphium elatum (Ascomycota) in Patagonia (Argentina). Bol. Soc. Argent. Bot. 2005, 40, 181–184. [Google Scholar]
- Réblová, M.; Seifert, K.A.; White, G.P. Chaetosphaeria tortuosa, the newly discovered teleomorph of Menispora tortuosa, with a key to known Menispora species. Mycol. Res. 2006, 110, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, M.; Münsterkötter, M.; Güldener, U.; Bruggmann, R.; Duò, A.; Hainaut, M.; Henrissat, B.; Sieber, C.M.K.; Hoffmeister, D.; Grünig, C.R. Globally distributed root endophyte Phialocephala subalpina links pathogenic and saprophytic lifestyles. BMC Genomics 2016, 17, 1015. [Google Scholar] [CrossRef]
- Estrada-de los Santos, P.; Rojas-Rojas, F.U.; Tapia-García, E.Y.; Vásquez-Murrieta, M.S.; Hirsch, A.M. To split or not to split: An opinion on dividing the genus Burkholderia. Ann. Microbiol. 2016, 66, 1303–1314. [Google Scholar] [CrossRef]
- Chauviat, A.; Meyer, T.; Favre-Bonté, S. Versatility of Stenotrophomonas maltophilia: Ecological roles of RND efflux pumps. Heliyon 2023, 9, e14639. [Google Scholar] [CrossRef]
- dos Santos, I.B.; Pereira, A.P.d.A.; de Souza, A.J.; Cardoso, E.J.B.N.; da Silva, F.G.; Oliveira, J.T.C.; Verdi, M.C.Q.; Sobral, J.K. Selection and Characterization of Burkholderia spp. for Their Plant-Growth Promoting Effects and Influence on Maize Seed Germination. Front. Soil Sci. 2022, 1, 805094. [Google Scholar] [CrossRef]
- Chen, L.; Hao, Z.; Li, K.; Sha, Y.; Wang, E.; Sui, X.; Mi, G.; Tian, C.; Chen, W. Effectsof growth-promoting rhizobacteria on maize growth and rhizosphere microbial community under conservation tillage in Northeast China. Microb. Biotechnol. 2021, 14, 535–550. [Google Scholar] [CrossRef]
- Costa-Gutierrez, S.B.; Adler, C.; Espinosa-Urgel, M.; de Cristóbal, R.E. Pseudomonas putida and its close relatives: Mixing and mastering the perfect tune for plants. Appl. Microbiol. Biotechnol. 2022, 106, 3351–3367. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Urón, P.; Glick, B.R.; Giachini, A.; Rossi, M.J. Genomic Analysis of the 1-Aminocyclopropane-1-Carboxylate Deaminase-Producing Pseudomonas thivervalensis SC5 Reveals Its Multifaceted Roles in Soil and in Beneficial Interactions with Plants. Front. Microbiol. 2021, 12, 752288. [Google Scholar] [CrossRef] [PubMed]
- Jung, B.K.; Ibal, J.C.; Pham, H.Q.; Kim, M.C.; Park, G.S.; Hong, S.J.; Jo, H.W.; Park, C.E.; Choi, S.D.; Jung, Y.; et al. Quorum Sensing System Affects the Plant Growth Promotion Traits of Serratia fonticola GS2. Front. Microbiol. 2020, 11, 536865. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Roseti, D.; Sharma, A.K. The evaluation of microbial diversity in a vegetable based cropping system under organic farming practices. Appl. Soil Ecol. 2007, 36, 116–123. [Google Scholar] [CrossRef]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriag, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Salazar, J.; Suárez, R.; Caballero-Mellado, J.; Iturriaga, G. Trehalose accumulation in Azospirillum brasilense improves drought tolerance and biomass in maize plants. FEMS Microbiol. Lett. 2009, 296, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Orozco, M.d.C.; Barraza, A.; Wong, A.; Suárez, R.; Iturriaga, G. A Rhizobium etli mutant in trehalose-6-phosphate synthase gene is stress sensitive and affects plant growth. In Biology of Plant-Microbe Interactions; International Society for Molecular Plant-Microbe Interactions: St. Paul, MN, USA, 2006; pp. 494–499. [Google Scholar]
- Schulte, C.C.M.; Borah, K.; Wheatley, R.M.; Terpolilli, J.J.; Saalbach, G.; Crang, N.; de Groot, D.H.; Ratcliffe, R.G.; Kruger, N.J.; Papachristodoulou, A.; et al. Metabolic control of nitrogen fixation in rhizobium-legume symbioses. Sci. Adv. 2021, 7, eabh2433. [Google Scholar] [CrossRef] [PubMed]
- Dunn, M.F.; Becerra-Rivera, V.A. The Biosynthesis and Functions of Polyamines in the Interaction of Plant Growth-Promoting Rhizobacteria with Plants. Plants 2023, 12, 2671. [Google Scholar] [CrossRef]
- Hernández-León, R.; Rojas-Solís, D.; Contreras-Pérez, M.; Orozco-Mosqueda, M.d.C.; Macías-Rodríguez, L.I.; Reyes-de la Cruz, H.; Valencia-Cantero, E.; Santoyo, G. Characterization of the antifungal and plant growth-promoting effects of diffusible and volatile organic compounds produced by Pseudomonas fluorescens strains. Biol. Control 2015, 81, 83–92. [Google Scholar] [CrossRef]
- Ortiz, M.; Hernández, J..; Valenzuela, B.; De Los Santo, S.; Del Carmen Rocha, M.; Santoyo, G. Diversity of cultivable endophytic bacteria associated with blueberry plants (Vaccinium corymbosum L.) cv. Biloxi with plant growth-promoting traits. Chil. J. Agric. Anim. Sci. 2018, 34, 140–151. [Google Scholar] [CrossRef]
- Mahuku, G.S. A Simple Extraction Method Suitable for PCR- Based Analysis of Plant, Fungal, and Bacterial DNA. Int. Soc. Plant Mol. Biol. Print. Can. 2004, 22, 71–81. [Google Scholar] [CrossRef]
- Cabanás, C.G.L.; Fernández-González, A.J.; Cardoni, M.; Valverde-Corredor, A.; López-Cepero, J.; Fernández-López, M.; Mercado-Blanco, J. The banana root endophytome: Differences between mother plants and suckers and evaluation of selected bacteria to control fusarium oxysporum f.sp. cubense. J. Fungi 2021, 7, 194. [Google Scholar] [CrossRef] [PubMed]
- Jing, G.; Sun, Z.; Wang, H.; Gong, Y.; Huang, S.; Ning, K.; Xu, J.; Su, X. Parallel-META 3: Comprehensive taxonomical and functional analysis platform for efficient comparison of microbial communities. Sci. Rep. 2017, 7, 40371. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.S.; Kirkegaard, R.H.; Karst, S.M.; Albertsen, M. ampvis2: An R package to analyse and visualise 16S rRNA amplicon data. bioRxiv 2018, 10–11. [Google Scholar] [CrossRef]
- Dixon, P. Computer program review VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Prieto-Barajas, C.M.; Alcaraz, L.D.; Valencia-Cantero, E.; Santoyo, G. Life in Hot Spring Microbial Mats Located in the Trans-Mexican Volcanic Belt: A 16S/18S rRNA Gene and Metagenomic Analysis. Geomicrobiol. J. 2018, 35, 704–712. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojas-Sánchez, B.; Castelán-Sánchez, H.; Garfias-Zamora, E.Y.; Santoyo, G. Diversity of the Maize Root Endosphere and Rhizosphere Microbiomes Modulated by the Inoculation with Pseudomonas fluorescens UM270 in a Milpa System. Plants 2024, 13, 954. https://doi.org/10.3390/plants13070954
Rojas-Sánchez B, Castelán-Sánchez H, Garfias-Zamora EY, Santoyo G. Diversity of the Maize Root Endosphere and Rhizosphere Microbiomes Modulated by the Inoculation with Pseudomonas fluorescens UM270 in a Milpa System. Plants. 2024; 13(7):954. https://doi.org/10.3390/plants13070954
Chicago/Turabian StyleRojas-Sánchez, Blanca, Hugo Castelán-Sánchez, Esmeralda Y. Garfias-Zamora, and Gustavo Santoyo. 2024. "Diversity of the Maize Root Endosphere and Rhizosphere Microbiomes Modulated by the Inoculation with Pseudomonas fluorescens UM270 in a Milpa System" Plants 13, no. 7: 954. https://doi.org/10.3390/plants13070954
APA StyleRojas-Sánchez, B., Castelán-Sánchez, H., Garfias-Zamora, E. Y., & Santoyo, G. (2024). Diversity of the Maize Root Endosphere and Rhizosphere Microbiomes Modulated by the Inoculation with Pseudomonas fluorescens UM270 in a Milpa System. Plants, 13(7), 954. https://doi.org/10.3390/plants13070954







