Channels of Evolution: Unveiling Evolutionary Patterns in Diatom Ca2+ Signalling
Abstract
1. Introduction
2. Results and Discussion
2.1. Four-Domain Voltage-Dependent Ca2+ Channel (Cav)
2.2. EukCats
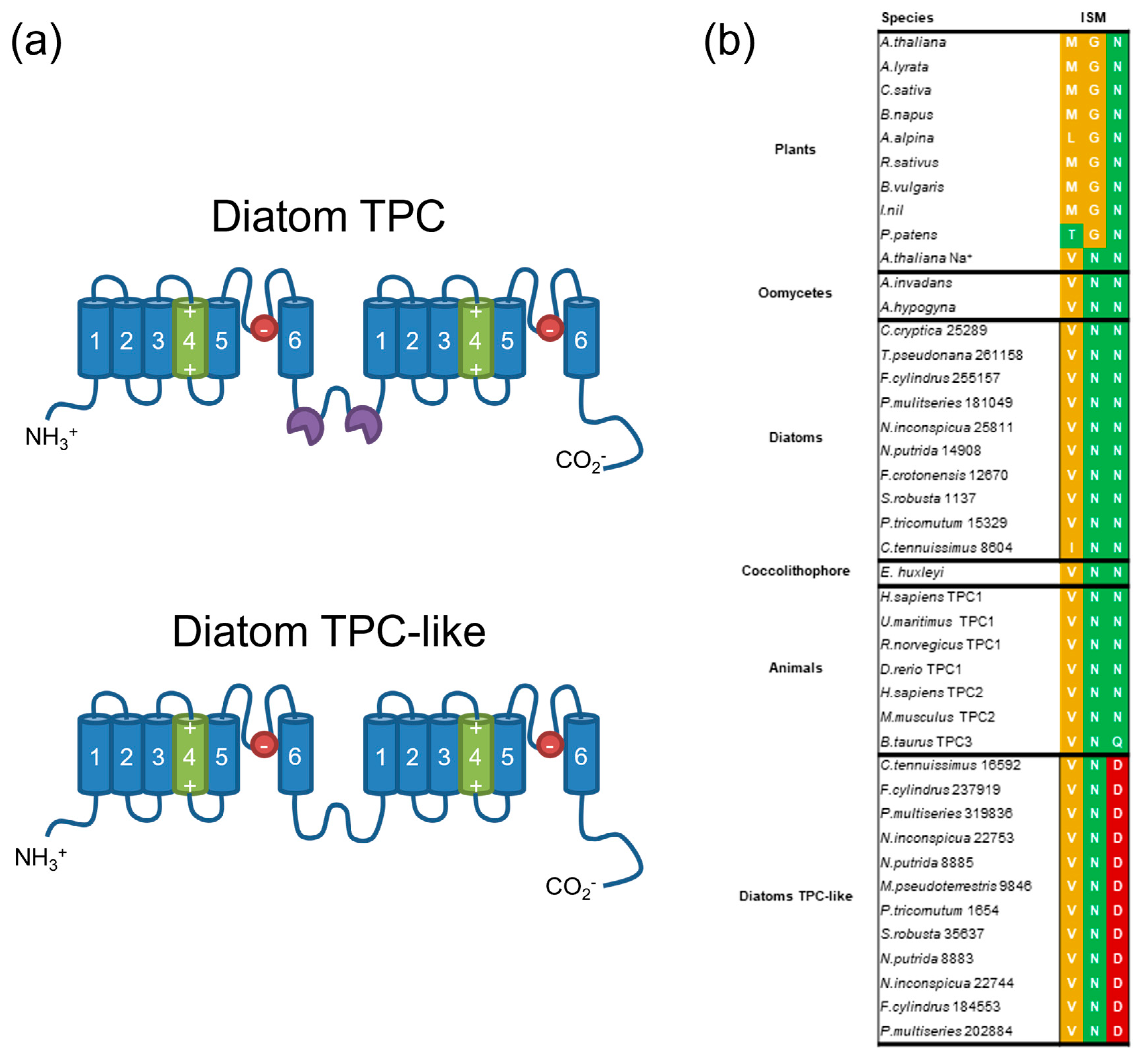
2.3. Two-Pore Channel (TPC)
2.4. Ionotropic Glutamate Receptor-like Channel (GLR)
2.5. Pentameric Ligand-Gated Ion Channel (pLGIC)
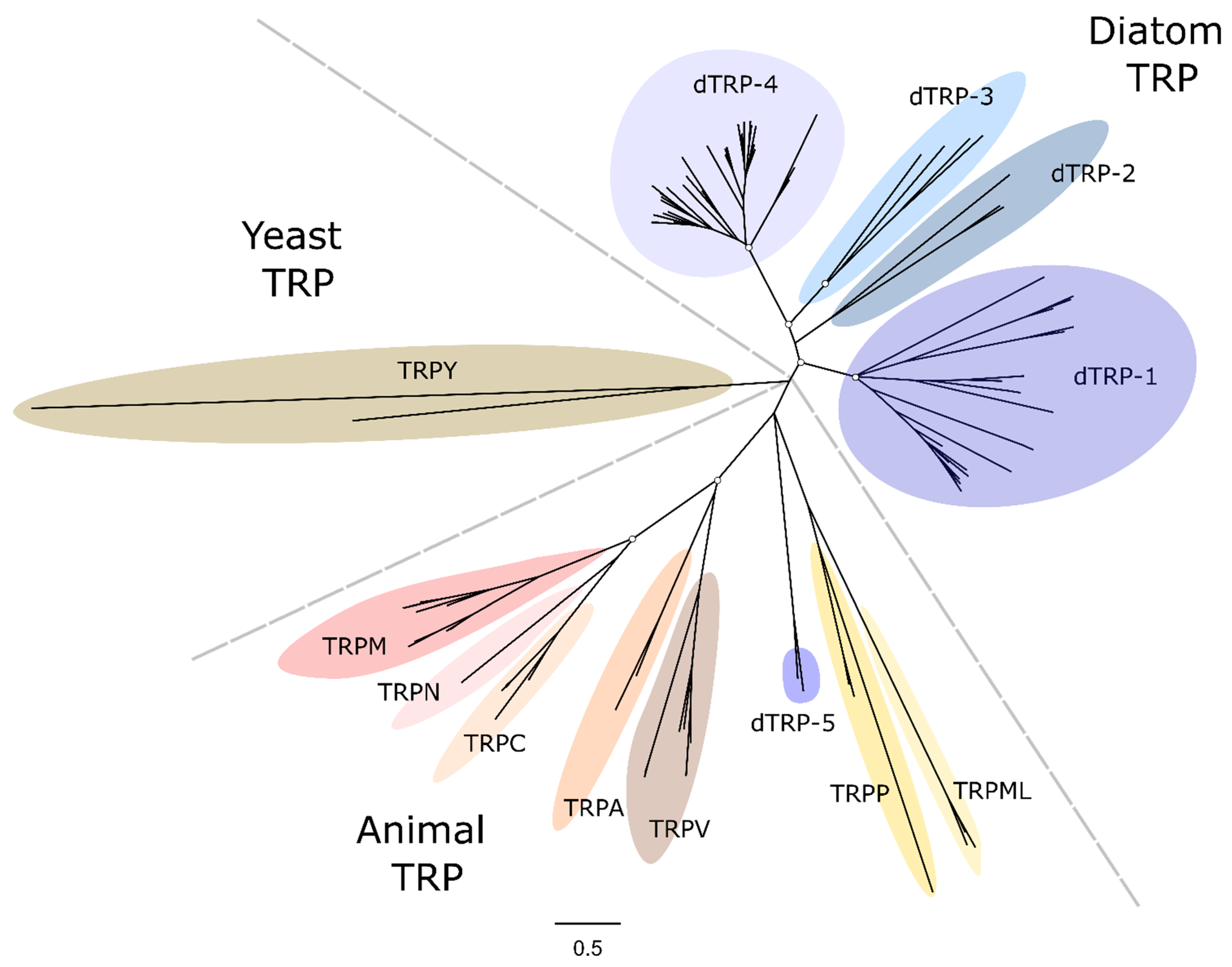
2.6. Transient Receptor Potential Channel (TRP)
2.7. Mechanosensitive Channel Small Conductance (MscS)
2.8. Reduced Hyperosmolarity-Induced [Ca2+] Increase Channel (OSCA)
2.9. P2X Purinoreceptor (P2XR)
2.10. Mitochondrial Uniporter (MCU)
2.11. Inositol 1,4,5-Triphosphate Receptor (IP3R)
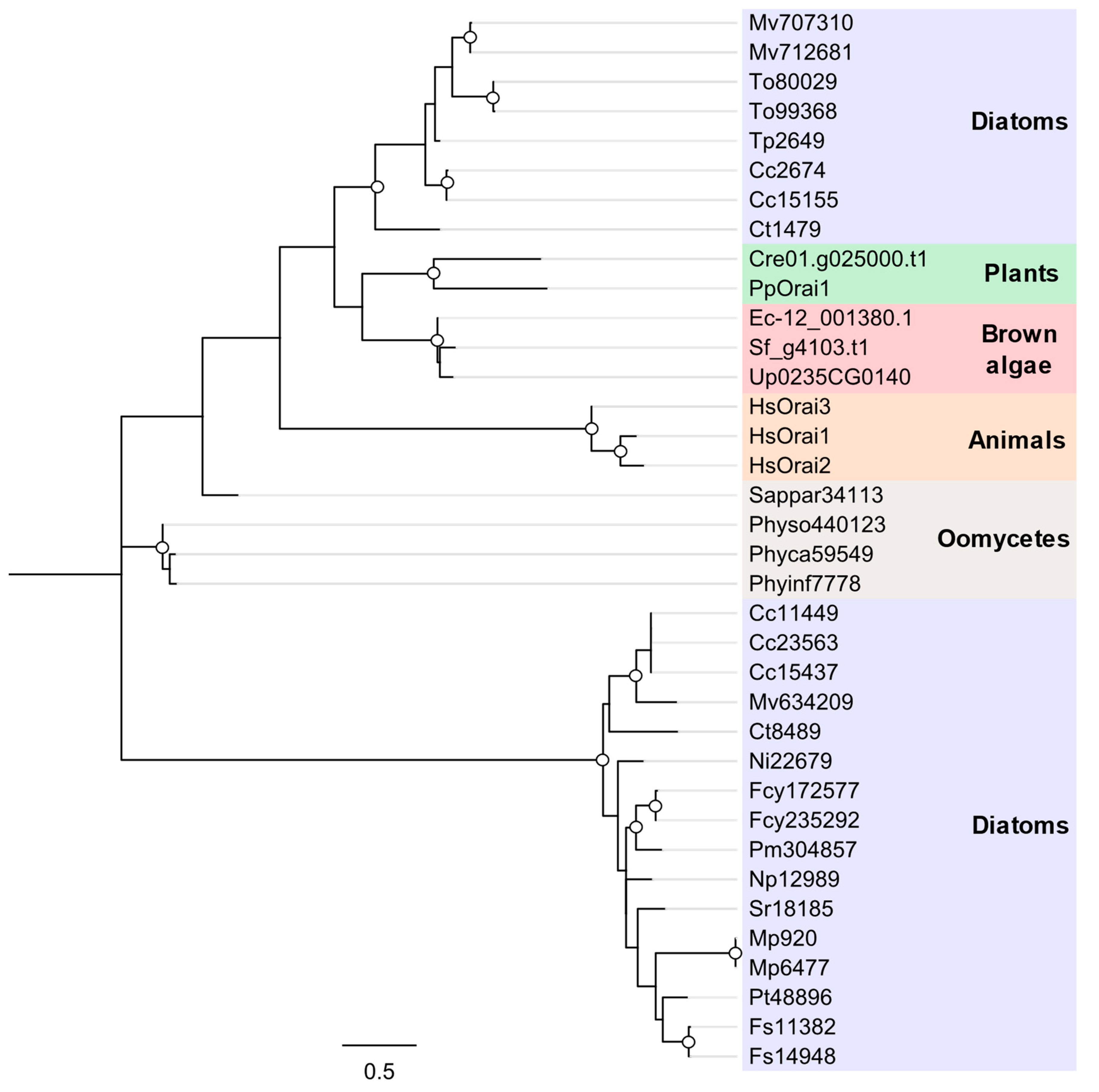
2.12. Ca2+Release-Activated Ca2+ Channel (Orai)
3. Materials and Methods
3.1. Bioinformatic Searches for Ca2+ Channels
3.2. Phylogenetic Analyses
3.3. Localisation of TPCL and OSCA1 in P. tricornutum
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cavalier-Smith, T. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae). Philos. Trans. R. Soc. B Biol. Sci. 2003, 358, 109–134. [Google Scholar] [CrossRef] [PubMed]
- Dorrell, R.G.; Gile, G.; McCallum, G.; Méheust, R.; Bapteste, E.P.; Klinger, C.M.; Brillet-Guéguen, L.; Freeman, K.D.; Richter, D.J.; Bowler, C. Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome. eLife 2017, 6, e23717. [Google Scholar] [CrossRef] [PubMed]
- Malviya, S.; Scalco, E.; Audic, S.; Vincent, F.; Veluchamy, A.; Poulain, J.; Wincker, P.; Iudicone, D.; de Vargas, C.; Bittner, L.; et al. Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. USA 2016, 113, E1516–E1525. [Google Scholar] [CrossRef] [PubMed]
- Benoiston, A.-S.; Ibarbalz, F.M.; Bittner, L.; Guidi, L.; Jahn, O.; Dutkiewicz, S.; Bowler, C. The evolution of diatoms and their biogeochemical functions. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160397. [Google Scholar] [CrossRef]
- Nakov, T.; Beaulieu, J.M.; Alverson, A.J. Accelerated diversification is related to life history and locomotion in a hyperdiverse lineage of microbial eukaryotes (Diatoms, Bacillariophyta). New Phytol. 2018, 219, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Rothpletz, A. Ueber die Flysch-Fucoiden und einige andere fossile Algen, sowie über liasische, Diatomeen führende Hornschwämme. Z. Dtsch. Geol. Ges. 1896, 48, 854–914. [Google Scholar]
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003, 4, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef] [PubMed]
- Braam, J.; Davis, R.W. Rain-, Wind-, and Touch-Induced Expression of Calmodulin and Calmodulin-Related Genes in Arabidopsis. Cell 1990, 60, 357–364. [Google Scholar] [CrossRef]
- Macrobbie, E.A.C. Signal Transduction and Ion Channels in Guard Cells. Trans. R. Soc. Lond. B 1998, 353, 1475–1488. [Google Scholar] [CrossRef]
- Knight, H.; Trewavas, A.J.; Knighta, M.R. Cold Calcium Signaling in Arabidopsis Lnvolves Two Cellular Pools and a Change in Calcium Signature after Acclimation. Plant Cell 1996, 8, 489–503. [Google Scholar] [PubMed]
- Falciatore, A.; D’Alcalà, M.R.; Croot, P.; Bowler, C. Perception of environmental signals by a marine diatom. Science 2000, 288, 2363–2366. [Google Scholar] [CrossRef] [PubMed]
- Vardi, A.; Formiggini, F.; Casotti, R.; De Martino, A.; Ribalet, F.; Miralto, A.; Bowler, C. A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol. 2006, 4, e60. [Google Scholar] [CrossRef] [PubMed]
- Rocha, P.R.F.; Silva, A.D.; Godinho, L.; Dane, W.; Estrela, P.; Vandamme, L.K.J.; Pereira-Leal, J.B.; de Leeuw, D.M.; Leite, R.B. Collective electrical oscillations of a diatom population induced by dark stress. Sci. Rep. 2018, 8, 5484. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, K.E.; Harrison, E.L.; Christie-Oleza, J.A.; Rees, A.P.; Kleiner, F.H.; Gaikwad, T.; Downe, J.; Aguilo-Ferretjans, M.M.; Al-Moosawi, L.; Brownlee, C.; et al. A Novel Ca2+ Signaling Pathway Coordinates Environmental Phosphorus Sensing and Nitrogen Metabolism in Marine Diatoms. Curr. Biol. 2021, 31, 978–989.e4. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, F.H.; Helliwell, K.E.; Chrachri, A.; Hopes, A.; Parry-Wilson, H.; Gaikwad, T.; Mieszkowska, N.; Mock, T.; Wheeler, G.L.; Brownlee, C. Cold-induced [Ca2+]cyt elevations function to support osmoregulation in marine diatoms. Plant Physiol. 2022, 190, 1384–1399. [Google Scholar] [CrossRef] [PubMed]
- Edel, K.H.; Kudla, J. Increasing complexity and versatility: How the calcium signaling toolkit was shaped during plant land colonization. Cell Calcium 2015, 57, 231–246. [Google Scholar] [CrossRef] [PubMed]
- Galione, A.; McDougall, A.; Busa, W.B.; Willmott, N.; Gillot, I.; Whitaker, M. Redundant mechanisms of calcium-induced calcium release underlying calcium waves during fertilization of sea urchin eggs. Science 1993, 261, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Webb, A.A.R.; McAinsh, M.R.; Taylor, J.E.; Hetherington, A.M. Calcium Ions as Intracellular Second Messengers in Higher Plants. In Advances in Botanical Research; Callow, J.A., Ed.; Academic Press: Cambridge, MA, USA, 1996; Volume 22, pp. 45–96. [Google Scholar]
- Wheeler, G.L.; Brownlee, C. Ca2+ signalling in plants and green algae—Changing channels. Trends Plant Sci. 2008, 13, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Edel, K.H.; Marchadier, E.; Brownlee, C.; Kudla, J.; Hetherington, A.M. The Evolution of Calcium-Based Signalling in Plants. Curr. Biol. 2017, 27, R667–R679. [Google Scholar] [CrossRef]
- Pozdnyakov, I.; Matantseva, O.; Skarlato, S. Diversity and evolution of four-domain voltage-gated cation channels of eukaryotes and their ancestral functional determinants. Sci. Rep. 2018, 8, 3539. [Google Scholar] [CrossRef] [PubMed]
- Verret, F.; Wheeler, G.; Taylor, A.R.; Farnham, G.; Brownlee, C. Calcium channels in photosynthetic eukaryotes: Implications for evolution of calcium-based signalling. New Phytol. 2010, 187, 23–43. [Google Scholar] [CrossRef]
- Fountain, S.J.; Burnstock, G. An evolutionary history of P2X receptors. Purinergic Signal. 2009, 5, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Hongo, Y.; Kimura, K.; Takaki, Y.; Yoshida, Y.; Baba, S.; Kobayashi, G.; Nagasaki, K.; Hano, T.; Tomaru, Y. The genome of the diatom Chaetoceros tenuissimus carries an ancient integrated fragment of an extant virus. Sci. Rep. 2021, 11, 22877. [Google Scholar] [CrossRef]
- Kamikawa, R.; Mochizuki, T.; Sakamoto, M.; Tanizawa, Y.; Nakayama, T.; Onuma, R.; Cenci, U.; Moog, D.; Speak, S.; Sarkozi, K.; et al. Genome Evolution of a Nonparasitic Secondary Heterotroph, the Diatom Nitzschia Putrida. Sci. Adv. 2022, 8, eabi5075. [Google Scholar] [CrossRef] [PubMed]
- Lommer, M.; Specht, M.; Roy, A.-S.; Kraemer, L.; Andreson, R.; Gutowska, M.A.; Wolf, J.; Bergner, S.V.; Schilhabel, M.B.; Klostermeier, U.C.; et al. Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biol. 2012, 13, R66. [Google Scholar] [CrossRef]
- Mock, T.; Otillar, R.P.; Strauss, J.; McMullan, M.; Paajanen, P.; Schmutz, J.; Salamov, A.; Sanges, R.; Toseland, A.; Ward, B.J.; et al. Evolutionary genomics of the cold-Adapted diatom Fragilariopsis cylindrus. Nature 2017, 541, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Ogura, A.; Akizuki, Y.; Imoda, H.; Mineta, K.; Gojobori, T.; Nagai, S. Comparative genome and transcriptome analysis of diatom, Skeletonema costatum, reveals evolution of genes for harmful algal bloom. BMC Genom. 2018, 19, 765. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; Podell, S.; Pinowska, A.; Traller, J.C.; Smith, S.R.; McClure, R.; Beliaev, A.; Bohutskyi, P.; Hill, E.A.; Rabines, A.; et al. Diploid genomic architecture of Nitzschia inconspicua, an elite biomass production diatom. Sci. Rep. 2021, 11, 15592. [Google Scholar] [CrossRef]
- Osuna-Cruz, C.M.; Bilcke, G.; Vancaester, E.; De Decker, S.; Bones, A.M.; Winge, P.; Poulsen, N.; Bulankova, P.; Verhelst, B.; Audoor, S.; et al. The Seminavis robusta genome provides insights into the evolutionary adaptations of benthic diatoms. Nat. Commun. 2020, 11, 3320. [Google Scholar] [CrossRef]
- Suzuki, S.; Ota, S.; Yamagishi, T.; Tuji, A.; Yamaguchi, H.; Kawachi, M. Rapid transcriptomic and physiological changes in the freshwater pennate diatom Mayamaea pseudoterrestris in response to copper exposure. DNA Res. 2022, 29, dsac037. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Maeda, Y.; Veluchamy, A.; Tanaka, M.; Abida, H.; Maréchal, E.; Bowler, C.; Muto, M.; Sunaga, Y.; Tanaka, M.; et al. Oil accumulation by the oleaginous diatom Fistulifera solaris as revealed by the genome and transcriptome. Plant Cell 2015, 27, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Traller, J.C.; Cokus, S.J.; Lopez, D.A.; Gaidarenko, O.; Smith, S.R.; McCrow, J.P.; Gallaher, S.D.; Podell, S.; Thompson, M.; Cook, O.; et al. Genome and methylome of the oleaginous diatom Cyclotella cryptica reveal genetic flexibility toward a high lipid phenotype. Biotechnol. Biofuels 2016, 9, 258. [Google Scholar] [CrossRef] [PubMed]
- Cronmiller, E.; Toor, D.; Shao, N.C.; Kariyawasam, T.; Wang, M.H.; Lee, J.-H. Cell wall integrity signaling regulates cell wall-related gene expression in Chlamydomonas reinhardtii. Sci. Rep. 2019, 9, 12204. [Google Scholar] [CrossRef] [PubMed]
- Teardo, E.; Carraretto, L.; Wagner, S.; Formentin, E.; Behera, S.; De Bortoli, S.; Larosa, V.; Fuchs, P.; Schiavo, F.L.; Raffaello, A.; et al. Physiological characterization of a plant mitochondrial Calcium uniporter in vitro and in vivo. Plant Physiol. 2017, 173, 1355–1370. [Google Scholar] [CrossRef] [PubMed]
- Arias-Darraz, L.; Cabezas, D.; Colenso, C.K.; Alegría-Arcos, M.; Bravo-Moraga, F.; Varas-Concha, I.; Almonacid, D.E.; Madrid, R.; Brauchi, S. A transient receptor potential ion channel in chlamydomonas shares key features with sensory transduction-associated trp channels in mammals. Plant Cell 2015, 27, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Catterall, W.A. Voltage-gated calcium channels. Cold Spring Harb. Perspect. Biol. 2011, 3, a003947. [Google Scholar] [CrossRef] [PubMed]
- Flavell, S.W.; Greenberg, M.E. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 2008, 31, 563–590. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, T.; Takeshima, H.; Mikami, A.; Flockerzi, V.; Takahashi, H.; Kangawa, K.; Kojima, M.; Matsuo, H.; Hirose, T.; Numa, S. Primary structure of the receptor for calcium channel blockers from skeletal muscle. Nature 1987, 328, 313–318. [Google Scholar] [CrossRef]
- Ertel, E.A.; Campbell, K.P.; Harpold, M.M.; Hofmann, F.; Mori, Y.; Perez-Reyes, E.; Schwartz, A.; Snutch, T.P.; Tanabe, T.; Birnbaumer, L.; et al. Nomenclature of Voltage-Gated Calcium Channels. Neuron 2000, 25, 533–535. [Google Scholar] [CrossRef]
- Brunet, T.; Arendt, D. From damage response to action potentials: Early evolution of neural and contractile modules in stem eukaryotes. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150043. [Google Scholar] [CrossRef] [PubMed]
- Fujiu, K.; Nakayama, Y.; Yanagisawa, A.; Sokabe, M.; Yoshimura, K. Chlamydomonas CAV2 encodes a voltage-dependent calcium channel required for the flagellar waveform conversion. Curr. Biol. 2009, 19, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, K.E.; Chrachri, A.; Koester, J.A.; Wharam, S.; Verret, F.; Taylor, A.R.; Wheeler, G.L.; Brownlee, C. Alternative Mechanisms for Fast Na+/Ca2+ Signaling in Eukaryotes via a Novel Class of Single-Domain Voltage-Gated Channels. Curr. Biol. 2019, 29, 1503–1511.e6. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Navarro, B.; Xu, H.; Yue, L.; Shi, Q.; Clapham, D.E. A Prokaryotic Voltage-Gated Sodium Channel. Science 2001, 294, 2372–2375. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, K.E.; Chrachri, A.; Koester, J.A.; Wharam, S.; Taylor, A.R.; Wheeler, G.L.; Brownlee, C. A novel single-domain Na1-selective voltage-gated channel in photosynthetic eukaryotes. Plant Physiol. 2020, 184, 1674–1683. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.R. A fast Na+/Ca2+-based action potential in a marine diatom. PLoS ONE 2009, 4, e4966. [Google Scholar] [CrossRef] [PubMed]
- Chepurnov, V.A.; Mann, D.G.; Sabbe, K.; Vyverman, W. Experimental Studies on Sexual Reproduction in Diatoms. Int. Rev. Cytol. 2004, 237, 91. [Google Scholar]
- Medina, D.L. Lysosomal calcium and autophagy. In International Review of Cell and Molecular Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2021; Volume 362, pp. 141–170. [Google Scholar]
- Calcraft, P.J.; Ruas, M.; Pan, Z.; Cheng, X.; Arredouani, A.; Hao, X.; Tang, J.; Rietdorf, K.; Teboul, L.; Chuang, K.-T.; et al. NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature 2009, 459, 596–600. [Google Scholar] [CrossRef]
- Castonguay, J.; Orth, J.H.; Müller, T.; Sleman, F.; Grimm, C.; Wahl-Schott, C.; Biel, M.; Mallmann, R.T.; Bildl, W.; Schulte, U.; et al. The two-pore channel TPC1 is required for efficient protein processing through early and recycling endosomes. Sci. Rep. 2017, 7, 10038. [Google Scholar] [CrossRef]
- Furuichi, T.; Cunningham, K.W.; Muto, S. A Putative Two Pore Channel AtTPC1 Mediates Ca2+ Flux in Arabidopsis Leaf Cells. Plant Cell Physiol. 2001, 42, 900–905. [Google Scholar] [CrossRef]
- Hedrich, R.; Neher, E. Cytoplasmic calcium regulates voltage-dependent ion channels in plant vacuoles. Nature 1987, 329, 833–836. [Google Scholar] [CrossRef]
- Peiter, E.; Maathuis, F.J.M.; Mills, L.N.; Knight, H.; Pelloux, J.; Hetherington, A.M.; Sanders, D. The vacuolar Ca2+-activated channel TPC1 regulates germination and stomatal movement. Nature 2005, 434, 404–408. [Google Scholar] [CrossRef]
- Hedrich, R.; Müller, T.D.; Marten, I.; Becker, D. TPC1 vacuole SV channel gains further shape–voltage priming of calcium-dependent gating. Trends Plant Sci. 2023, 28, 673–684. [Google Scholar] [CrossRef] [PubMed]
- She, J.; Guo, J.; Jiang, Y. Structure and Function of Plant and Mammalian TPC Channels. In Endolysosomal Voltage-Dependent Cation Channels; Springer International Publishing: Cham, Switzerland, 2022; pp. 155–180. [Google Scholar] [CrossRef]
- Guo, J.; Zeng, W.; Jiang, Y. Tuning the ion selectivity of two-pore channels. Proc. Natl. Acad. Sci. USA 2017, 114, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Vassileva, K.; Marsh, M.; Patel, S. Two-pore channels as master regulators of membrane trafficking and endocytic well-being. Curr. Opin. Physiol. 2020, 17, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Gerndt, S.; Chen, C.C.; Chao, Y.K.; Yuan, Y.; Burgstaller, S.; Scotto Rosato, A.; Krogsaeter, E.; Urban, N.; Jacob, K.; Nguyen, O.N.P.; et al. Agonist-mediated switching of ion selectivity in TPC2 differentially promotes lysosomal function. eLife 2020, 9, e54712. [Google Scholar] [CrossRef] [PubMed]
- Larisch, N.; Schulze, C.; Galione, A.; Dietrich, P. An N-Terminal Dileucine Motif Directs Two-Pore Channels to the Tonoplast of Plant Cells. Traffic 2012, 13, 1012–1022. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, V.; Dersch, J.; Puzik, K.; Bäcker, O.; Liu, X.; Stork, S.; Schulz, J.; Heimerl, T.; Klingl, A.; Zauner, S.; et al. The Central Vacuole of the Diatom Phaeodactylum tricornutum: Identification of New Vacuolar Membrane Proteins and of a Functional Di-leucine-based Targeting Motif. Protist 2017, 168, 271–282. [Google Scholar] [CrossRef]
- Reiner, A.; Levitz, J. Glutamatergic Signaling in the Central Nervous System: Ionotropic and Metabotropic Receptors in Concert. Neuron 2018, 98, 1080–1098. [Google Scholar] [CrossRef]
- Lisman, J.; Yasuda, R.; Raghavachari, S. Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 2012, 13, 169–182. [Google Scholar] [CrossRef]
- Moroz, L.L.; Nikitin, M.A.; Poličar, P.G.; Kohn, A.B.; Romanova, D.Y. Evolution of glutamatergic signaling and synapses. Neuropharmacology 2021, 199, 108740. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Vicente, D.; Bayés, À. AMPA receptor auxiliary subunits emerged during early vertebrate evolution by neo/subfunctionalization of unrelated proteins. Open Biol. 2020, 10, 200234. [Google Scholar]
- Lam, H.-M.; Chiu, J.; Hsieh, M.-H.; Meisel, L.; Oliveira, I.C.; Shin, M.; Coruzzi, G. Glutamate-receptor genes in plants. Nature 1998, 396, 125–126. [Google Scholar] [CrossRef]
- Wudick, M.M.; Portes, M.T.; Michard, E.; Rosas-Santiago, P.; Lizzio, M.A.; Nunes, C.O.; Campos, C.; Damineli, D.S.C.; Carvalho, J.C.; Lima, P.T.; et al. CORNICHON sorting and regulation of GLR channels underlie pollen tube Ca2+ homeostasis. Science 2018, 360, 533–536. [Google Scholar] [CrossRef]
- Michard, E.; Lima, P.T.; Borges, F.; Silva, A.C.; Portes, M.T.; Carvalho, J.E.; Gilliham, M.; Liu, L.H.; Obermeyer, G.; Feijó, J.A. Glutamate Receptor–like Genes Form Ca2+ Channels in Pollen Tubes and Are Regulated by Pistil d-Serine. Science 2011, 332, 434–437. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Chien, C.T.; Chang, I.F. The Arabidopsis glutamate receptor-like gene GLR3.6 controls root development by repressing the Kip-related protein gene KRP4. J. Exp. Bot. 2016, 67, 1853–1869. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Ju, C.; Parihar, A.; Kim, S.; Cho, D.; Kwak, J.M. Arabidopsis glutamate receptor homolog3.5 modulates cytosolic Ca2+ level to counteract effect of abscisic acid in seed germination. Plant Physiol. 2015, 167, 1630–1642. [Google Scholar] [CrossRef]
- Mousavi, S.A.R.; Chauvin, A.; Pascaud, F.; Kellenberger, S.; Farmer, E.E. Glutamate receptor-like genes mediate leaf-to-leaf wound signalling. Nature 2013, 500, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Wudick, M.M.; Michard, E.; Oliveira Nunes, C.; Feijó, J.A. Comparing plant and animal glutamate receptors: Common traits but different fates? J. Exp. Bot. 2018, 69, 4151–4163. [Google Scholar] [CrossRef]
- Meyerhoff, O.; Müller, K.; Roelfsema, M.R.G.; Latz, A.; Lacombe, B.; Hedrich, R.; Dietrich, P.; Becker, D. AtGLR3.4, a glutamate receptor channel-like gene is sensitive to touch and cold. Planta 2005, 222, 418–427. [Google Scholar] [CrossRef]
- Teardo, E.; Formentin, E.; Segalla, A.; Giacometti, G.M.; Marin, O.; Zanetti, M.; Schiavo, F.L.; Zoratti, M.; Szabò, I. Dual localization of plant glutamate receptor AtGLR3.4 to plastids and plasmamembrane. Biochim. Biophys. Acta (BBA) Bioenerg. 2011, 1807, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Vincill, E.D.; Bieck, A.M.; Spalding, E.P. Ca2+ Conduction by an Amino Acid-Gated Ion Channel Related to Glutamate Receptors. Plant Physiol. 2012, 159, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Teardo, E.; Carraretto, L.; De Bortoli, S.; Costa, A.; Behera, S.; Wagner, R.; Lo Schiavo, F.; Formentin, E.; Szabo, I. Alternative splicing-Mediated targeting of the arabidopsis GLUTAMATE RECEPTOR 3.5 to mitochondria affects organelle morphology. Plant Physiol. 2015, 167, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Vincill, E.D.; Clarin, A.E.; Molenda, J.N.; Spalding, E.P. Interacting glutamate receptor-like proteins in phloem regulate lateral root initiation in Arabidopsis. Plant Cell 2013, 25, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Tapken, D.; Anschütz, U.; Liu, L.-H.; Huelsken, T.; Seebohm, G.; Becker, D.; Hollmann, M. A Plant Homolog of Animal Glutamate Receptors Is an Ion Channel Gated by Multiple Hydrophobic Amino Acids. Sci. Signal. 2013, 6, ra47. [Google Scholar] [CrossRef] [PubMed]
- Corrie, J.B.; Baenziger, J.E. Gating of Pentameric Ligand-Gated Ion Channels: Structural Insights and Ambiguities. Structure 2013, 21, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Tasneem, A.; Iyer, L.M.; Jakobsson, E.; Aravind, L. Open Access Identification of the Prokaryotic Ligand-Gated Ion Channels and Their Implications for the Mechanisms and Origins of Animal Cys-Loop Ion Channels. Genome Biol. 2004, 6, R4. [Google Scholar] [CrossRef]
- Lynagh, T.; Pless, S.A. Principles of agonist recognition in Cys-loop receptors. Front. Physiol. 2014, 5, 160. [Google Scholar] [CrossRef] [PubMed]
- Jaiteh, M.; Taly, A.; Hénin, J. Evolution of pentameric ligand-gated ion channels: Pro-loop receptors. PLoS ONE 2016, 11, e0151934. [Google Scholar] [CrossRef]
- Samanta, A.; Hughes, T.E.T.; Moiseenkova-Bell, V.Y. Transient receptor potential (TRP) channels. In Subcellular Biochemistry; Springer: New York, NY, USA, 2018; Volume 87, pp. 141–165. [Google Scholar]
- Ramsey, I.S.; Delling, M.; Clapham, D.E. An introduction to TRP channels. Annu. Rev. Physiol. 2006, 68, 619–647. [Google Scholar] [CrossRef]
- Nilius, B.; Owsianik, G. The transient receptor potential family of ion channels. Genome Biol. 2011, 12, 218. [Google Scholar] [CrossRef] [PubMed]
- Fujiu, K.; Nakayama, Y.; Iida, H.; Sokabe, M.; Yoshimura, K. Mechanoreception in motile flagella of Chlamydomonas. Nat. Cell Biol. 2011, 13, 630–632. [Google Scholar] [CrossRef] [PubMed]
- Booth, I.R. Bacterial mechanosensitive channels: Progress towards an understanding of their roles in cell physiology. Curr. Opin. Microbiol. 2014, 18, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Wilson, M.E.; Richardson, R.A.; Haswell, E.S. Genetic and physical interactions between the organellar mechanosensitive ion channel homologs MSL1, MSL2, and MSL3 reveal a role for inter-organellar communication in plant development. Plant Direct 2019, 3, e00124. [Google Scholar] [CrossRef] [PubMed]
- Haswell, E.S.; Meyerowitz, E.M. MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr. Biol. 2006, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, Y.; Fujiu, K.; Sokabe, M.; Yoshimura, K. Molecular and electrophysiological characterization of a mechanosensitive channel expressed in the chloroplasts of Chlamydomonas. Proc. Natl. Acad. Sci. USA 2007, 104, 5883–5888. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Murthy, S.E.; Dubin, A.E.; Whitwam, T.; Jojoa-Cruz, S.; Cahalan, S.M.; Mousavi, S.A.R.; Ward, A.B.; Patapoutian, A. OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. eLife 2018, 7, e41844. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Ye, C.; Wu, D.; Zang, Y.-Y.; Zhang, L.; Chen, C.; He, X.-Y.; Yang, J.-J.; Hu, P.; Xu, Z.; et al. The Cation Channel TMEM63B Is an Osmosensor Required for Hearing. Cell Rep. 2020, 31, 107596. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yuan, F.; Wang, X.; Zhu, S.; Pei, Z.-M. Evolution of osmosensing OSCA1 Ca2+ channel family coincident with plant transition from water to land. Plant Genome 2022, 15, e20198. [Google Scholar] [CrossRef]
- Ovide, C.; Kiefer-Meyer, M.C.; Bérard, C.; Vergne, N.; Lecroq, T.; Plasson, C.; Burel, C.; Bernard, S.; Driouich, A.; Lerouge, P.; et al. Comparative in depth RNA sequencing of P. tricornutum’s morphotypes reveals specific features of the oval morphotype. Sci. Rep. 2018, 8, 14340. [Google Scholar] [CrossRef] [PubMed]
- Sabir, J.S.M.; Theriot, E.C.; Manning, S.R.; Al-Malki, A.L.; Khiyami, M.A.; Al-Ghamdi, A.K.; Sabir, M.J.; Romanovicz, D.K.; Hajrah, N.H.; El Omri, A.; et al. Phylogenetic analysis and a review of the history of the accidental phytoplankter, Phaeodactylum tricornutum Bohlin (Bacillariophyta). PLoS ONE 2018, 13, e0196744. [Google Scholar] [CrossRef] [PubMed]
- De Martino, A.; Meichenin, A.; Shi, J.; Pan, K.; Bowler, C. Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions. J. Phycol. 2007, 43, 992–1009. [Google Scholar] [CrossRef]
- Helliwell, K.E.; Kleiner, F.H.; Hardstaff, H.; Chrachri, A.; Gaikwad, T.; Salmon, D.; Smirnoff, N.; Wheeler, G.L.; Brownlee, C. Spatiotemporal patterns of intracellular Ca2+ signalling govern hypo-osmotic stress resilience in marine diatoms. New Phytol. 2021, 230, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Fountain, S.J.; Cao, L.; Young, M.T.; North, R.A. Permeation properties of a P2X receptor in the green algae Ostreococcus tauri. J. Biol. Chem. 2008, 283, 15122–15126. [Google Scholar] [CrossRef] [PubMed]
- Rizzuto, R.; Simpson, A.W.M.; Brini, M.; Pozzan, T. Rapid Changes of Mitochondrial Ca2+ Revealed by Specifically Targeted Recombinant Aequorin. Nature 1992, 358, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Kevin Foskett, J.; Philipson, B. The mitochondrial Ca2+ uniporter complex. J. Mol. Cell. Cardiol. 2015, 78, 3–8. [Google Scholar] [CrossRef] [PubMed]
- De Stefani, D.; Patron, M.; Rizzuto, R. Structure and function of the mitochondrial calcium uniporter complex. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1853, 2006–2011. [Google Scholar] [CrossRef] [PubMed]
- Bick, A.G.; Calvo, S.E.; Mootha, V.K. Evolutionary diversity of the mitochondrial calcium uniporter. Science 2012, 336, 886. [Google Scholar] [CrossRef]
- Pittis, A.A.; Goh, V.; Cebrian-Serrano, A.; Wettmarshausen, J.; Perocchi, F.; Gabaldón, T. Discovery of EMRE in fungi resolves the true evolutionary history of the mitochondrial calcium uniporter. Nat. Commun. 2020, 11, 4031. [Google Scholar] [CrossRef]
- Berridge, M.J. Inositol trisphosphate and calcium signalling mechanisms. Biochim. Biophys. Acta Mol. Cell Res. 2009, 1793, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Prole, D.L.; Taylor, C.W. Structure and function of ip3 receptors. Cold Spring Harb. Perspect. Biol. 2019, 11, a035063. [Google Scholar] [CrossRef]
- Prole, D.L.; Taylor, C.W. Identification of intracellular and plasma membrane calcium channel homologues in pathogenic parasites. PLoS ONE 2011, 6, e26218. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.; Brownlee, C. Calcium influx, fertilisation potential and egg activation in Fucus serratus. Zygote 1995, 3, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Prakriya, M.; Feske, S.; Gwack, Y.; Srikanth, S.; Rao, A.; Hogan, P.G. Orai1 is an essential pore subunit of the CRAC channel. Nature 2006, 443, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Yeromin, A.V.; Zhang, S.L.; Jiang, W.; Yu, Y.; Safrina, O.; Cahalan, M.D. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature 2006, 443, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.R.; Meyer, T. Evolutionary origins of STIM1 and STIM2 within ancient Ca2+ signaling systems. Trends Cell Biol. 2011, 21, 202–211. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, J.; Sun, Y.; Li, S.; Gao, Y.I.; Yu, Y.; Liu, C.; Wang, Q.; Lv, X.; Zhang, X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef] [PubMed]
- Gemmell, B.J.; Oh, G.; Buskey, E.J.; Villareal, T.A. Dynamic sinking behaviour in marine phytoplankton: Rapid changes in buoyancy may aid in nutrient uptake. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161126. [Google Scholar] [CrossRef]
- Armbrust, E.V.; Berges, J.A.; Bowler, C.; Green, B.R.; Martinez, D.; Putnam, N.H.; Zhou, S.; Allen, A.E.; Apt, K.E.; Bechner, M.; et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 2004, 306, 79–86. [Google Scholar] [CrossRef]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef]
- Chen, X.; Wang, N.; Liu, J.W.; Zeng, B.; Chen, G.L. TMEM63 mechanosensitive ion channels: Activation mechanisms, biological functions and human genetic disorders. Biochem. Biophys. Res. Commun. 2023, 683, 149111. [Google Scholar] [CrossRef]
- Cock, J.M.; Sterck, L.; Rouze, P.; Scornet, D.; Allen, A.E.; Amoutzias, G.; Anthouard, V.; Artiguenave, F.; Aury, J.M.; Badger, J.H.; et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 2010, 465, 617–621. [Google Scholar] [CrossRef]
- Jones, A.K.; Sattelle, D.B. The cys-loop ligand-gated ion channel gene superfamily of the nematode, Caenorhabditis elegans. Invert. Neurosci. 2008, 8, 41–47. [Google Scholar] [CrossRef]
- Maksaev, G.; Haswell, E.S. MscS-Like10 is a stretch-activated ion channel from Arabidopsis thaliana with a preference for anions. Proc. Natl. Acad. Sci. USA 2012, 109, 19015–19020. [Google Scholar] [CrossRef]
- Nagata, T.; Iizumi, S.; Satoh, K.; Ooka, H.; Kawai, J.; Carninci, P.; Hayashizaki, Y.; Otomo, Y.; Murakami, K.; Matsubara, K.; et al. Comparative analysis of plant and animal calcium signal transduction element using plant full-length cDNA data. Mol. Biol. Evol. 2004, 21, 1855–1870. [Google Scholar] [CrossRef]
- Pivato, M.; Ballottari, M. Chlamydomonas reinhardtii cellular compartments and their contribution to intracellular calcium signalling. J. Exp. Bot. 2021, 72, 5312–5335. [Google Scholar] [CrossRef]
- Ruberti, C.; Feitosa-Araujo, E.; Xu, Z.; Wagner, S.; Grenzi, M.; Darwish, E.; Lichtenauer, S.; Fuchs, P.; Parmagnani, A.S.; Balcerowicz, D.; et al. MCU proteins dominate in vivo mitochondrial Ca2+ uptake in Arabidopsis roots. Plant Cell 2022, 34, 4428–4452. [Google Scholar] [CrossRef]
- Shan, T.; Yuan, J.; Su, L.; Li, J.; Leng, X.; Zhang, Y.; Gao, H.; Pang, S. First Genome of the Brown Alga Undaria pinnatifida: Chromosome-Level Assembly Using PacBio and Hi-C Technologies. Front. Genet. 2020, 11, 140. [Google Scholar] [CrossRef]
- Wang, S.; Lin, L.; Shi, Y.; Qian, W.; Li, N.; Yan, X.; Zou, H.; Wu, M. First Draft Genome Assembly of the Seaweed Sargassum fusiforme. Front. Genet. 2020, 11, 590065. [Google Scholar] [CrossRef]
- Zepernick, B.N.; Truchon, A.R.; Gann, E.R.; Wilhelm, S.W. Draft Genome Sequence of the Freshwater Diatom Fragilaria crotonensis SAG 28.96. Microbiol. Resour. Announc. 2022, 11, e0028922. [Google Scholar] [CrossRef] [PubMed]





| Species | Diatom Group | TPC1 | TPCL |
|---|---|---|---|
| Chaetoceros tenuissimus | Centric | 2 | 1 |
| Minidiscus variabilis | Centric | - | - |
| Thalassiosira pseudonana | Centric | 1 | - |
| Thalassiosira oceanica | Centric | 1 | - |
| Fragilaria crotonensis | Araphid pennate | 1 | 1 |
| Pseudo-Nitzschia multiseries | Raphid pennate | 1 | 1 |
| Fragilariopsis cylindrus | Raphid pennate | 1 | 2 |
| Nitzschia inconspicua | Raphid pennate | 1 | 2 |
| Nitzschia putrida | Raphid pennate | 1 | 2 |
| Phaeodactylum tricornutum | Raphid pennate | 1 | 1 |
| Seminavis robusta | Raphid pennate | 1 | 1 |
| Fistulifera solaris | Raphid pennate | 2 | 2 |
| Mayamaea pseudoterrestris | Raphid pennate | - | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murphy, E.A.; Kleiner, F.H.; Helliwell, K.E.; Wheeler, G.L. Channels of Evolution: Unveiling Evolutionary Patterns in Diatom Ca2+ Signalling. Plants 2024, 13, 1207. https://doi.org/10.3390/plants13091207
Murphy EA, Kleiner FH, Helliwell KE, Wheeler GL. Channels of Evolution: Unveiling Evolutionary Patterns in Diatom Ca2+ Signalling. Plants. 2024; 13(9):1207. https://doi.org/10.3390/plants13091207
Chicago/Turabian StyleMurphy, Eleanor A., Friedrich H. Kleiner, Katherine E. Helliwell, and Glen L. Wheeler. 2024. "Channels of Evolution: Unveiling Evolutionary Patterns in Diatom Ca2+ Signalling" Plants 13, no. 9: 1207. https://doi.org/10.3390/plants13091207
APA StyleMurphy, E. A., Kleiner, F. H., Helliwell, K. E., & Wheeler, G. L. (2024). Channels of Evolution: Unveiling Evolutionary Patterns in Diatom Ca2+ Signalling. Plants, 13(9), 1207. https://doi.org/10.3390/plants13091207







