Molecular Phylogenetics and the Evolution of Morphological Complexity in Aytoniaceae (Marchantiophyta)
Abstract
1. Introduction
2. Results
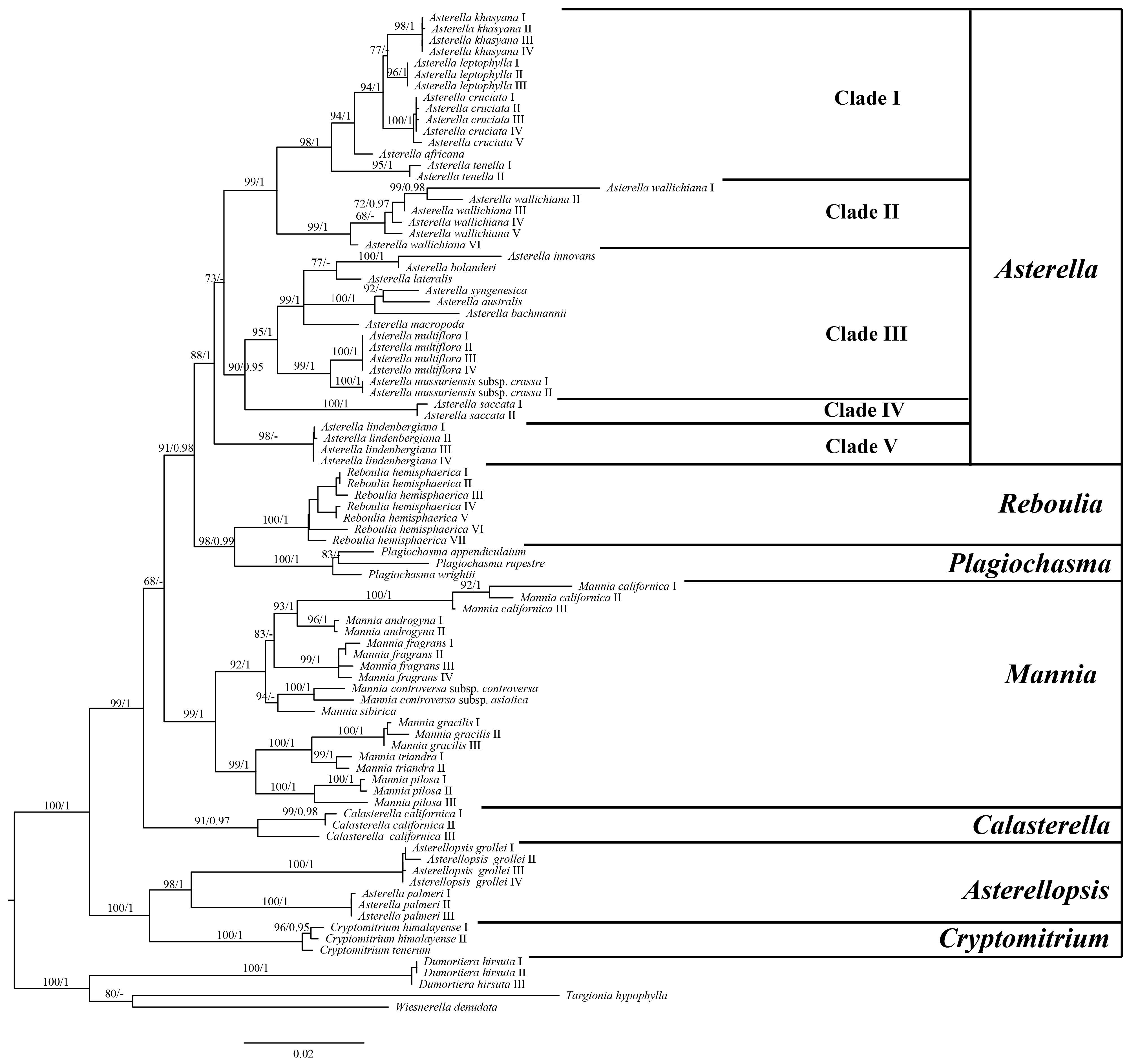
2.1. Phylogenetic Analyses
2.2. P-Distances
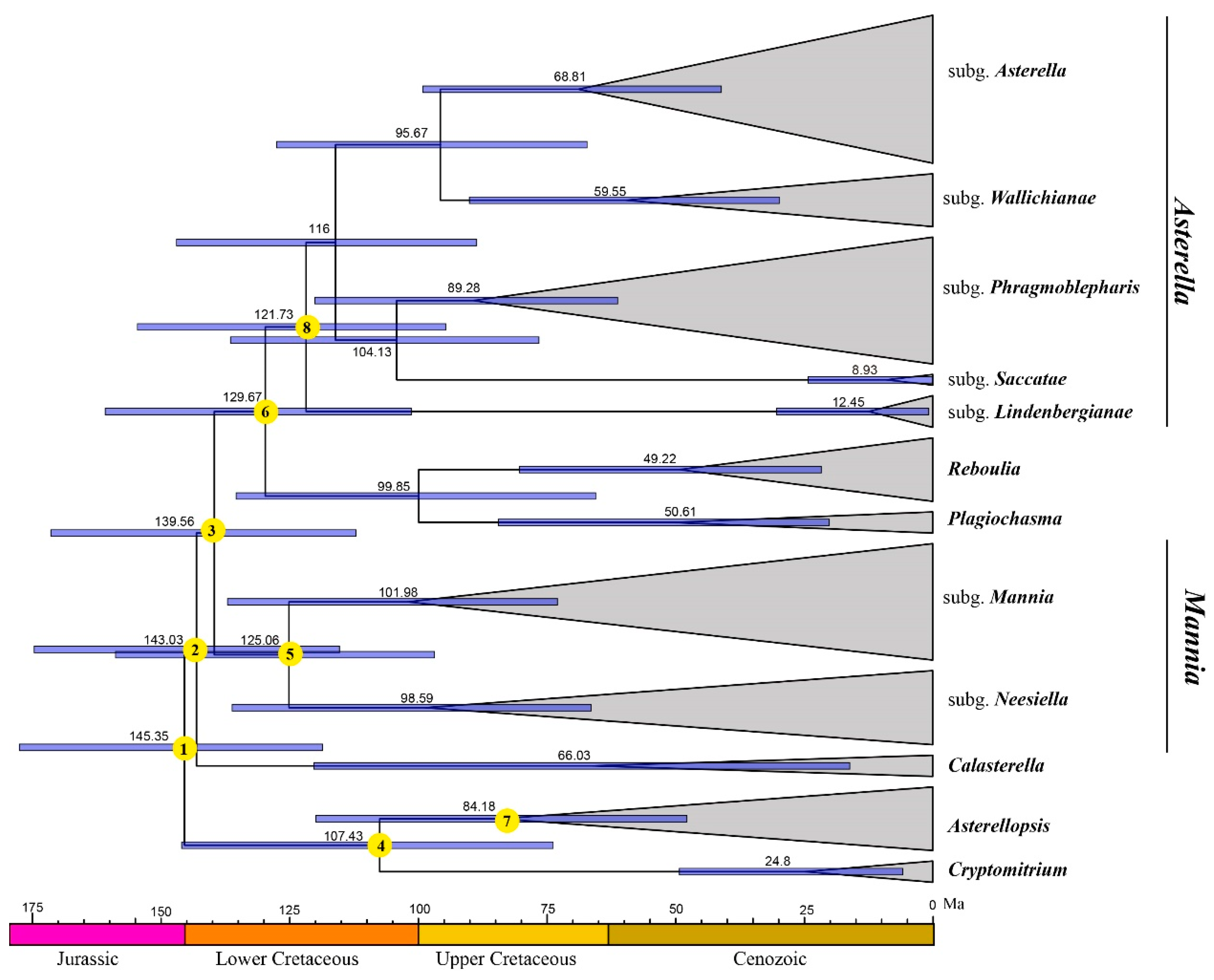
2.3. Divergence Time Estimations
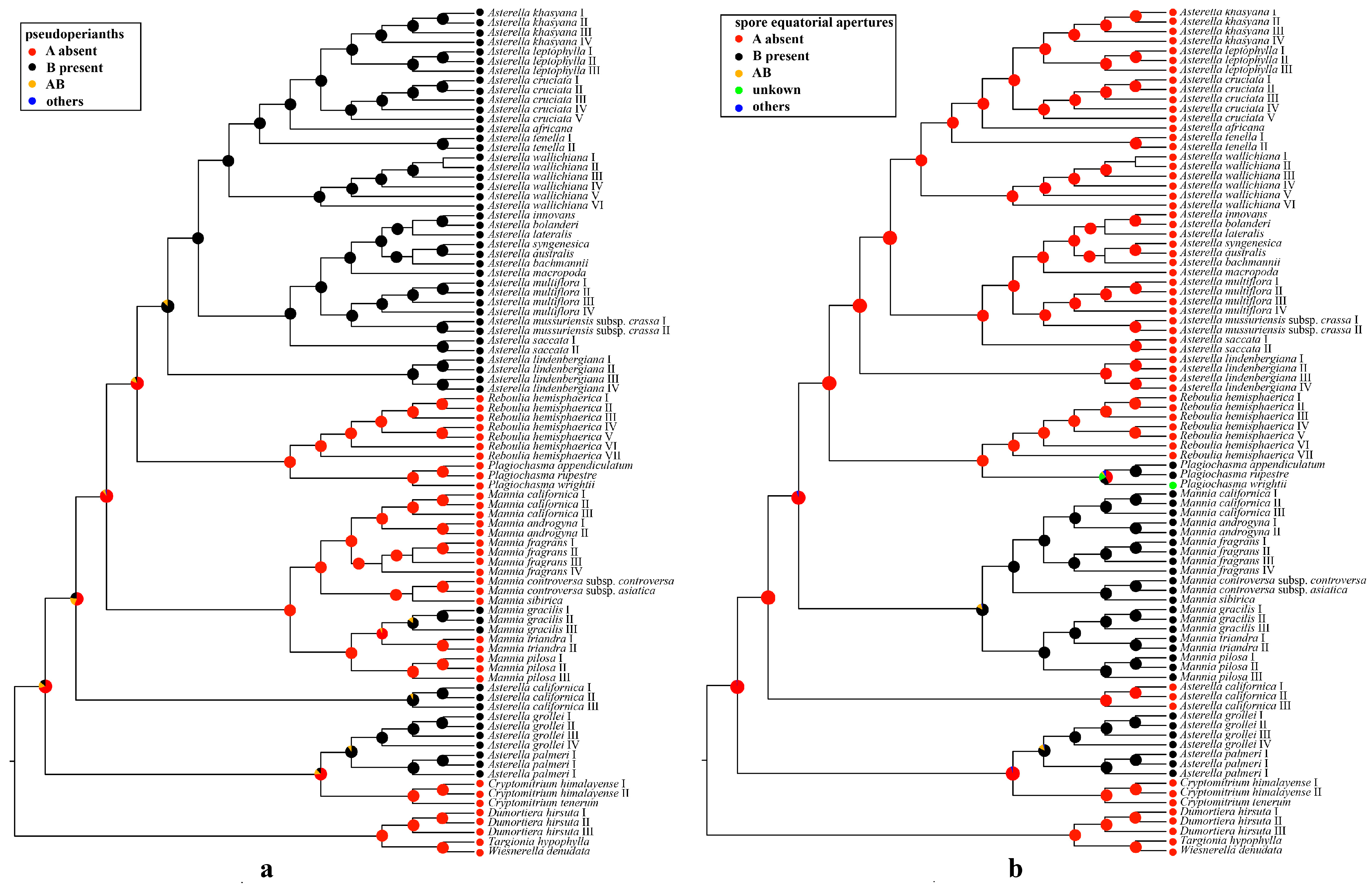
2.4. Ancestral State Reconstructions of Complex Traits
3. Discussion
3.1. Phylogenetic Relationships within Aytoniaceae
3.2. Evolution of the Pseudoperianth, Spore Equatorial Apertures, and Sexual Conditions in Aytoniaceae
3.3. Taxonomy
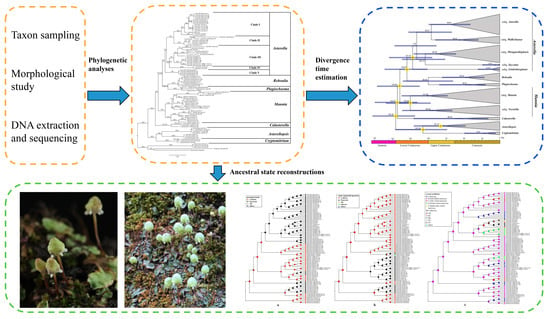
4. Materials and Methods
4.1. Taxon Sampling
4.2. Morphological Study
4.3. DNA Extraction and Sequencing
4.4. Phylogenetic Analyses
4.5. P-Distances
4.6. Divergence Time Estimation
4.7. Ancestral State Reconstructions of Complex Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xiang, Y.L.; Jin, X.J.; Shen, C.; Cheng, X.F.; Shu, L.; Zhu, R.L. New insights into the phylogeny of the complex thalloid liverworts (Marchantiopsida) based on chloroplast genomes. Cladistics 2022, 38, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, J.C.; Crandall-Stotler, B.J.; Hart, M.L.; Long, D.G.; Forrest, L.L. Divergence times and the evolution of morphological complexity in an early land plant lineage (Marchantiopsida) with a slow molecular rate. New Phytol. 2016, 209, 1734–1746. [Google Scholar] [CrossRef] [PubMed]
- Long, D.G.; Zheng, T.X. A new subfamily Calasterelloideae and new genus Calasterella for a phylogenetically and morphologically distinct member of the Aytoniaceae. Phytotaxa 2023, 606, 225–230. [Google Scholar] [CrossRef]
- Borovichev, E.A.; Bakalin, V.A.; Vilnet, A.A. Revision of the Russian Marchantiales. II. A review of the genus Asterella P. Beauv. Arctoa 2015, 24, 294–313. [Google Scholar] [CrossRef]
- Long, D.G.; Moelller, M.; Preston, J. Phylogenetic relationships of Asterella (Aytoniaceae, Marchantiopsida) inferred from chloroplast DNA sequences. Bryologist 2000, 103, 625–644. [Google Scholar] [CrossRef]
- Underwood, L.M. Notes on our Hepaticae III. The distribution of North American Marchantiaceae. Bot. Gaz. 1895, 20, 59–71. [Google Scholar] [CrossRef]
- Long, D.G. Studies on the genus Asterella (Aytoniaceae). VI. Infrageneric classification in Asterella. J. Hattori Bot. Lab. 2005, 97, 249–261. [Google Scholar]
- Long, D.G. Revision of the genus Asterella P. Beauv. in Eurasia. Bryophyt. Biblioth. 2006, 63, 1–299. [Google Scholar]
- Söderström, L.; Hagborg, A.; von Konrat, M.; Bartholomew-Began, S.; Bell, D.; Briscoe, L.; Brown, E.; Cargill, D.C.; Cooper, E.D.; Costa, D.P.; et al. World checklist of hornworts and liverworts. PhytoKeys 2016, 59, 1–828. [Google Scholar]
- Forrest, L.L.; Davis, E.C.; Long, D.G.; Crandall-Stotler, B.J.; Hollingsworth, M.L.; Clark, A. Unravelling the evolutionary history of the liverworts (Marchantiophyta)–multiple taxa, genomes and analyses. Bryologist 2006, 109, 303–334. [Google Scholar] [CrossRef]
- He-Nygrén, X.; Juslén, A.; Ahonen, I.; Glenny, D.; Piippo, S. Illuminating the evolutionary history of liverworts (Marchantiophyta)—Towards a natural classification. Cladistics 2006, 22, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Grolle, R. Verzeichnis der Lebermoose Europas und benachbarter Gebiete. Feddes Repert. 1976, 87, 171–279. [Google Scholar] [CrossRef]
- Schill, D.B. Taxonomy and Phylogeny of the Liverwort Genus Mannia (Aytoniaceae, Marchantiales). Doctoral Thesis, The University of Edinburgh, Royal Botanic Garden Edinburgh, Edinburgh, UK, 2006; pp. 1–261. [Google Scholar]
- Hedenäs, L.; Bisang, I. The overlooked dwarf males in mosses–Unique among green land plants. Perspect. Plant Ecol. Evol. Syst. 2011, 13, 121–135. [Google Scholar] [CrossRef]
- Ando, H. Evolution of bryophytes in relation to their sexuality. Proc. Bryol. Soc. Jpn. 1980, 9, 129–130. [Google Scholar]
- Stotler, R.E.; Crandall-Stotler, B. A Synopsis of the Liverwort Flora of North America North of Mexico. Ann. Mo. Bot. Gard. 2017, 102, 574–709. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.L.; Ma, X.Y.; Shen, C.; Lu, S.H.; Huang, W.Z.; Tian, G.Q.; Zhu, R.L. The Macaronesian liverwort Riccia boumanii Dirkse, Losada & M.Stech (Marchantiophyta: Ricciaceae) confirmed new to Asia by morphological and molecular evidence. Cryptog. Bryol. 2022, 43, 201–209. [Google Scholar]
- Xiang, Y.L.; Zhu, R.L. Morphological and molecular evidence for a new species, Mannia gradsteinii sp. nov. (Aytoniaceae) from southwestern China. Cryptog. Bryol. 2023, 44, 237–245. [Google Scholar] [CrossRef]
- Xiang, Y.L.; Zhu, R.L. Sphaerocarpales (Marchantiophyta) new to China, with special references to a new species of Sphaerocarpos from Hengduan Mountains. Bryologist 2019, 122, 586–596. [Google Scholar] [CrossRef]
- Müller, J.; Müller, K.; Neinhuis, C.; Quandt, D. PhyDE: Phylogenetic Data Editor v0.9971. 2010. Available online: www.phyde.de (accessed on 23 November 2010).
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Meth. 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Flores, J.R.; Bippus, A.C.; de Ullivarri, C.F.; Suárez, G.M.; Hyvönen, J.; Tomescu, A.M.F. Dating the evolution of the complex thalloid liverworts (Marchantiopsida): Total-evidence dating analysis supports a Late Silurian-Early Devonian origin and post-Mesozoic morphological stasis. New Phytol. 2023, 240, 2137–2150. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Schuster, R.M. The Hepaticae and Anthocerotae of North America. VI; Columbia University Press: New York, NY, USA, 1992; pp. 1–937. [Google Scholar]
- Yu, Y.; Harris, A.J.; Blair, C.; He, X.J. RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenet. Evol. 2015, 87, 46–49. [Google Scholar] [CrossRef]


| No. and Taxon | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| 1. Asterella | – | – | – | – | – | – | – | – |
| 2. Asterella palmeri | 3.5/9.8/3.5 | – | – | – | – | – | – | – |
| 3. Asterellopsis | 3.4/10.6/3.7 | 2.1/8.4/3.2 | – | – | – | – | – | – |
| 4. Calasterella | 3.3/5.6/4.2 | 2.6/5.4/4.4 | 2.4/7.0/4.1 | – | – | – | – | – |
| 5. Cryptomitrium | 3.5/8.9/4.0 | 3.0/7.0/2.4 | 2.7/10.3/3.4 | 3.1/6.2/4.0 | – | – | – | – |
| 6. Mannia | 2.6/7.2/3.6 | 3.2/9.0/3.7 | 3.1/10.5/4.3 | 2.8/5.0/4.0 | 3.1/8.9/3.9 | – | – | – |
| 7. Plagiochasma | 2.4/7.9/3.9 | 3.6/9.9/4.0 | 3.1/11.3/4.3 | 3.1/6.0/3.9 | 3.4/7.9/3.4 | 2.5/7.5/3.3 | – | – |
| 8. Reboulia | 2.0/7.2/2.6 | 3.1/10.3/4.4 | 3.1/11.4/4.0 | 2.8/4.8/3.4 | 3.3/9.1/3.2 | 2.2/7.0/3.5 | 1.5/6.0/2.3 | – |
| Nodes | Taxon | Mean Age (Ma) | 95% HPD Intervals (Ma) |
|---|---|---|---|
| 1 | Aytoniaceae | 145.35 | 118.52–177.46 |
| 2 | - | 143.03 | 115.26–174.55 |
| 3 | - | 139.56 | 112.05–171.32 |
| 4 | - | 107.43 | 73.71–145.92 |
| 5 | Mannia | 125.06 | 96.77–158.76 |
| 6 | - | 129.67 | 101.27–160.76 |
| 7 | Asterellopsis | 84.18 | 47.75–119.83 |
| 8 | Asterella | 121.73 | 94.63–154.49 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, Y.-L.; Shen, C.; Ma, W.-Z.; Zhu, R.-L. Molecular Phylogenetics and the Evolution of Morphological Complexity in Aytoniaceae (Marchantiophyta). Plants 2024, 13, 1053. https://doi.org/10.3390/plants13081053
Xiang Y-L, Shen C, Ma W-Z, Zhu R-L. Molecular Phylogenetics and the Evolution of Morphological Complexity in Aytoniaceae (Marchantiophyta). Plants. 2024; 13(8):1053. https://doi.org/10.3390/plants13081053
Chicago/Turabian StyleXiang, You-Liang, Chao Shen, Wen-Zhang Ma, and Rui-Liang Zhu. 2024. "Molecular Phylogenetics and the Evolution of Morphological Complexity in Aytoniaceae (Marchantiophyta)" Plants 13, no. 8: 1053. https://doi.org/10.3390/plants13081053
APA StyleXiang, Y.-L., Shen, C., Ma, W.-Z., & Zhu, R.-L. (2024). Molecular Phylogenetics and the Evolution of Morphological Complexity in Aytoniaceae (Marchantiophyta). Plants, 13(8), 1053. https://doi.org/10.3390/plants13081053